Introduction
The armadillo family of proteins is growing rapidly
in number. This family exhibits diverse functions, such as
embryogenesis and tumorigenesis, by interacting with multiple
binding partners at their characteristic armadillo repeat (1). Among these proteins, β-catenin has
been intensively investigated as a crucial integrator of the Wnt
signaling pathway as well as cell-cell adhesions (2). To date, a unique subfamily whose
chromosomal localization has been identified as chromosome region
Xq has been discovered; this subfamily is referred to as the
armadillo repeat containing, X-linked (ARMCX) subfamily.
ARMCX1, ARMCX2 and ARMCX3, also known as
ALEX1, ALEX2 and ALEX3, respectively, are
located at the chromosome region Xq21.33-q22.2 and have been
implicated in tumorigenesis (3).
Their amino (N)-terminal region harbors a transmembrane domain,
suggesting that these proteins may localize at membrane structures
in cells. Recently, ARMCX3 was revealed as an integral membrane
protein of the mitochondrial outer membrane that functionally
interacts with the transcriptional regulator Sox10 (4). In addition, ARMCX4,
ARMCX5 and ARMCX6, the localizations of which have
been mapped to chromosome regions Xq22.1, Xq22.1-q22.3 and
Xq21.33-q22.3, respectively, were also identified. Among them,
ARMCX5 was found to be activated and bound by oncogene ZNF217
(5). An oligonucleotide array
identified ARMCX6 as having an at least 2-fold increase in
mRNA expression in peripheral blood mononuclear cells from patients
with rheumatoid arthritis when compared with control subjects
(6). We previously identified
ARMCX6 as an up-regulated gene in both p16INK4A- and
p14ARF-silenced HeLa cells (7).
Both p16INK4A and p14ARF were found to be frequently deleted or
inactivated in various types of cancer (8).
In the present study, we revealed that ARMCX6
was exclusively localized in the cytoplasm of HeLa cells. Tissue
distribution of ARMCX6 was highly detected in the pancreas
and spleen. Taken together, these findings suggest that
ARMCX6 may be useful as a diagnostic marker for the
carcinogenesis of specific cells or tissues.
Materials and methods
Bioinformatics
Pfam and PROSITE (pattern/profile) (http://motif.genome.jp/), CLUSTAL W (ver. 1.81)
Multiple Sequence Alignments (http://align.genome.jp/), SOSUI (ver. 1.11)
(http://bp.nuap.nagoya-u.ac.jp/sosui/)
and PSORT II and iPSORT (http://psort.ims.u-tokyo.ac.jp/) predictions were
performed using web-based free software. The GenBank accession
numbers that were used were ARMCX1 (NM_016608),
ARMCX2 (NM_014782), ARMCX3 (NM_016607), ARMCX4
(AK292543), ARMCX5 (NM_022838), ARMCX6 (NM_019007),
armadillo repeat containing 10 (ARMC10) (alias
SVH) (NM_031905), G-protein coupled receptor-associated
sorting protein 1 (GPRASP1) (NM_014710) and
GPRASP2 (NM_001004051).
Cell cultures, plasmid, transfection and
fluorescent microscopy
Human cervical carcinoma (HeLa) cells were cultured
in Earle's modified Eagle's medium (MEM) supplemented with 10%
fetal bovine serum, 1% non-essential amino acids and
antibiotic-antimycotics (all from Invitrogen, Carlsbad, CA, USA).
The ARMCX6 EST (expressed sequence tag) cDNA clone (GenBank
no. BC007677; IMAGE 3609980; Open Biosystems, Huntsville, AL, USA)
was used as a template to amplify the ARMCX6-coding region
using a polymerase chain reaction (PCR), and the coding region was
ligated into the respective enzyme EcoRI and BamHI
sites of the pEGFP-N2 vector (Clontech, Mountain View, CA, USA).
The PCR primer set was as follows (underlines indicate the flanking
enzyme site): CGGAATTCGCCACCATGGGCCGGGCTCGGGAAGTG
and CGGGATCCATGGGGCAGGGGTTTCCAG. The
plasmid sequence was verified by sequencing at the Takara facility
(Mie, Japan). The cells were transfected using Lipofectamine Plus
(Invitrogen) or the Neon Transfection System (Invitrogen) according
to the manufacturer's instructions. Briefly, Neon electroporation
was performed in a 24-well plate with a pulse voltage of 1,200 and
a pulse width of 40. After 24–48 h of transfection, the cells were
washed using phosphate-buffered saline (PBS) and photographed using
a fluorescent microscope DM IRB (Leica, Wetzlar, Germany), equipped
with a digital single-lens reflex camera (Camedia E-20; Olympus,
Tokyo, Japan).
Reverse transcription (RT)-PCR
The total RNA samples were prepared using an RNeasy
Mini-Spin Column (Qiagen, Valencia, CA, USA) in accordance with the
manufacturer's instructions. Prior to RT-PCR, total RNA was treated
with deoxyribonuclease I (Invitrogen) according to the
manufacturer's instructions. One or two micrograms of total RNA was
reverse transcribed using High-Capacity cDNA Reverse Transcription
Kits (Applied Biosystems, Foster City, CA, USA). The PCR was
carried out in 25 μl of a mix consisting of 1X buffer, 200
μM of dNTPs, 400 nM of primers, 1 mM of MgSO4, 5%
DMSO and 1 unit of KOD plus DNA polymerase (Toyobo, Osaka, Japan).
Hot-start PCR was then performed as follows: denaturation for 3 min
at 94°C, followed by ad libitum cycles at 94°C for 15 sec, a
gene-specific annealing temperature [ARMCX3, 56°C;
ARMCX4, 56°C; ARMCX5, 57°C; ARMCX6, 54°C;
ARMC10, 67°C; glyceraldehyde-3-phosphate dehydrogenase
(GAPDH), 60°C] for 30 sec and 68°C for 30 sec, followed by
an extension step of 3 min at 68°C. The PCR results were verified
by varying the number of PCR cycles for each cDNA and set of
primers. The target gene primer pairs were as follows:
ARMCX3, GGGCTGTCCAGA AACGGGCT and CCCTGAGCAGTTCCCTAGTC for
540 bp; ARMCX4, GCAAGAAGTGGGCCTAGGGC and TTCCCT
GGGTACTGCCAAGG for 420 bp; ARMCX5, GGCCTAAT CCGAAGGCCTGC and
CCAGCTGCACAGGGGAGTTC for 360 bp; ARMCX6,
GGGCTCAATCCAGGACCACA and GTTCACTATCCATCAGGCGC for 350 bp;
ARMC10, TCAC CTGCCAACCTGACCAT and GCGCTATCTCAGCTCAC TGC for
460 bp; and GAPDH (NM_002046), CCATGGC AAATTCCATGGCA and
GTCCTTCCACGATACCAAAG for 365 bp. The amplified products were
separated on 1.0% agarose gels and visualized under ultraviolet
transillumination. For the cDNA panel analysis, 2.5 μl of
cDNA purchased from Clontech was used (Human MTC panels I and II
cDNA panel). The GAPDH primer in the kit was used as a
control to amplify 983 bp.
Western blotting
The cells were harvested and lysed in modified RIPA
lysis buffer [50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% Nonidet P40,
0.5% sodium deoxycholate and 1 mM EDTA] containing a protease
inhibitor cocktail (Sigma, St. Louis, MO, USA) for 20 min on ice.
The cell lysates were centrifuged, and the protein concentrations
were determined using the Bio-Rad Protein Assay Kit (Bio-Rad
Laboratories, Hercules, CA, USA). The protein lysates were loaded
onto each lane of a gel. Before performing sodium dodecyl sulfate
polyacrylamide gel electrophoresis (SDS-PAGE), the reaction was
terminated by the addition of Laemmli sample buffer containing 100
mM of dithiothreitol (DTT). Equal amounts of cellular protein were
electrophoresed on NuPAGE 4–12% Bis-Tris gel with MES running
buffer (Invitrogen) and transferred to a Hybond-PVDF membrane (GE
Healthcare, Piscataway, NJ, USA). The membrane was first blocked
using PBS containing 0.1% Tween-20 and 5% non-fat dried milk, then
incubated with GFP (JL-8; Clontech) and GAPDH (Applied Biosystems)
antibodies. Alkaline phosphatase (AP)-labeled secondary antibodies
were purchased from Promega (Madison, WI, USA). A Western
Blue-stabilized substrate was used to detect the signals, according
to the manufacturer's protocol (Promega).
Results
Characteristic features of ARMCXs
To determine the functional aspects of ARMCX6, we
predicted the motifs or domains of the amino acids (aa) composing
ARMCX6 using a Pfam and PROSITE (pattern and profile) search. Pfam
revealed that a protein of unknown function (referred to as DUF634)
is a characteristic feature of the carboxyl (C)-terminal of the
ARMCX6 protein (9–300 aa of full-length 300 aa). In addition to
ARMCX6, DUF634 was observed in other proteins including ARMCX1
(184–447 aa of full-length 453 aa), ARMCX2 (365–626 aa of
full-length 632 aa), ARMCX3 (100–363 aa of full-length 379 aa),
ARMCX5 (288–552 aa of full-length 558 aa) and ARMC10 (74–337 aa of
full-length 343 aa). Among the ARMCXs, DUF634 was not identified in
ARMCX4. DUF634 was also recognized in the C-terminus of GPRASP1
(1,132–1,384 aa of full-length 1,395 aa) and GPRASP2 (574–836 aa of
full-length 838 aa). In contrast, Arm (Armadillo/β-catenin-like
repeat) was exclusively predicted in ARMCX1 (239–276 aa), ARMCX2
(419–457 aa), ARMCX3 (155–192 aa) and ARMC10 (127–167 aa). Based on
PROSITE, ARM_REPEAT (Armadillo/plakoglobin ARM repeat motif) was
only noted in ARMCX1 (247–284 aa) and ARMCX3 (163–200 aa).
Next, we performed CLUSTAL W multiple sequence
alignments using full-length ARMCX1 (453 aa), ARMCX2 (632 aa),
ARMCX3 (379 aa), ARMCX4 (348 aa), ARMCX5 (558 aa), ARMCX6 (300 aa)
and ARMC10 (343 aa). GPRASP1 and GPRASP2 apparently had longer
coding regions compared to ARMCXs and ARMC10; therefore, these two
proteins were excluded from further analysis. Consistent with the
DUF634 domain, the C-terminals of each of the proteins were similar
(data not shown), suggesting the evolutionarily conserved
functional importance of this region. Previously, ARMCX1, ARMCX2
and ARMCX3 were predicted to possess similar N-terminal
transmembrane domain and armadillo repeats (3). Based on the alignment of ARMCX1-6 and
ARMC10, conserved amino acids were detected in the N-terminal
transmembrane domain (data not shown); however, SOSUI prediction
revealed that one trans-membrane helix was identified in the
N-terminal of ARMCX1 (5–27 aa), ARMCX2 (5–27 aa) and ARMCX3 (7–29
aa) as well as the central region of ARMCX4 (96–118 aa) but not in
any region of ARMCX5 or ARMCX6. SOSUI also predicted that ARMCX5
and ARMCX6 may be soluble proteins. On the other hand, iPSORT
predicted that the N-terminal (1–30 aa) of ARMCX6 as well as
ARMCX1–3 and ARMC10 may be a signal peptide. Apparently, ARMCX4 and
ARMCX5 do not possess a signal peptide.
To predict the subcellular localization of ARMCXs,
PSORT II prediction was performed. ARMCX6 had a 44.4% probability
of localizing in an extracellular compartment, including the cell
wall, whereas the other ARMCXs did not have a high probability of
being localized extracellularly (Table
I). Based on published information, PSORT II prediction is a
reliable bioinformatics tool. Indeed, ARMCX3 has been shown to be
an integral membrane protein of the mitochondrial outer membrane
(4). In addition to the ARMCXs,
ARMC10 also possesses a transmembrane domain and has been shown to
localize in the endoplasmic reticulum (10). PSORT II predicted that
ARMC10 is localized in vacuolar regions (22.2%), extracellular
regions including the cell wall (22.2%), the Golgi apparatus
(22.2%), mitochondrial regions (22.2%) and the endoplasmic
reticulum (11.1%). These cellular regions are functionally proximal
to the endoplasmic reticulum, as they all are membrane-enclosed
organelles. To determine the cellular localization of ARMCX6, we
constructed a C-terminal green fluorescent protein (GFP)-tagged
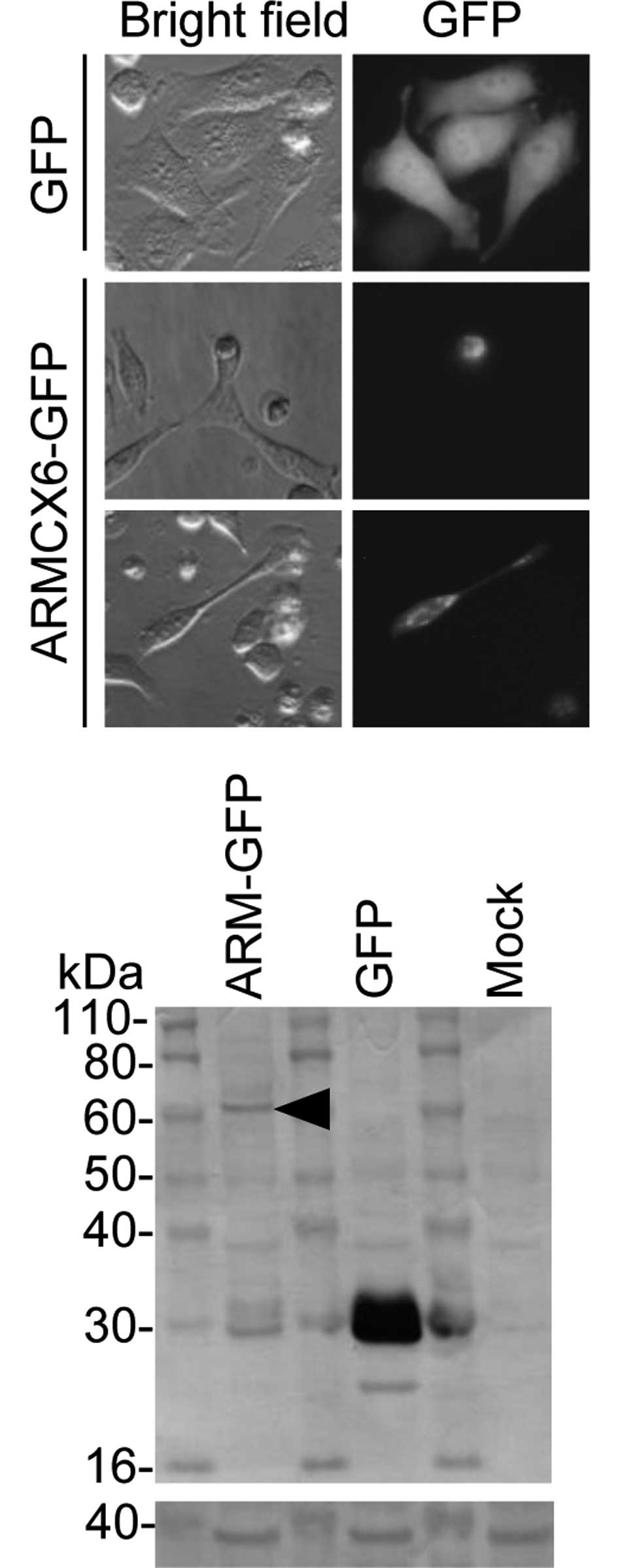
ARMCX6 expression plasmid. Fluorescent microscopy revealed a few
GFP-positive cells in a GFP-tagged ARMCX6-transfected well, whereas
abundant GFP-positive cells were detected in a control
GFP-transfected well (data not shown). In the GFP-tagged
ARMCX6-transfected well, the GFP-positive cells were all round in
shape and had shrunken, but the control GFP-transfected cells
showed a typical extended shape (Fig.
1A, middle panel vs. upper panel). Exceptionally, GFP-positive
intact cells were detected in the GFP-tagged ARMCX6-transfected
well. In it, GFP-tagged ARMCX6 was diffusely spread throughout the
cells, with the probable exception of the nuclei (Fig. 1A, lower panel).
 | Table I.PSORT II prediction of subcellular
localization of ARMCXs. |
Table I.
PSORT II prediction of subcellular
localization of ARMCXs.
| ARMCX1 | ARMCX2 | ARMCX3 | ARMCX4 | ARMCX5 | ARMCX6 |
|---|
| Mit (39.1) | Cyt (39.1) | Mit (34.8) | Cyt (69.6) | Nuclear (56.5) | Ext (44.4) |
| Cyt (21.7) | Mit (26.1) | Cyt (26.1) | Nuclear (17.4) | Mit (30.4) | Cyt (22.2) |
| ER (17.4) | Nuclear (17.4) | Vacuolar (8.7) | Mit (8.7) | Cyt (8.7) | Vacuolar (11.1) |
| Golgi (8.7) | Vacuolar (8.7) | Ext (8.7) | Per (4.3) | Cytoskeletal
(4.3) | Nuclear (11.1) |
| Nuclear (8.7) | Per (4.3) | Nuclear (8.7) | | | ER (11.1) |
| Vacuolar (4.3) | ER (4.3) | ER (8.7) | | | |
| | Golgi (4.3) | | | |
After 48 h of transfection, using an anti-GFP
antibody, an ARMCX6-GFP fusion protein was detected as a faint band
with an estimated molecular weight of ∼60 kDa, compared with the
abundantly expressed 27-kDa GFP (Fig.
1B, upper panel). GAPDH was consistently detected in all lanes
(Fig. 1B, lower panel). The ARMCX6
protein seems to be unstable, since another expression system
FLAG-tagged ARMCX6 again produced a faint band (data not shown).
Furthermore, we constructed N-terminal deleted FLAG-tagged ARMCX6
constructs to exclude the signal peptide but failed to detect a
reasonable protein level using immunoblotting (data not shown).
Taken together, these results suggest that ARMCX6 is not a secreted
protein and cellular toxicity is induced by ectopic ARMCX6
expression.
Comparative expression analysis of
ARMCXs
Next, we focused on the transcriptional regulation
of ARMCXs, the chromosomal localizations of which are uniquely
mapped to the same region of Xq. ARMC10 is localized at 7q22.1 but
was included in this investigation. To date, the mRNA distributions
of ARMCX1 and ARMCX2 in human tissues have been
reported (3), therefore
ARMCX1 and ARMCX2 were omitted from the present
study. The primer set used to detect ARMCX3–6 and
ARMC10 was able to specifically amplify their mRNAs in the
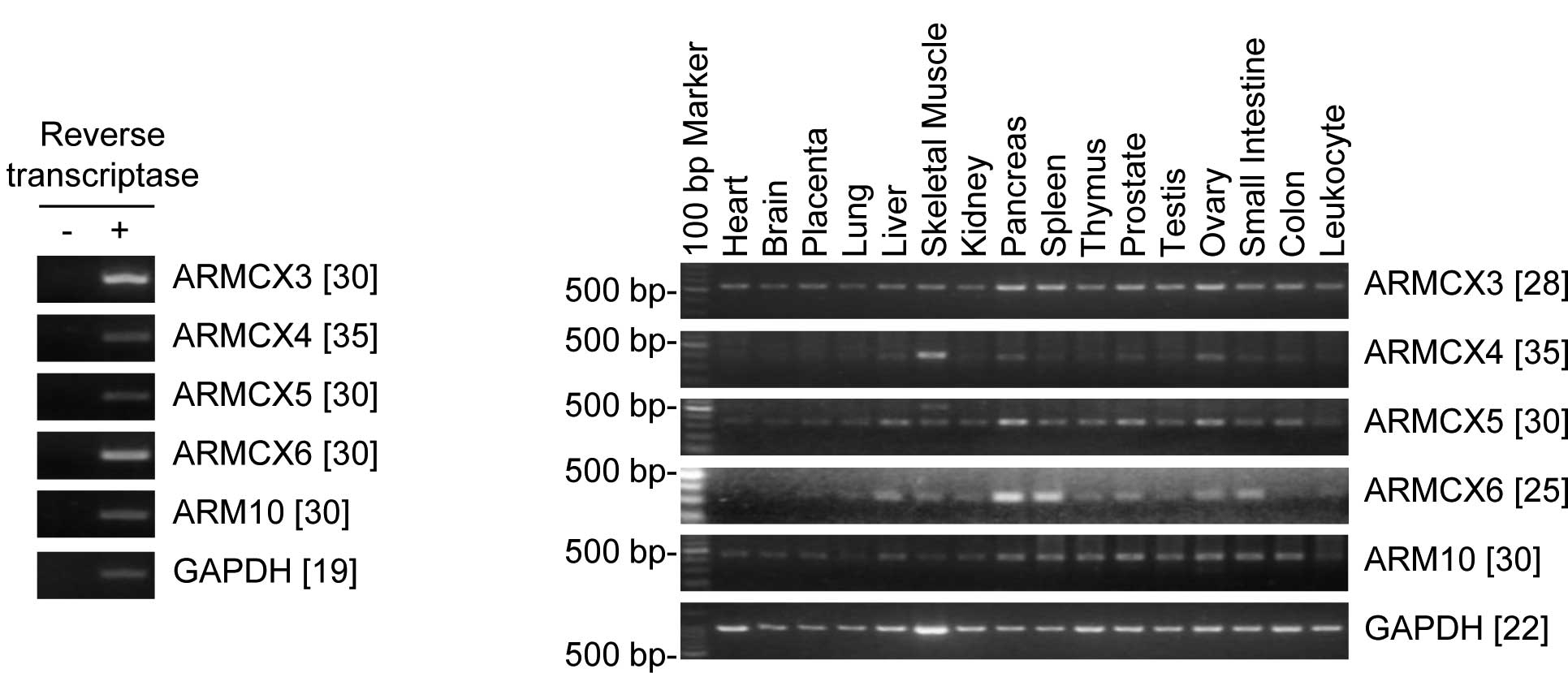
case of RT plus- but not RT minus-derived cDNAs (Fig. 2A). The expression levels of
ARMCX3–6 and ARMC10 were examined using a multiple
tissue-derived cDNA pool that included 16 adult tissues.
ARMCX6 mRNA was predominantly detected in the pancreas and
spleen, moderately detected in the liver, skeletal muscle, kidney,
thymus, prostate, testis, ovary and small intestine, and scarcely
detected in the placenta and leukocytes; ARMCX6 mRNA was not
detected in the heart, brain, or colon (Fig. 2B). In contrast to ARMCX6,
ARMCX3–5 and ARMC10 showed a relatively ubiquitous
expression pattern (Fig. 2B).
Discussion
A unique aspect of the ARMCX subfamily of
armadillo family proteins is their chromosomal localization, which
is mapped to Xq, and their presumed involvement in tumorigenesis
(1,3). We previously identified ARMCX6
as an up-regulated gene in p16INK4a and p14ARF knocked down HeLa
cells (7). The cyclin-dependent
kinase inhibitor 2A (CDKN2A) gene alternatively produces two
protein products, p16INK4A and p14ARF, and both proteins have
crucial roles in cell fate determination as upstream regulators of
pRb and p53, respectively (8). In
the present study, we aimed to reveal the functional aspect of
ARMCX6 and the expression pattern of ARMCXs.
Based on predictions made using their amino acid
compositions, ARMCXs tend to be membrane-associated proteins. Based
on sequence homology, ARMCX6 seems to be localized or associated
with the membrane structure of cells. We investigated the ARMCX6
subcellular localization using an ARMCX6-GFP expression plasmid.
ARMCX6-expressing cells tended to become atrophic compared to
control cells, indicating that ARMCX6 may cause cellular toxicity.
In addition, the number of ectopically ARMCX6-expressing cells was
very low, compared to that of the control GFP-transfected cells.
The ARMCX6 protein may have a short half-life. A future functional
study examining ARMCX6 is needed.
The mRNA expression patterns of ARMCX1 and
ARMCX2 have been reported in various human tissues (3). These patterns were remarkably
similar; namely, high expression levels were observed in the ovary,
heart, testis, prostate, brain, spleen and colon; faint expression
levels were observed in the liver and thymus; and no expression was
observed in leukocytes. In contrast, the ARMCX6 mRNA levels
were high in the pancreas and spleen, and no expression was
observed in the heart, brain and colon. ARMCX1 and
ARMCX2 were originally described as genes that are
down-regulated in epithelial cancers (3). In addition, ARMC10 is known to be
up-regulated in hepatocellular carcinomas (9). These results support the idea that
ARMCX6 may be a novel biomarker of cell fate determination
in specific tissues or neoplasms derived from certain tissues.
Abbreviations:
|
aa
|
amino acid;
|
|
ARMCX
|
armadillo repeat containing,
X-linked;
|
|
GAPDH
|
glyceraldehyde-3-phosphate
dehydrogenase;
|
|
GFP
|
green fluorescent protein;
|
|
PCR
|
polymerase chain reaction;
|
|
RT
|
reverse transcription
|
Acknowledgements
This work was supported by a Research
Project Grant (B) from the Institute of Science and Technology,
Meiji University, and by a Grant from the Daiwa Securities Health
Foundation. We thank the members of the Yoshida laboratory for the
technical assistance.
References
|
1.
|
Hatzfeld M: The armadillo family of
structural proteins. Int Rev Cytol. 186:179–224. 1999. View Article : Google Scholar
|
|
2.
|
Brembeck FH, Rosário M and Birchmeier W:
Balancing cell adhesion and Wnt signaling, the key role of
beta-catenin. Curr Opin Genet Dev. 16:51–59. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Kurochkin IV, Yonemitsu N, Funahashi SI
and Nomura H: ALEX1, a novel human armadillo repeat protein that is
expressed differentially in normal tissues and carcinomas. Biochem
Biophys Res Commun. 280:340–347. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Mou Z, Tapper AR and Gardner PD: The
armadillo repeat-containing protein, ARMCX3, physically and
functionally interacts with the developmental regulatory factor
Sox10. J Biol Chem. 284:13629–13640. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Krig SR, Jin VX, Bieda MC, O'Geen H,
Yaswen P, Green R and Farnham PJ: Identification of genes directly
regulated by the oncogene ZNF217 using chromatin
immunoprecipitation (ChIP)-chip assays. J Biol Chem. 282:9703–9712.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Edwards CJ, Feldman JL, Beech J, Shields
KM, Stover JA, Trepicchio WL, Larsen G, Foxwell BM, Brennan FM,
Feldmann M and Pittman DD: Molecular profile of peripheral blood
mononuclear cells from patients with rheumatoid arthritis. Mol Med.
13:40–58. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Sato K, Kusama Y, Tategu M and Yoshida K:
FBXL16 is a novel E2F1-regulated gene commonly upregulated in
p16INK4A- and p14ARF-silenced HeLa cells. Int J Oncol. 36:479–490.
2010.PubMed/NCBI
|
|
8.
|
Sharpless NE and DePinho RA: The INK4A/ARF
locus and its two gene products. Curr Opin Genet Dev. 9:22–30.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Huang R, Xing Z, Luan Z, Wu T, Wu X and Hu
G: A specific splicing variant of SVH, a novel human armadillo
repeat protein, is up-regulated in hepatocellular carcinomas.
Cancer Res. 63:3775–3782. 2003.PubMed/NCBI
|