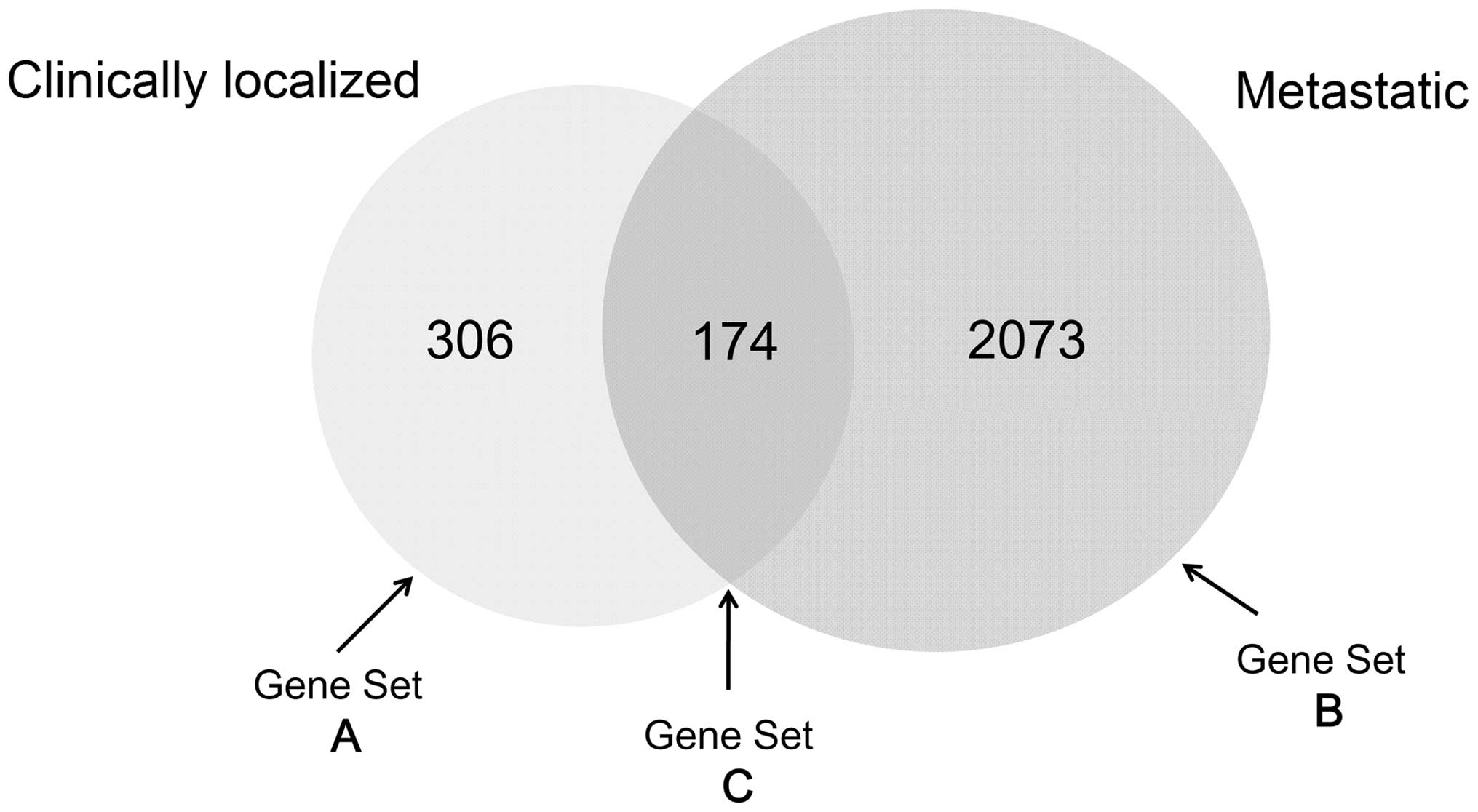
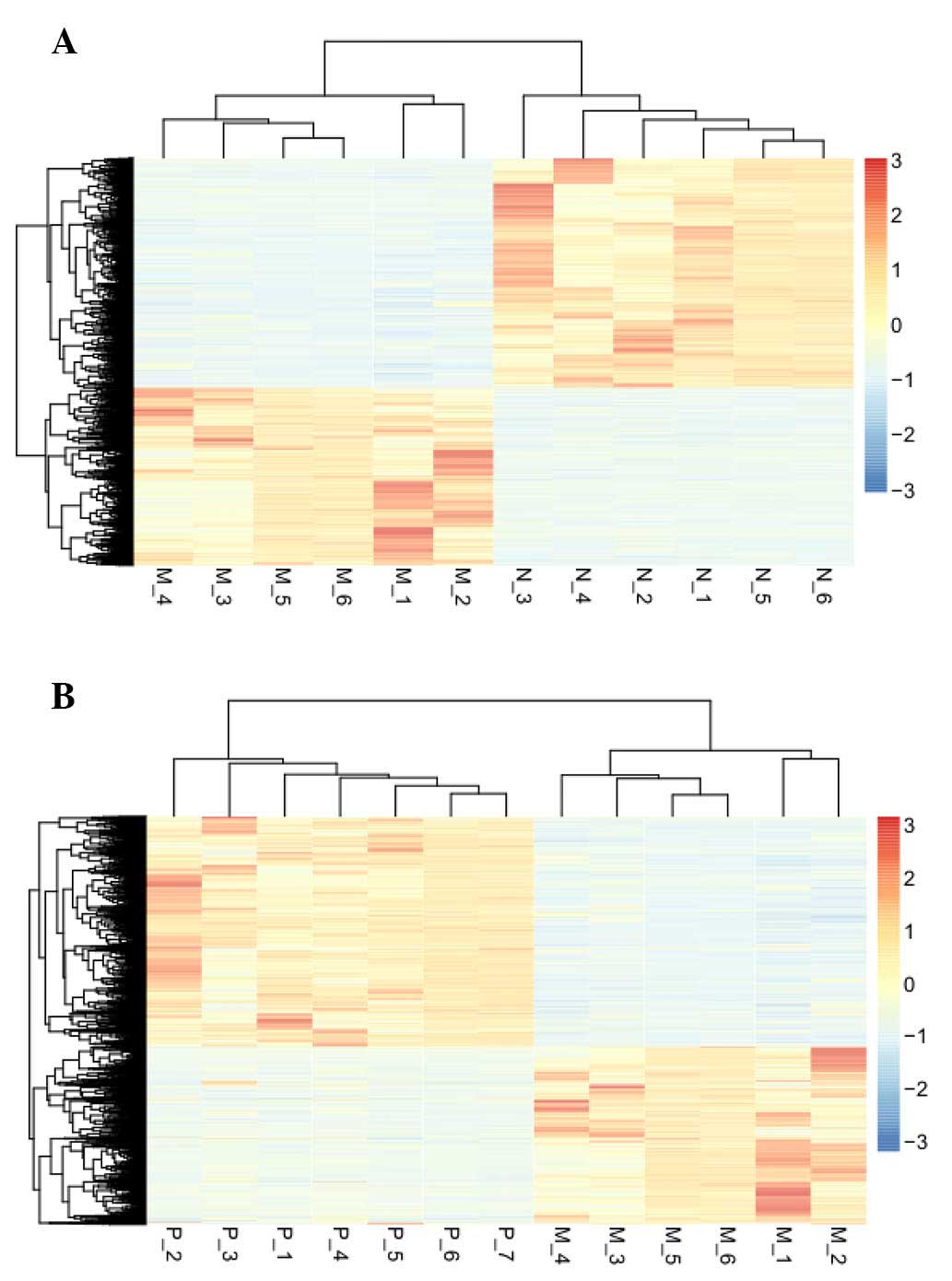
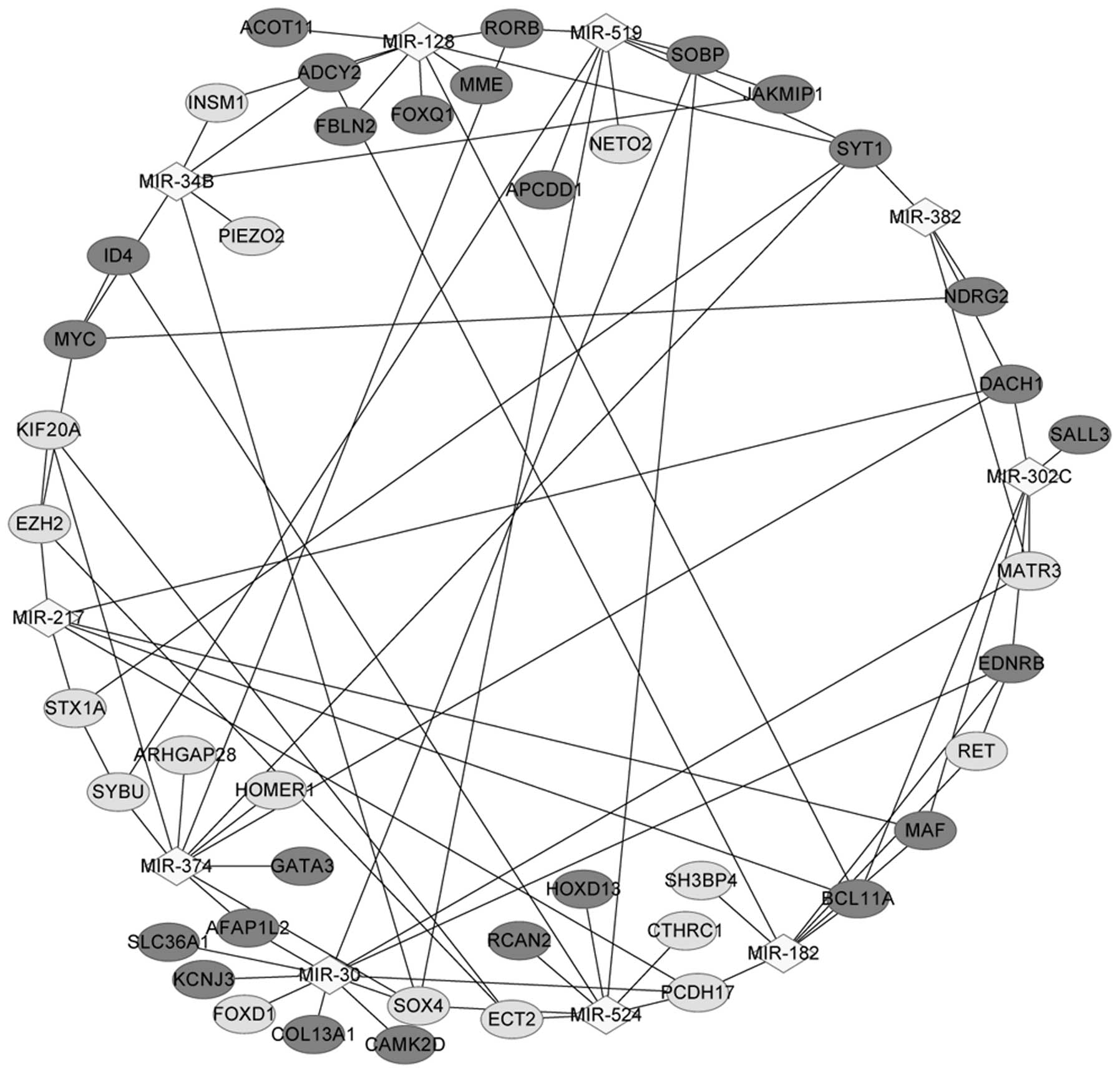
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Parkin DM and Steliarova-Foucher
E: Estimates of cancer incidence and mortality in Europe in 2008.
Eur J Cancer. 46:765–781. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Watahiki A and Wang Y, Morris J, Dennis K,
O'Dwyer HM, Gleave M, Gout PW and Wang Y: MicroRNAs associated with
metastatic prostate cancer. PLoS One. 6:e249502011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sun YX, Schneider A, Jung Y, Wang J, Dai
J, Wang J, Cook K, Osman NI, Koh-Paige AJ, Shim H, et al: Skeletal
localization and neutralization of the SDF-1 (CXCL12)/CXCR4 axis
blocks prostate cancer metastasis and growth in osseous sites in
vivo. J Bone Miner Res. 20:318–329. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cheng L, Nagabhushan M, Pretlow TP, Amini
SB and Pretlow TG: Expression of E-cadherin in primary and
metastatic prostate cancer. Am J Pathol. 148:1375–1380.
1996.PubMed/NCBI
|
|
6
|
Tripathi V, Popescu NC and Zimonjic DB:
DLC1 induces expression of E-cadherin in prostate cancer cells
through Rho pathway and suppresses invasion. Oncogene. 33:724–733.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hu L, Shi Y, Hsu JH, Gera J, Van Ness B
and Lichtenstein A: Downstream effectors of oncogenic ras in
multiple myeloma cells. Blood. 101:3126–3135. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hall A: Rho family GTPases. Biochem Soc
Trans. 40:1378–1382. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Khafagy R, Stephens T, Hart C, Ramani V,
Brown M and Clarke N: In vitro effects of the prenyl transferase
inhibitor AZD3409 on prostate cancer epithelial cells. J Clin
Oncol. 22:47442004.
|
|
10
|
Toren P and Zoubeidi A: Targeting the
PI3K/Akt pathway in prostate cancer: Challenges and opportunities
(review). Int J Oncol. 45:1793–1801. 2014.PubMed/NCBI
|
|
11
|
Zhang J, Patel L and Pienta KJ: CC
chemokine ligand 2 (CCL2) promotes prostate cancer tumorigenesis
and metastasis. Cytokine Growth Factor Rev. 21:41–48. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Luo JL, Tan W, Ricono JM, Korchynskyi O,
Zhang M, Gonias SL, Cheresh DA and Karin M: Nuclear
cytokine-activated IKKalpha controls prostate cancer metastasis by
repressing Maspin. Nature. 446:690–694. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cai H, Smith DA, Memarzadeh S, Lowell CA,
Cooper JA and Witte ON: Differential transformation capacity of Src
family kinases during the initiation of prostate cancer. Proc Natl
Acad Sci USA. 108:6579–6584. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Salagierski M and Schalken JA: Molecular
diagnosis of prostate cancer: PCA3 and TMPRSS2: ERG gene fusion. J
Urol. 187:795–801. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Saini S, Majid S, Yamamura S, Tabatabai L,
Suh SO, Shahryari V, Chen Y, Deng G, Tanaka Y and Dahiya R:
Regulatory role of miR-203 in prostate cancer progression and
metastasis. Clin Cancer Res. 17:5287–5298. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schaefer A, Jung M, Mollenkopf HJ, Wagner
I, Stephan C, Jentzmik F, Miller K, Lein M, Kristiansen G and Jung
K: Diagnostic and prognostic implications of microRNA profiling in
prostate carcinoma. Int J Cancer. 126:1166–1176. 2010.PubMed/NCBI
|
|
17
|
Gandellini P, Folini M and Zaffaroni N:
Towards the definition of prostate cancer-related microRNAs: Where
are we now? Trends Mol Med. 15:381–390. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Szczyrba J, Löprich E, Wach S, Jung V,
Unteregger G, Barth S, Grobholz R, Wieland W, Stöhr R, Hartmann A,
et al: The microRNA profile of prostate carcinoma obtained by deep
sequencing. Mol Cancer Res. 8:529–538. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Varambally S, Yu J, Laxman B, Rhodes DR,
Mehra R, Tomlins SA, Shah RB, Chandran U, Monzon FA, Becich MJ, et
al: Integrative genomic and proteomic analysis of prostate cancer
reveals signatures of metastatic progression. Cancer Cell.
8:393–406. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Smyth GK: Linear models and empirical
bayes methods for assessing differential expression in microarray
experiments. Stat Appl Genet Mol Biol. 3:2004.PubMed/NCBI
|
|
24
|
Verhoeven KJ, Simonsen KL and McIntyre LM:
Implementing false discovery rate control: Increasing your power.
Oikos. 108:643–647. 2005. View Article : Google Scholar
|
|
25
|
Olson CF: Parallel algorithms for
hierarchical clustering. Parallel Computing. 21:1313–1325. 1995.
View Article : Google Scholar
|
|
26
|
Kolde R: Pheatmap: Pretty Heatmaps. R
package version 0.7. 7. Journal. 2012.
|
|
27
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord G, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID Gene functional classification tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang DW, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Visel A, Alvarez-Bolado G, Thaller C and
Eichele G: Comprehensive analysis of the expression patterns of the
adenylate cyclase gene family in the developing and adult mouse
brain. J Comp Neurol. 496:684–679. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hagemann D, Bohlender J, Hoch B, Krause EG
and Karczewski P: Expression of
Ca2+/calmodulin-dependent protein kinase II
delta-subunit isoforms in rats with hypertensive cardiac
hypertrophy. Mol Cell Biochem. 220:69–76. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Prevarskaya N, Skryma R and Shuba Y:
Calcium in tumour metastasis: New roles for known actors. Nat Rev
Cancer. 11:609–618. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liao J, Schneider A, Datta NS and McCauley
LK: Extracellular calcium as a candidate mediator of prostate
cancer skeletal metastasis. Cancer Res. 66:9065–9073. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rajan P, Sudbery IM, Villasevil ME, Mui E,
Fleming J, Davis M, Ahmad I, Edwards J, Sansom OJ, Sims D, et al:
Next-generation sequencing of advanced prostate cancer treated with
androgen-deprivation therapy. Eur Urol. 66:32–39. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Whitfield ML, Sherlock G, Saldanha AJ,
Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM,
Brown PO and Botstein D: Identification of genes periodically
expressed in the human cell cycle and their expression in tumors.
Mol Biol Cell. 13:1977–2000. 2002. View Article : Google Scholar PubMed/NCBI
|
|
35
|
White R and Kung H: miR-30 as a tumor
suppressor connects EGF/Src signal to ERG and EMT. Oncogene.
33:2495–2503. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kao C, Martiniez A, Shi X, Yang J, Evans
C, Dobi A, Devere White R and Kung H: miR-30 as a tumor suppressor
connects EGF/Src signal to ERG and EMT. Oncogene. 33:2495–2503.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Rasheed SA, Teo CR, Beillard EJ, Voorhoeve
PM and Casey PJ: MicroRNA-182 and microRNA-200a control G-protein
subunit α-13 (GNA13) expression and cell invasion synergistically
in prostate cancer cells. J Biol Chem. 288:7986–7995. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu R, Li J, Teng Z, Zhang Z and Xu Y:
Overexpressed microRNA-182 promotes proliferation and invasion in
prostate cancer PC-3 cells by down-regulating N-myc downstream
regulated gene 1 (NDRG1). PLoS One. 8:e689822013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nagasawa H, Uto Y, Sasaki H, Okamura N,
Murakami A, Kubo S, Kirk KL and Hori H: Gc protein (vitamin
D-binding protein): Gc genotyping and GcMAF precursor activity.
Anticancer Res. 25:3689–3695. 2005.PubMed/NCBI
|
|
40
|
Yamamoto N, Naraparaju VR and Asbell SO:
Deglycosylation of serum vitamin D3-binding protein leads to
immunosuppression in cancer patients. Cancer Res. 56:2827–2831.
1996.PubMed/NCBI
|
|
41
|
Yamamoto N, Suyama H and Yamamoto N:
Immunotherapy for prostate cancer with Gc protein-derived
macrophage-activating factor, GcMAF. Transl Oncolo. 1:65–72. 2008.
View Article : Google Scholar
|
|
42
|
Sharad S, Srivastava A, Ravulapalli S,
Parker P, Chen Y, Li H, Petrovics G and Dobi A: Prostate cancer
gene expression signature of patients with high body mass index.
Prostate Cancer Prostatic Dis. 14:22–29. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Coppola V, De Maria R and Bonci D:
MicroRNAs and prostate cancer. Endocr Relat Cancer. 17:F1–F17.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang K and Waxman DJ: PC3 prostate
tumor-initiating cells with molecular profile
FAM65Bhigh/MFI2low/LEF1low increase tumor angiogenesis. Mol Cancer.
9:3192010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Dunlevy JR, Koppelman ED and Kolberg JB:
The expression of a SH3BP4-related protein in retinal cells. Invest
Ophthalmol Vis Sci. 46:2996. 2005.
|
|
46
|
Dunlevy JR, Berryhill BL, Vergnes JP,
SundarRaj N and Hassell JR: Cloning, chromosomal localization and
characterization of cDNA from a novel gene, SH3BP4, expressed by
human corneal fibroblasts. Genomics. 62:519–524. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lin YL, Xie PG, Wang L and Ma JG: Aberrant
methylation of protocadherin 17 and its clinical significance in
patients with prostate cancer after radical prostatectomy. Med Sci
Monit. 20:1376–1382. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tang X, Tang X, Gal J, Kyprianou N, Zhu H
and Tang G: Detection of microRNAs in prostate cancer cells by
microRNA array. Methods Mol Biol. 732:69–88. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ma S, Chan YP, Kwan PS, Lee TK, Yan M,
Tang KH, Ling MT, Vielkind JR, Guan XY and Chan KW: MicroRNA-616
induces androgen-independent growth of prostate cancer cells by
suppressing expression of tissue factor pathway inhibitor TFPI-2.
Cancer Res. 71:583–592. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
He HC, Han ZD, Dai QS, Ling XH, Fu X, Lin
ZY, Deng YH, Qin GQ, Cai C, Chen JH, et al: Global analysis of the
differentially expressed miRNAs of prostate cancer in Chinese
patients. BMC Genomics. 14:7572013. View Article : Google Scholar : PubMed/NCBI
|