Introduction
Tuberculosis (TB), which is a chronic infectious
disease caused by Mycobacterium tuberculosis, is a global
burden, with 8.7 million new cases and 1.4 million mortalities
reported in 2011 (1). Anti-TB
drug-induced liver injury (ADLI) is a severe adverse effect of TB
treatment and may negatively affect treatment compliance.
Isoniazid (INH) is a first-line therapy for the
treatment of TB, however, hepatotoxicity is a frequently observed
side effect of INH that may progress into liver cirrhosis (2).
The current biomarkers for liver injury [serum
alanine aminotransferase (ALT) and aspartate aminotransferase
(AST)] are adequate indicators of damage or altered liver function
(3). However, the aforementioned
parameters are unable to conclusively identify liver injury. In
addition, serum ALT and AST activity also increase following the
injury of other organs, and therefore are not selective for liver
injury (4). Thus, gene expression
variations are preferable for the classification of hepatotoxicants
(5). Our previous study used
epigenetics to identify that DNA methylation is a more sensitive
marker for the detection of ADLI (6). Furthermore, recent studies have
revealed that microRNA (miRNA/miR) may be used as a novel biomarker
for mRNA regulatory genes (7,8).
miRNAs are small (18–25 nt) endogenous, non-coding
RNA molecules that regulate post-transcriptional gene expression
through RNA interference, or through inhibition of translational
initiation and progression (9). Over
30% of mammalian genes are regulated by miRNA (10); therefore, miRNAs are important in a
wide variety of physiological and pathological processes (11). It has been reported that miRNA
expression levels differ significantly in various diseases,
indicating the potential for their use as biomarkers (12–14).
Among miRNAs, miR-122, which accounts for ~70% of
the total miRNA in the adult liver, is associated with liver
biology and disease, including cell cycle progression,
hepatocellular carcinogenesis (15),
lipid metabolism (16) and fibrosis
(17). Furthermore, miR-122 has been
confirmed as a potential biomarker for the diagnosis of
hepatotoxicity caused by acetaminophen (18) and alcohol (19). In addition, miR-155 is a
multi-functional miRNA known to have a regulatory role in numerous
biological processes, including immunity (20), inflammation (21), atherosclerosis (22) and cancer (23). However, it has been demonstrated that
liver tissue miR-155 expression levels are increased in
non-alcoholic steatohepatitis and hepatocellular carcinoma, and the
expression levels were associated with disease severity (24,25).
These findings suggest a strong association between the miR-122/155
ratio and liver injury. Although much is known concerning the
effects of liver injury, information on miR-122/155 expression and
ADLI remains limited.
miR-122/155 regulate a large number of genes and
consequently are involved in numerous biological processes,
including the response to environmental chemicals. Therefore, the
present study hypothesized that the levels of miR-122/155 in the
liver tissue and blood may serve as a biomarker for ADLI.
Consequently, miR-122/155 expression levels in the liver tissue of
mice with INH-induced ADLI were analyzed to identify changes in
miR-122/155 expression levels over a 24-h period. The aim of this
was to confirm the effectiveness of the miR-122/155 ratio as
quantitative marker for ADLI.
Materials and methods
Animals
A total of 64 Kunming mice (32 males and 32 females;
5 weeks old; body weight, 18–22 g; certificate no. 2009–0004) were
obtained from Beijing HFK Bioscience Co., Ltd. (Beijing, China).
The present study was approved by the ethics committee of North
China University of Science and Technology (Tangshan, China). To
establish the ADLI models, mice were orally administered INH (0.2
ml/mouse; 180 mg/kg body weight; Shenyang Hongqi Pharmaceutical
Co., Ltd., Liaoning, China; batch no. 1204081) dissolved in
double-distilled water (ddH20) to a final concentration
of 18 mg/ml. Blood and liver tissue samples were collected at 0.25,
0.75, 1.5, 6, 12, 18 and 24 h following the administration of INH
or ddH20 (control group; n=8 per time point/group). In
order to harvest the liver, the mice were sacrificed by cervical
dislocation. The blood samples were centrifuged at 1,370 × g for 10
min at 4°C, and the serum was collected and stored at −80°C until
use. A section of the liver from the porta hepatis was fixed in 10%
neutral buffered formalin (Tianjin Kemiou Chemical Reagent Co.,
Ltd., Tianjin, China) and the remaining liver was preserved at
−80°C.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from the livers of mice using
a benchtop homogenizer (no. TGL-16B; Shanghai Anting Scientific
Instrument Factory, Shanghai, China) with TRIzol®
reagent (cat no. 15596-018; Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA), according to the manufacturer's protocol.
The total RNA concentration was quantified using a UV-VIS
spectrophotometer (product no. TU-1901; Beijing Purkinje General
Instrument Co., Ltd. Beijing, China). The RNA quality and integrity
was assessed by measuring the absorbance ratio at 260 and 280 nm,
and by performing 1% agarose gel electrophoresis. RNA was reverse
transcribed into cDNA using miR-122/155 and U6-specific RT primers
and the TaqMan MicroRNA Reverse Transcription kit (product no.
4366596; Applied Biosystems; Thermo Fisher Scientific, Inc.) on a
C1000 Touch Thermo Cycler (Bio-Rad Laboratories, Inc., Hercules,
CA, USA), according to the manufacturer's protocols. The cycling
conditions for the RT reaction were as follows: 16°C for 30 min,
42°C for 30 min, 85°C for 5 min and 4°C for 1 min. The presence of
miR-122/155 was confirmed using Platinum SYBR Green qPCR
SuperMix-UDG (cat no. 11733-038; Invitrogen; Thermo Fisher
Scientific, Inc, Waltham, MA, USA) on a StepOnePlus Real-Time PCR
System (Applied Biosystems; Thermo Fisher Scientific, Inc.). U6
small nuclear RNA (snRNA) was used as the miRNA internal control.
The primers were designed using Primer Premier software (version
5.0; Premier Biosoft International, Palo Alto, CA, USA) and were
synthesized by Invitrogen (Thermo Fisher Scientific, Inc.). The
primer and probe sequences for the RT-qPCR are presented in
Table I. The PCR cycling conditions
were as follows: 50°C for 1 min, followed by 40 cycles of 95°C for
15 sec and 60°C for 1 min. Relative miRNA production was calculated
using the 2−∆∆Cq method (26), where Cq is the
quantification cycle. All reactions were run in triplicate, and the
results were normalized to U6 snRNA.
 | Table I.RT-qPCR primer and probe
sequences. |
Table I.
RT-qPCR primer and probe
sequences.
| Gene | Primer/Probe |
|---|
| RT |
|
|
miR-122 |
5′-ACAATGGTGTTTGTGTCCAAACCACAAACACCATTGTCA-3′ |
|
miR-155 |
5′-ATAGGGGTTTTGGCCTCTGACTGACTCCTAATCACAATTAGC-3′ |
| U6 |
5′-ATGGAACGCTTCACGAATTTGCGTGTCATCC-3′ |
| qPCR |
|
|
miR-122 | F:
5′-GCAGCTGTGGAGTGACAATGG-3′ |
|
miR-155 | F:
5′-GCCTGTTAAGCTAATTGTGAT-3′ |
|
miR-122/155 | R:
5′-GTGCAGGGTCCGAGGT-3′ |
| U6 | F:
5′-GCAAGGATGACACGCAAT-3′ |
|
| R:
5′-ATGGAACGCTTCACGAAT-3′ |
Biochemical assay and pathological
examination
Serum ALT and AST levels were determined using an
automatic biochemical analyzer (no. 7180; Hitachi, Tokyo, Japan),
according to the manufacturer's protocol. Formalin-fixed samples
were embedded in paraffin, sectioned (4-µm thickness), and stained
with hematoxylin and eosin (H&E; Beijing Solarbio Science &
Technology Co., Ltd., Beijing, China) for microscopic examination
(CX21; Olympus Corporation, Tokyo, Japan). The degree of liver
tissue injury was determined using a semi-quantitative method
(27), in which the liver lesion
severity number was multiplied by the weighted coefficient. The
liver lesion severity numbers were as follows: Hepatocellular
congestion, hemorrhage, 1; hepatocyte degeneration water, 1;
inflammatory cell infiltration, 1; and hepatocyte necrosis, 3. The
degree of liver injury was then calculated.
Statistical analysis
The data are expressed as the mean ± standard
deviation. Statistical differences between groups were determined
using Students t-tests. Multiple group comparisons were performed
using analysis of variance in combination with a Tukey's or
Dunnett's post-hoc test. Statistical analyses were conducted using
SPSS software, version 17 (SPSS, Inc., Chicago, IL, USA). P<0.05
was considered to indicate a statistically significant
difference.
Results
Histopathology and biochemistry of
mouse liver
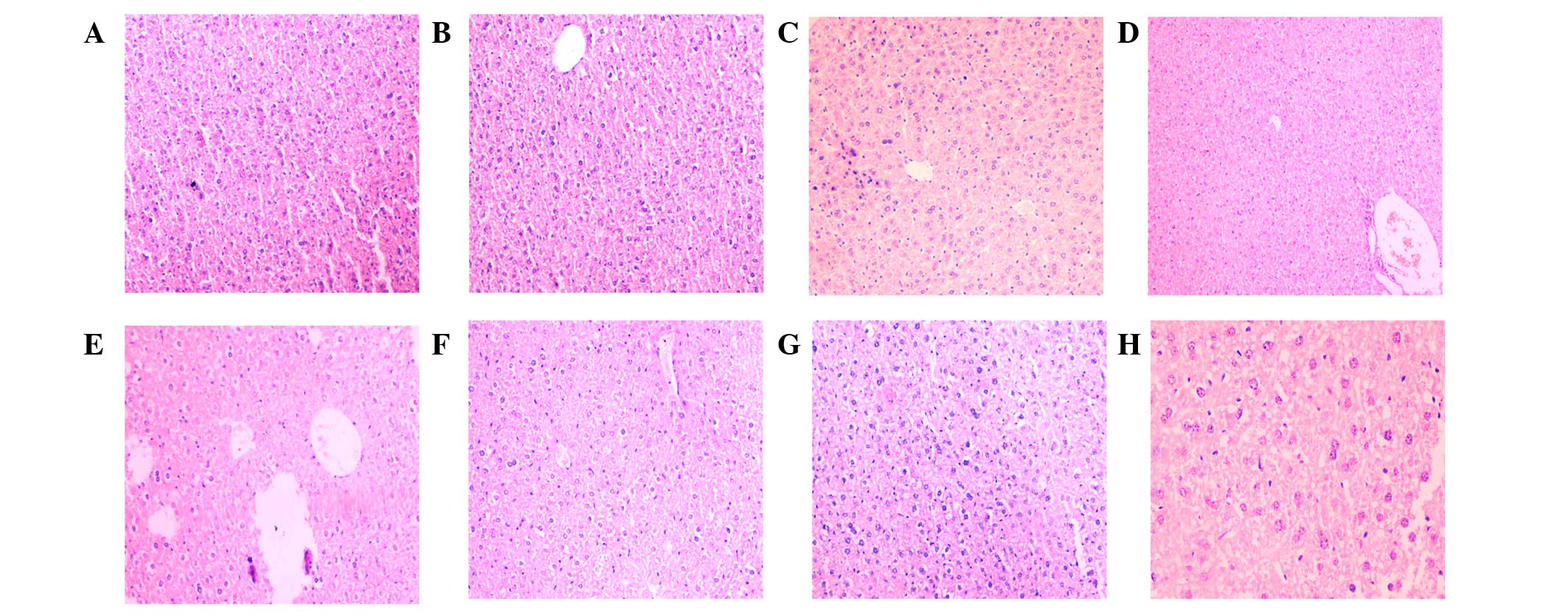
Histological examinations were performed on liver
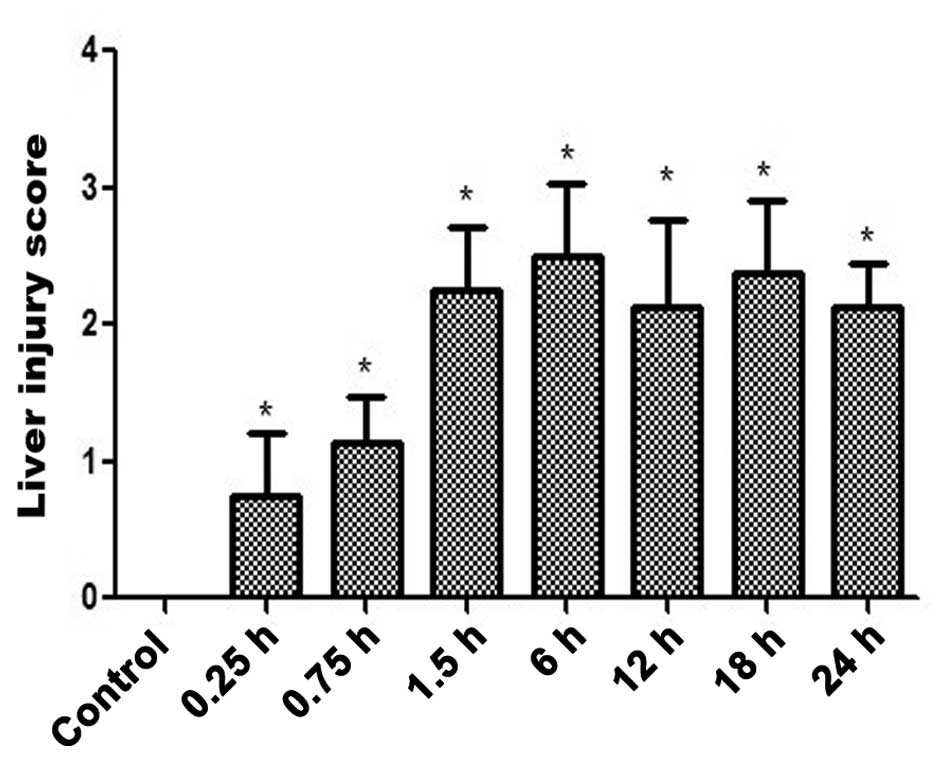
specimens (Fig. 1A-H) and the
associated scores of the liver injury are displayed in Fig. 2. The livers of the INH-treated mice
exhibited evidence of damaged cells from the 0.25 h time point
(Figs. 1B and 2). Thus, an INH-induced liver injury model
was established. Furthermore, at the 0.75 h time point, evidence of
inflammatory cell infiltration was observed (Fig. 1C).
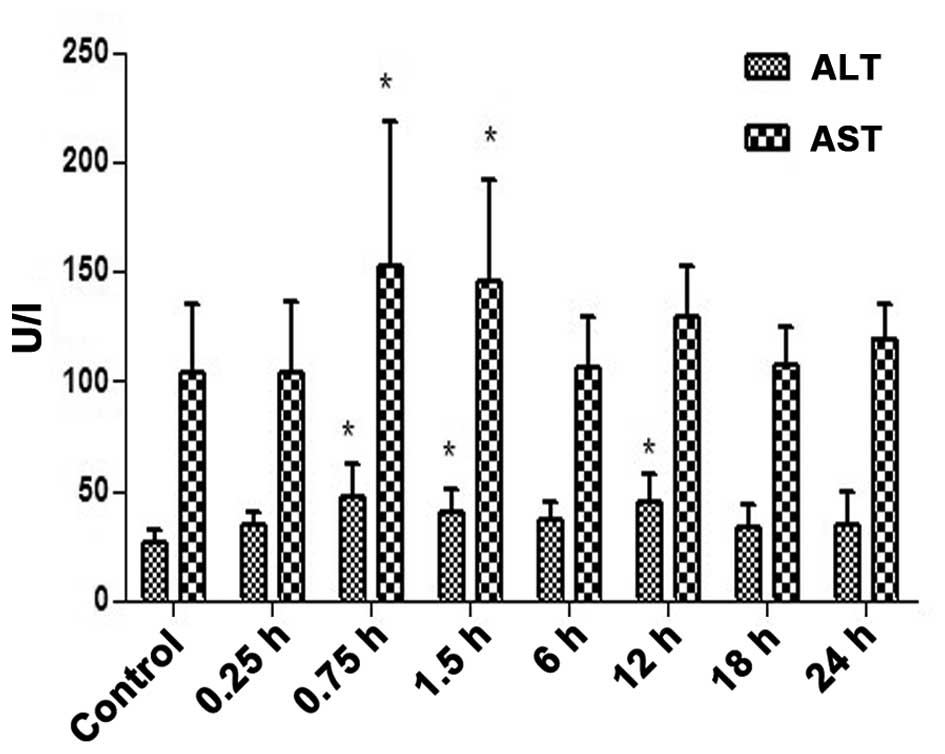
The serum ALT and AST levels (Fig. 3) were increased at different time
points of INH-induced liver injury (Fig.
3). The serum ALT and AST levels were significantly higher 0.75
and 1.5 h after INH administration compared with the control group
(P<0.05; Fig. 3), however, liver
injury was observed from 0.25 h onwards (Fig. 2).
Thus, the present study determined that the
variations in serum ALT and AST occur subsequent to the
histopathological changes caused by INH-induced liver injury.
Changes in tissue miR-122/155
expression in mice with INH-induced liver injury
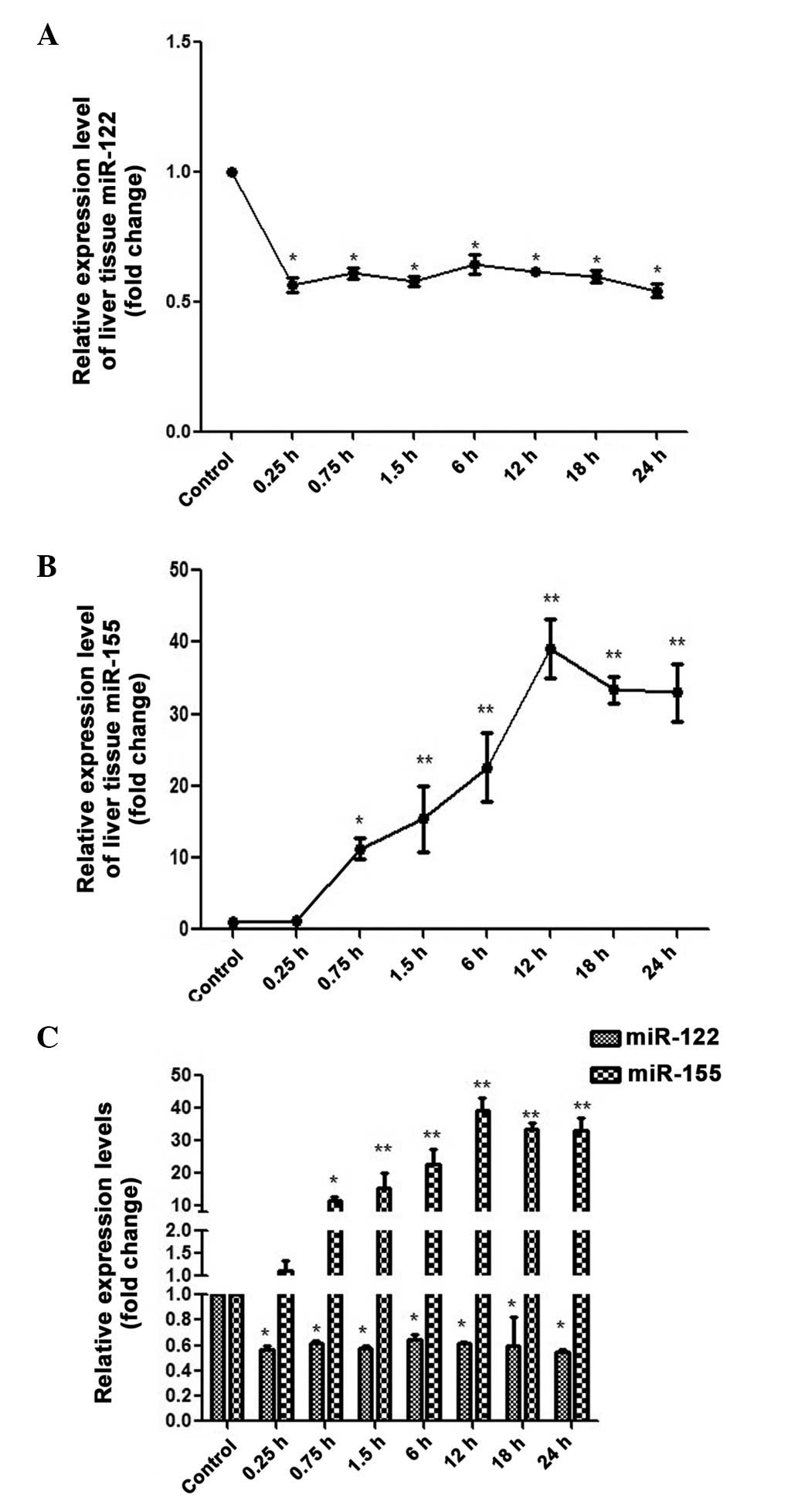
miR-122/155 expression levels were observed in the
mice liver the tissues over a 24-h period (Fig. 4). The miR-122 levels were
significantly declined at 0.25 h, with a 56.50±27.77%-fold decrease
observed compared with the control (P<0.05). Although the
expression levels of miR-122 were marginally increased at the other
time points, they were immediately followed by small declines in
expression, and the expression levels remained lower, as compared
with the control (Fig. 4A). These
results suggest that miR-122 expression levels decline during
INH-induced liver injury. Conversely, miR-155 levels were
significantly increased at 0.75 h (11.25±1.43%-fold increase;
P<0.05) and reached peak expression levels at the 12-h time
point (39.04±4.10%-fold increase; P<0.01; Fig. 4B). These results suggest that tissue
miR-155 expression levels are elevated during INH-induced liver
injury. Notably, the expression levels of miR-122 were altered more
rapidly in response to INH-induced liver injury, as compared with
miR-155 (Fig. 4C).
Correlation of pathological and
biochemical changes with INH-induced liver injury
Liver injury score was correlated to changes in
biochemistry over the 24-h period during INH-induced liver injury.
An association was observed between liver injury score and the
following: miR-122 expression, miR-155 expression, the ratio of
miR-122/155 and the ratio of miR-155/122, in which the ratio of
miR-122/155 exhibited the most significant correlation (r=−0.779;
P<0.001). However, liver injury scores revealed no correlation
with ALT or AST. In conclusion, the ratio of miR-122/155 has a
greater degree of correlation with liver damage compared with ALT,
AST, miR-122 and miR-155 expression (Table II).
 | Table II.Association between pathological
changes and liver injury score. |
Table II.
Association between pathological
changes and liver injury score.
| Liver injury score
correlation | ALT | AST | miR-122 | miR-155 |
miR-122/miR-155 |
|---|
| r-value | 0.157 | 0.053 | −0.592 | 0.678 | −0.779 |
| P-value | 0.215 | 0.678 | 0.001a | 0.001a | 0.001a |
Discussion
miRNAs are important in a wide variety of
physiological and pathological processes (28). Recently, miRNAs have been revealed as
potential biomarkers for the diagnosis of several diseases. In the
present study, tissue miR-122 was examined as a potential marker of
hepatocyte damage, and miR-155 as a marker of inflammation in
INH-induced liver injury. A mouse model of INH-induced liver injury
was established and changes in tissue miR-122/155 expression levels
at different time points during liver injury were analyzed.
Changes in miR-122/155 expression levels in
INH-induced liver injury were recorded at seven time points (0.25,
0.75, 1.5, 6, 12, 18 and 24 h) based on the known pharmacokinetics
of INH in mice (29). Double the
recommended dosage for humans for was administered to replicate the
initial dosing administered to humans in clinical treatment. The
present study identified that tissue miR-122/155 expression levels
significantly changed during liver injury. Compared with the
control group, tissue miR-122 expression levels decreased during
INH-induced liver injury. In a previous study involving the use of
a mouse model administered with acetaminophen, miR-122 expression
levels in the tissue were also observed to decrease (30). The aforementioned study also
indicated that the damaged cells within the liver tissue resulted
in the transport or release of cellular miRNAs into the peripheral
circulation (30). Notably, in the
present study, tissue miR-122 expression levels initially decreased
and initiated two upward trends after 0.25 h and 1.5 h. The
aforementioned change may be a result of the pharmacokinetics of
INH, as the peak concentration times of INH and its metabolite
hydrazine, which are the primary chemical substances that give rise
to INH-induced liver injury, are 0.25 h and 1.5 h, respectively
(29). As the peak concentration
times correspond to the increase in miR-122 expression levels, we
hypothesize that there may be an association between miR-122 and
the peak concentration times of INH; however, further investigation
is required.
The target genes of miR-155, including interleukin-1
and v-ets avian erythroblastosis virus E26 oncogene homolog 1, are
associated with the immune and hematopoietic systems (31,32). To
investigate the role of miR-155 during liver injury, the present
study examined the dynamic changes in miR-155 expression levels in
the livers of mice with INH-induced liver injury. Liver tissues
were observed under a microscope with H&E staining to
investigate the inflammatory response at 0.25 h. miR-155 expression
levels displayed an overall upward trend in the present study,
suggesting that tissue miR-155 expression levels increase during
inflammatory responses, reaching peak levels at 12 h and declining
thereafter. Notably, the changes in miR-155 expression occur later
than those of miR-122, which may be a result of the complex
internal environment. An association between miR-155 and
inflammatory factors was also observed in the present study.
miR-155 has previously been demonstrated to promote autoimmune
inflammation in inflammatory responses (33) and is upregulated in macrophages
following stimulation by affecting inflammatory mediators (34). A further study determined that
miR-155 was able to influence tumor necrosis factor-α expression
levels to increase inflammatory injury through adjusting
fas-associated death domain and inhibitor of κB expression
(35). Furthermore, it was revealed
that inflammatory mediators and miR-155 can influence one another's
expression levels (36).
Currently, elevated serum levels of ALT and AST are
used as a diagnosis of liver injury; however, their levels are also
increased in other diseases, and differences in the levels of ALT
and AST may occur later in the serum, as compared with the liver
(37). Previous studies have
demonstrated that miR-122 expression levels have a greater
association with liver injury than serum ALT and AST levels; thus,
miR-122 is a potential biomarker for the diagnosis of liver injury
(38). In the current study, changes
in miR-122 expression levels occurred prior to changes in serum ALT
and AST levels. Previous evidence revealed that miR-122 is more
sensitive and specific for liver injury compared with ALT and AST
(18), which is consistent with the
results of the present study. In addition, where the levels of ALT
and AST were returned to normal at 6 h, the expression levels of
miR-122 and histopathological changes of liver tissue remained
unchanged. Therefore, the levels of ALT and AST are unable to
accurately reveal the histopathological changes of the liver tissue
(39).
At present, the ratio of ALT/AST is used to
determine the degree of liver injury. In the present study, the
ratio of miR-122/155 was determined by dividing miR-122 expression
levels by those of miR-155. The ratio of miR-122/155 was more
closely associated with the degree of liver injury than the ALT/AST
ratio. miR-122 may indicate the presence of damaged hepatocytes,
possibly because miR-122 is expressed by liver cells under normal
physiological conditions but declines in the event of liver cell
injury (40). miR-155, as an
inflammation-specific miRNA, can regulate the expression of
inflammatory cells (41). The
current study also reported that miR-155 expression levels increase
when inflammatory cells invade the liver.
In conclusion, the present study indicates that the
ratio of miR-122/155 may be a more accurate biomarker of liver
damage in INH-induced acute liver injury than the ratio of ALT/AST.
Further studies involving various anti-TB drugs are, however,
required to consolidate upon this finding.
Acknowledgements
The present study was supported by the Key Lab of
Tangshan (grant no. 08150201A-1-8).
References
|
1
|
Glaziou P, Falzon D, Floyd K and
Raviglione M: Global epidemiology of tuberculosis. Semin Respir
Crit Care Med. 34:3–16. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shih TY, Young TH, Lee HS, Hsieh CB and Hu
OY: Protective effects of kaempferol on isoniazid- and
rifampicin-induced hepatotoxicity. AAPS J. 15:753–762. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shi Q, Hong H, Senior J and Tong W:
Biomarkers for drug-induced liver injury. Expert Rev Gastroenterol
Hepatol. 4:225–234. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zelber-Sagi S, Toker S, Armon G, Melamed
S, Berliner S, Shapira I, Halpern Z, Santo E and Shibolet O:
Elevated alanine aminotransferase independently predicts new onset
of depression in employees undergoing health screening
examinations. Psychol Med. 43:2603–2613. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang X, Greenhaw J, Shi Q, Su Z, Qian F,
Davis K, Mendrick DL and Salminen WF: Identification of urinary
microRNA profiles in rats that may diagnose hepatotoxicity. Toxicol
Sci. 125:335–344. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang B, Sun S, Shen L, Zu X, Chen Y, Hao
J, Huang X and Feng F: DNA methylation in the rat livers induced by
low dosage isoniazid treatment. Environ Toxicol Pharmacol.
32:486–490. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Duskova K, Nagilla P, Le HS, Iyer P,
Thalamuthu A, Martinson J, Bar-Joseph Z, Buchanan W, Rinaldo C and
Ayyavoo V: MicroRNA regulation and its effects on cellular
transcriptome in human immunodeficiency virus-1 (HIV-1) infected
individuals with distinct viral load and CD4 cell counts. BMC
Infect Dis. 3:2502013. View Article : Google Scholar
|
|
8
|
Mi SL, Zhang J, Zhang W and Huang RS:
Circulating microRNAs as biomarkers for inflammatory diseases.
Microrna. 2:64–72. 2013. View Article : Google Scholar
|
|
9
|
Henderson MC and Azorsa DO: The genomic
and proteomic content of cancer cell-derived exosomes. Front Oncol.
2:382012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu D and Murashov AK: Molecular mechanisms
of peripheral nerve regeneration: Emerging roles of microRNAs.
Front. 4:552013.
|
|
12
|
Ulivi P, Foschi G, Mengozzi M, Scarpi E,
Silvestrini R, Amadori D and Zoli W: Peripheral blood miR-328
expression as a potential biomarker for the early diagnosis of
NSCLC. Int J Mol Sci. 14:10332–10342. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sita-Lumsden A, Dart DA, Waxman J and
Bevan CL: Circulating microRNAs as potential new biomarkers for
prostate cancer. Br J Cancer. 108:1925–1930. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Giusti I, D'Ascenzo S and Dolo V:
Microvesicles as potential ovarian cancer biomarkers. BioMed Res
Int. 2013:7030482013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang B, Wang H and Yang Z: MiR-122
inhibits cell proliferation and tumorigenesis of breast cancer by
targeting IGF1R. PLoS One. 7:e470532012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ,
Shen R, Huang Y, Chen HC, Lee CH, Tsai TF, et al: MicroRNA-122
plays a critical role in liver homeostasis and
hepatocarcinogenesis. J Clin Invest. 22:2884–2897. 2012. View Article : Google Scholar
|
|
17
|
Arataki K, Hayes CN, Akamatsu S, Akiyama
R, Abe H, Tsuge M, Miki D, Ochi H, Hiraga N, Imamura M, et al:
Circulating microRNA-22 correlates with microRNA-122 and represents
viral replication and liver injury in patients with chronic
hepatitis B. J Med Virol. 85:789–798. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Antoine DJ, Dear JW, Lewis PS, Platt V,
Coyle J, Masson M, Thanacoody RH, Gray AJ, Webb DJ, Moggs JG, et
al: Mechanistic biomarkers provide early and sensitive detection of
acetaminophen-induced acute liver injury at first presentation to
hospital. Hepatology. 58:777–787. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang Y, Jia Y, Zheng R, Guo Y, Wang Y,
Guo H, Fei M and Sun S: Plasma microRNA-122 as a biomarker for
viral-, alcohol-, and chemical-related hepatic diseases. Clin Chem.
56:1830–1838. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lind EF and Ohashi PS: Mir-155, a central
modulator of T-cell responses. Eur J Immunol. 44:11–15. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
McDaniel K, Herrera L, Zhou T, Francis H,
Han Y, Levine P, Lin E, Glaser S, Alpini G and Meng F: The
functional role of microRNAs in alcoholic liver injury. J Cell Mol
Med. 18:197–207. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wei Y, Nazari-Jahantigh M, Neth P, Weber C
and Schober A: MicroRNA-126, −145, and −155: A therapeutic triad in
atherosclerosis? Arterioscler Thromb Vasc Biol. 33:449–454. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Farooqi AA, Qureshi MZ, Coskunpinar E,
Naqvi SK, Yaylim I and Ismail M: miR-421, miR-155 and miR-650:
Emerging trends of regulation of cancer and apoptosis. Asian Pac J
Cancer Prev. 15:1909–1912. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang B, Majumder S, Nuovo G, Kutay H,
Volinia S, Patel T, Schmittgen TD, Croce C, Ghoshal K and Jacob ST:
Role of microRNA-155 at early stages of hepatocarcinogenesis
induced by choline-deficient and amino acid-defined diet in C57BL/6
mice. Hepatology. 50:1152–1161. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pogribny IP, Starlard-Davenport A,
Tryndyak VP, Han T, Ross SA, Rusyn I and Beland FA: Difference in
expression of hepatic microRNAs miR-29c, miR-34a, miR-155, asnd
miR-200b is associated with strain-specific susceptibility to
dietary nonalcoholic steatohepatitis in mice. Lab Invest.
90:1437–1446. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guan TM, Liu DC, Wang JF, Cai S, Lin MJ,
Lu RR, Wu KF, Ma XL, Wu T and Li WD: Protective effect of extract
from fruit of Clausena lansium (Lour.) Skeels against acute
alcohol-induced hepatotoxicity in mice. Zhong Guo Yao Li Xue Yu Du
Li Xue Za Zhi. 26:829–834. 2012.(In Chinese).
|
|
28
|
Le HS and Bar-Joseph Z: Integrating
sequence, expression and interaction data to determine
condition-specific miRNA regulation. Bioinformatics. 29:i89–i97.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hou YN, Zhang XL and Wu HH: Effect of
rifampicin and isoniazid coadministration on the metabolism and
pharmacokinetics of isoniazid in mice. Jie Fang Jun Yao Xue Xue
Bao. 27:23–26. 2011.(In Chinese).
|
|
30
|
Wang K, Zhang S, Marzolf B, Troisch P,
Brightman A, Hu Z, Hood LE and Galas DJ: Circulating microRNAs,
potential biomarkers for drug-induced liver injury. Proc Natl Acad
Sci USA. 106:4402–4407. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ceppi M, Pereira PM, Dunand-Sauthier I,
Barras E, Reith W, Santos MA and Pierre P: MicroRNA-155 modulates
the interleukin-1 signaling pathway in activated human
monocyte-derived dendritic cells. Proc Natl Acad Sci USA.
106:2735–2740. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lu F, Weidmer A, Liu CG, Volinia S, Croce
CM and Lieberman PM: Epstein-Barr virus-induced miR-155 attenuates
NF-kappaB signaling and stabilizes latent virus persistence. J
Virol. 82:10436–10443. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kuo YC, Li YS, Zhou J, Shih YR, Miller M,
Broide D, Lee OK and Chien S: Human mesenchymal stem cells suppress
the stretch-induced inflammatory miR-155 and cytokines in bronchial
epithelial cells. PLoS One. 8:e713422013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nazari-Jahantigh M, Wei Y, Noels H, Akhtar
S, Zhou Z, Koenen RR, Heyll K, Gremse F, Kiessling F, Grommes J, et
al: MicroRNA-155 promotes atherosclerosis by repressing Bcl6 in
macrophages. J Clin Invest. 122:4190–4202. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tili E, Michaille JJ, Cimino A, Costinean
S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA and
Croce CM: Modulation of miR-155 and miR-125b levels following
lipopolysaccharide/TNF-alpha stimulation and their possible roles
in regulating the response to endotoxin shock. J Immunol.
179:5082–5089. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Miller AM, Gilchrist DS, Nijjar J, Araldi
E, Ramirez CM, Lavery CA, Fernández-Hernando C, McInnes IB and
Kurowska-Stolarska M: MiR-155 has a protective role in the
development of non-alcoholic hepatosteatosis in mice. PLoS One.
8:e723242013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Senior JR: Alanine aminotransferase: A
clinical and regulatory tool for detecting liver injury-past,
present, and future. Clin Pharmacol Ther. 92:332–339. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yamaura Y, Nakajima M, Takagi S, Fukami T,
Tsuneyama K and Yokoi T: Plasma microRNA profiles in rat models of
hepatocellular injury, cholestasis, and steatosis. PLoS One.
7:e302502012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
An FM, Yu DS, Xie Q, Gong BD, Wang H, Guo
Q and Yu H: The role of miR-122 expression during the acute liver
failure in mice induced by D-GalN/LPS. Zhonghua Gan Zang Bing Za
Zhi. 18:527–532. 2010.(In Chinese). PubMed/NCBI
|
|
40
|
Jopling C: Liver-specific microRNA-122:
Biogenesis and function. RNA Biol. 9:137–142. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wagner AE, Boesch-Saadatmandi C, Dose J,
Schultheiss G and Rimbach G: Anti-inflammatory potential of
allyl-isothiocyanate - role of Nrf2, NF-(k) B and microRNA-155. J
Cell Mol Med. 16:836–843. 2012. View Article : Google Scholar : PubMed/NCBI
|