Introduction
Mesenchymal stem cells (MSCs) possess a strong
proliferative capacity and are able to differentiate into
endothelium (1), adipocytes
(1–3), neurons (2,4),
osteoblasts (1,3,5) and
myoblasts (5,6), which were first identified in bone
marrow. In recent years, MSCs have been discovered in amniotic
fluid, muscles, umbilical cord blood, dermis and other tissues
(2,7–9).
Although the most abundant source of MSCs is bone marrow, they can
be obtained from other tissues, including the kidneys (10). Prior studies have indicated that
metanephric mesenchyme (MM) contains embryonic renal stem cells
which can generate all the epithelial cells of the nephron
(11–13).
Renal disease is a common clinical disease and is
growing worldwide. There are no effective therapy modes to treat
renal disease, but continuous renal replacement therapy is
typically performed at the end stage of the disease (14). MSCs may be capable of reversing
kidney injury (15). Transplanted
MSCs are localized at targeted tissue supporting the notion that
the efficacy of MSCs in treating diseases is independent of
engraftment and differentiation (16); however, more research is
required.
The kidneys originate from the intermediate mesoderm
(IM), located between the paraxial and lateral mesoderms, of the
early embryonic germ layer (17). In
mammals, the IM finally develops three types of kidneys:
Pronephros, mesonephros and the metanephros (18). The human definitive kidneys begin to
develop early in the fifth week and become functional at
approximately four weeks later (19). The mammalian adult kidney is
developed from the MM and the ureteric bud, which forms from the IM
(10). The reciprocal interaction
between the MM and the ureteric bud is a central step in kidney
development. The MM is differentiated into epithelialized nephrons
and interstitium by the ureteric bud induction, while the MM
induces the ureteric bud to differentiate into the lower urinary
tract (10).
Current studies of stem cells focus on humans,
mouse, rat and other mammals, but limited research has been
conducted involving sheep (10,12,13,17,19). The
purpose of the present study was to investigate the isolation and
culture process of MMSCs in vitro, which were isolated from
six-week-old embryonic sheep kidney tissues, and to study their
biological characterization.
Materials and methods
Experimental materials
A total of 2 one-year-old demale Ovis aries sheep,
weighing 74 and 76 kg, were supplied by the Chinese Academy of
Agriculture Science farm. All animal experiments were performed in
accordance with the guidelines established by the Institutional
Animal Care and Use Committee at Chinese Academy of Agriculture
Sciences. All cell culture media and supplements were obtained from
Sigma-Aldrich (Merck KGaA, Darmstadt, Germany), unless stated
otherwise. All cell culture plates were provided by Wuxi Nest
Biotechnology Co., Ltd. (Wuxi, China). Animals were sacrificed by
intraperitoneal injection of sodium pentobarbital (100 mg/kg;
Sigma-Aldrich; Merck Millipore). The study was approved by the
Institutional Animal Care and Use Committee of the Chinese Academy
of Agricultural Sciences (Beijing, China).
Isolation and culture of the MMSCs
from fetal kidney
The sheep fetuses (n=4; n=2 at six weeks-old, n=2 at
ten weeks-old) were collected after caesarean section (1–2 month)
under sterile conditions. The kidney tissues were isolated from the
fetus and washed three times by phosphate-buffered saline (PBS)
without calcium and magnesium. The kidney tissues were cut into ~1
cm2 pieces, then digested with 0.2% (w/v) collagenase
(Gibco; Thermo Fisher Scientific, Inc., Grand Island, NY, USA) in
DMEM/F12 with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.) for 30 min at 37°C. Then the undigested tissues
were incubated with 0.25% (w/v) trypsin and 0.02% (w/v) EDTA
(Gibco) for 15 min. In addition, neutralized the enzymatic activity
was neutralized using an equal volume of FBS. The digested tissues
and digestive juice were filtrated using a 74-µm cell strainer, and
the cell suspension was centrifuged at 200 × g for 8 min at
room temperature. The bottom cells were resuspended in complete
medium [DMEM/F12, 10% (v/v) FBS and 104 IU/ml
penicillin/streptomycin]. After counting, cells were plated into a
60-mm culture dish and incubated at 37°C in 5% CO2.
Cells were washed twice with PBS to remove non-adherent cells after
24 h later. When cultures reached 80% confluence, cells were
digested with warmed trypsin (0.25% trypsin and 0.02% EDTA) and
then subcultured onto fresh culture plates. After 3–4 passages (P)
cells were purified.
Immunofluorescent detection of cell
markers
Cultures of MMSCs at different passages were seeded
(0.5×105 cells per 1 cm2 glass coverslip) on
glass coverslips coated with poly-L-lysine. For immunofluorescent
assay analysis, the sheep MMSCs were fixed with 4% paraformaldehyde
for 15 min and washed three times (5 min per wash) with PBS. After
cultures reached 70% confluence, the cells were permeabilized with
0.25 % Triton X-100 for 10 min. Cells were washed three times (5
min per wash) with PBS, then blocking with 10% goat serum was
performed in 1 h prior to the incubation with primary antibodies.
The cells were incubated with the following primary antibodies:
Anti-Oct-4 (cat. no. ab27985), anti-CD44 (cat. no. ab157107) (both
purchased from Abcam, Cambridge, MA, USA), anti-FN1 (cat. no.
bs-4859R), anti-PAX2 (cat. no. bs-1187R), anti-VIM (cat. no.
bs-3472R) and anti-CD34 (cat. no. bs-8996R) (all 1:100; Beijing
Biosynthesis Biotechnology Co., Ltd., Beijing, China) at 4°C
overnight. Cells were washed three times (5 min per wash) with PBS,
followed by with fluorescein isothiocyanate-conjugated goat
anti-rabbit (cat. no. ZF-0314) or Cy5-conjugated goat anti-rabbit
(cat. no. ZF-0516) secondary antibodies (1:100; Beijing Zhongshan
Golden Bridge Biotechnology, Co., Ltd., Beijing) in dark for 1 h at
room temperature. Finally, 10 µg/ml DAPI (Solarbio Biotechnology
Co., Ltd., Beijing, China) was used to label the cell nuclei for 15
min, then images were captured using a fluorescence microscope
(TE-2000-E; Nikon Corporation, Tokyo, Japan). PBS was used as a
substitute for primary antibodies for technical controls.
Reverse transcription-polymerase chain
reaction (RT-PCR) analysis
MMSCs at different passages were collected and total
RNA was extracted using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc., Carlsbad, CA, USA). DNase I (Solarbio
Biotechnology Co., Ltd.) was used to digest the potential
contamination of genomic DNA. Total RNA (2.0 µg) was reverse
transcribed into cDNA using an RNA PCR kit, version 3.0 (Takara
Biotechnology, Co., Ltd., Dalian, China). Primer sequences are
listed in Table I. The PCR reaction
was performed by the PCR Master Mix Kit (Takara Biotechnology, Co.,
Ltd.), according to the manufacturer's instructions. PCR analyses
were performed in 50-µl volumes containing 25 µl 2X PCR Mix, 15 µl
ddH2O, 5 µl template cDNA and 2.5 µl each of forward and
reverse primers. Cycling conditions consisted of an initialization
step at 94°C for 5 min, then 35 cycles of a denaturation step at
94°C for 30 sec, annealing step at 50–60°C for 30 sec, elongation
step at 72°C for 30 sec and final elongation step at 72°C for 5
min. The PCR products were visualized by electrophoresis using 2.0%
(w/v) agarose gels.
 | Table I.Primer sequences used for reverse
transcription-polymerase chain reaction. |
Table I.
Primer sequences used for reverse
transcription-polymerase chain reaction.
| Gene | Primer
sequence | Product length
(bp) | Temperature
(°C) |
|---|
| OCT4 | F:
5′-GGACACCTCGCTTCTGACT-3′ | 386 | 59.03 |
|
| R:
5′-GGCTCCAGCTTCTCCTTGT-3′ |
| 59.32 |
| VIM | F:
5′-CAGGAGGAGATGCTTCAGAGA-3′ | 356 | 58.61 |
|
| R:
5′-GCGTCGTTGTTGCGGTTA-3′ |
| 59.07 |
| PAX2 | F:
5′-CAGAGTGGCGTGGACAGTT-3′ | 438 | 59.93 |
|
| R:
5′-GAATCTCCAAGCCTCGTTGTAG-3′ |
| 58.81 |
| FN1 | F:
5′-CCGAGGAAGGAGAGCAGAAT-3′ | 113 | 58.88 |
|
| R:
5′-CCAGCGAACGACAATAGAAGT-3′ |
| 58.39 |
| CD44 | F:
5′-TGGAGAAGAATGGTCGCTACA-3′ | 228 | 58.82 |
|
| R:
5′-AGGTGTTGGATGTGAGGATGT-3′ |
| 59.01 |
| PPARG | F:
5′-CCACTATGGAGTTCATGCTTGT-3′ | 183 | 58.39 |
|
| R:
5′-AACCTGATGGCATTATGAGACA-3′ |
| 57.49 |
| LPL | F:
5′-CCGACAGGATTACAGGAGGAA-3′ | 141 | 58.89 |
|
| R:
5′-TGTGGTTGAAGTGACAGTTAGC-3′ |
| 58.80 |
| AFP | F:
5′-GACCTTCCGAGCCATAAC-3′ | 243 | 54.81 |
|
| R:
5′-CAATGACCAAGTTCAAGTGT-3′ |
| 54.03 |
| ALB | F:
5′-ATCTCACTAAGGTCCACAAG-3′ | 145 | 54.08 |
|
| R:
5′-TCCAACACAGGCTTATCAC-3′ |
| 54.79 |
| SOX9 | F: 5′-
ACCGCCTTGTCGTTAGACTG-3′ | 116 | 60.04 |
|
| R:
5′-GAATCTCCATCGTCCTCCAC-3′ |
| 57.48 |
| COL-2 | F:
5′-CTCAAGTCCCTCAACAACCAG-3′ | 118 | 58.50 |
|
| R:
5′-AGTCTCCGCTCTTCCACTCA-3′ |
| 60.25 |
| GAPDH | F:
5′-CACTGTCCACGCCATCACT-3′ | 442 | 60.00 |
|
| R:
5′-CCTGTTGCTGTAGCCGAATT-3′ |
| 58.55 |
Karyotype analysis
Chromosomes were prepared, fixed and stained
according to a method described previously (20). The sheep MMSCs were incubated in 0.5
µg/ml colcemid (Sigma-Aldrich; Merck Millipore) at 37.5°C for 6 h,
then the cells were digested with 0.25% trypsin. The centrifuged
cells (200 × g, 8 min at room temperature) were resuspended
in 0.075 M KCl solution at 37°C for 30 min. Finally, the cells were
fixed in 3:1 methanol: glacial acetic acid and dropped in the
frozen glass slides, stained with Giemsa. The cell chromosome
numbers were counted under an oil immersion objective. Relative
length, centromeric index and arm ratio were analyzed according to
the protocol of Sun et al (21) and Kawarai et al (22).
Colony-forming cell analysis
The sheep MMSCs from P4, 10 and 16 were cultured in
60-mm plates at a density of 100 cells/well for two weeks and the
numbers of colony-forming units were counted. The cloning
efficiencies were calculated as follows: Colony-forming unit
number/100 × 100% (2).
Growth kinetics
The sheep MMSCs from P5, 10 and 16 were plated in
24-well microplates at a density of 1×104 cells/well for
growth kinetics analysis for seven days. Growth curves were plotted
according to the mean values (1).
The population doubling time (PDT) was computed as follows: PDT =
(t-t0)
log2/(logNt-logN0), where
t=termination time of culture, t0=start time of
culture, Nt=the final number of cells in culture and
N0=initial number of cells in culture.
Adipogenic differentiation
The sheep MMSCs were seeded in six-well plates at a
density of 2×104 cells/well. Upon reaching 60–70%
confluence, cells in the control group were maintained in culture
medium. While the cells in the induced group were induced to
differentiate into adipogenic by culturing in DMEM/F12 supplemented
with 10% FBS, 1 mM dexamethasone, 200 µM indomethacin, 0.5 mM IBMX
and 10 µM insulin. Two weeks later, cells in the two groups were
detected by staining with Oil Red O.
Hepatocellular differentiation
The sheep MMSCs were seeded and separated into two
groups, as described above. Upon reaching 60–70% confluence, cells
in the control group were maintained in culture medium. While the
cells in the induced group were induced to differentiate into
hepatocellular by culturing in DMEM/F12 supplemented with 5% FBS,
100 µg/ml streptomycin, 100 IU/ml penicillin, 20 ng/ml hepatocyte
growth factor, 40 nmol/ml dexamethasone, 1% ITS liquid media
supplement, 10 ng/ml interleukin-6 and 20 ng/ml epidermal growth
factor. Two weeks later, cells in the two groups were detected by
staining with periodic acid-Schiff stain (PAS).
Chondrogenic differentiation
The sheep MMSCs were seeded and separated into two
groups as described above. Upon reaching 50–60% confluence, cells
in the control group were maintained in culture medium. While the
cells in the induced group were induced to differentiate into
chondrogenic cells by culturing in DMEM/F12 supplemented with 10%
FBS, 1% ITS, 50 µg/ml L-proline, 0.1 µm dexamethasone, 0.9 mM
sodium pyruvate, 50 ug/ml L-ascorbic acid and 10 ng/ml transforming
growth factor-β3. Two weeks later, cells in the two groups were
detected by staining with Alcian blue.
Statistical analysis
Statistical analyses of the data were performed with
a one-way analysis of variance, followed by the Tukey-Kramer honest
significant difference test for the three sets of results.
P<0.01 was considered to indicate a statistically significant
difference. Statistical analyses were performed using
JMP® Statistical Discovery Software (version 9.2; SAS
Institute, Inc., Cary, NC, USA).
Results
Isolation, culture and morphology of
MMSCs
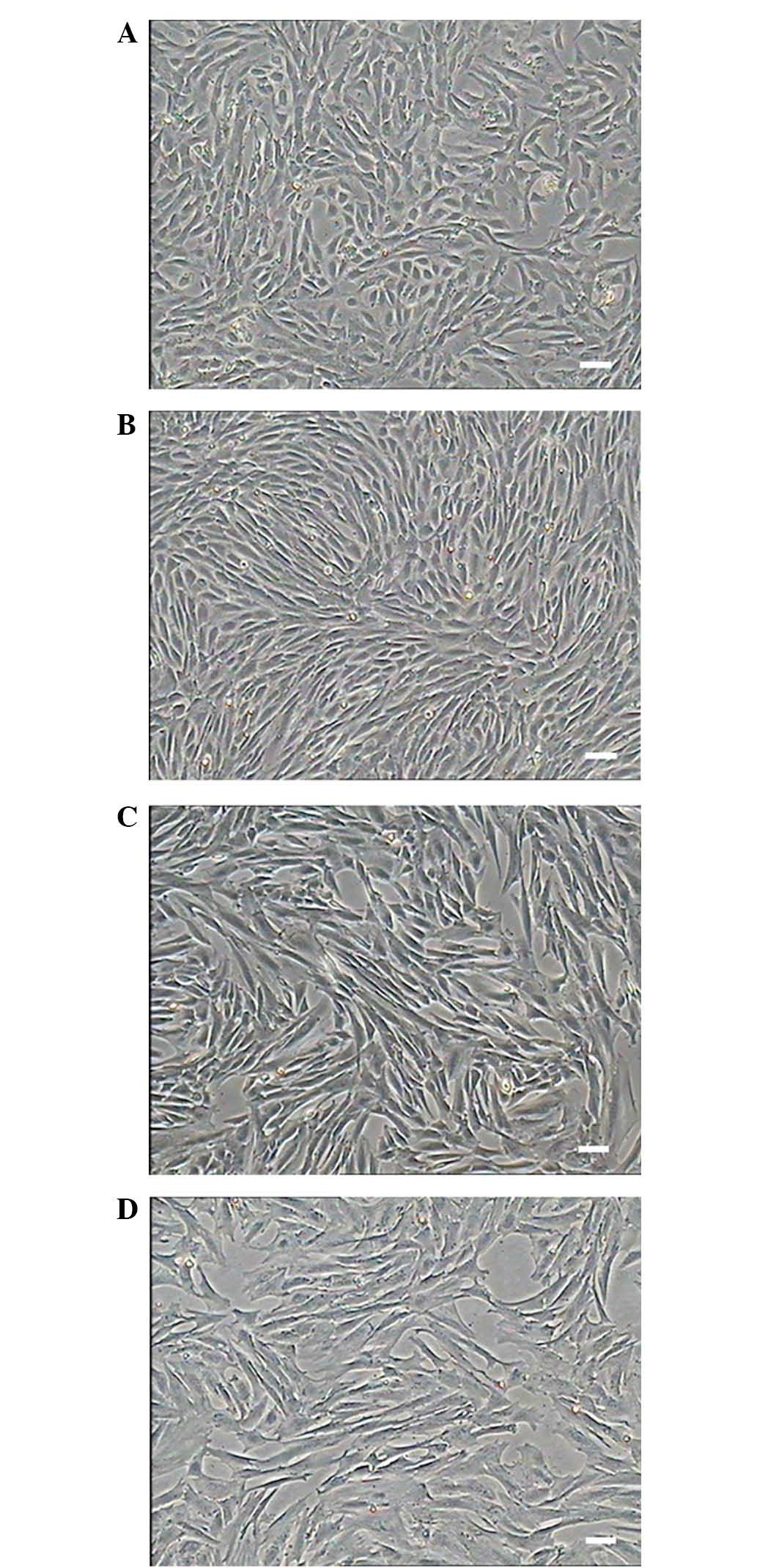
The cells isolated from the kidney adhered to the
culture plates with the initially morphology of round or irregular
shape and started to elongate after 48 h. The cells displayed a
fibroblast-like morphology (Fig. 1)
and grew to 80–90% confluence after five days (Fig. 1A). The isolated primary MMSCs were
mixed with blood cells, fibroblast cells and epithelial cells and
proliferated readily, after P3-4, other cells detached and were
eliminated from the population and which displayed a unique vortex
shape (Fig. 1B). The cells showed no
obvious morphological differences, and cellular morphology remained
stable among successive passages (Fig.
1C). The cells were cultured to passage 16, with the majority
of cells displaying signs of senescence, such as slow proliferation
and blebbing. These phenomena were consistent with the PDT
statistical analysis and growth curve (Fig. 2). As the increased passage, nearly
all the cells flat grown on the culture plates without replete
cytoplasm (Fig. 1D).
Self-renewal and proliferation
assays
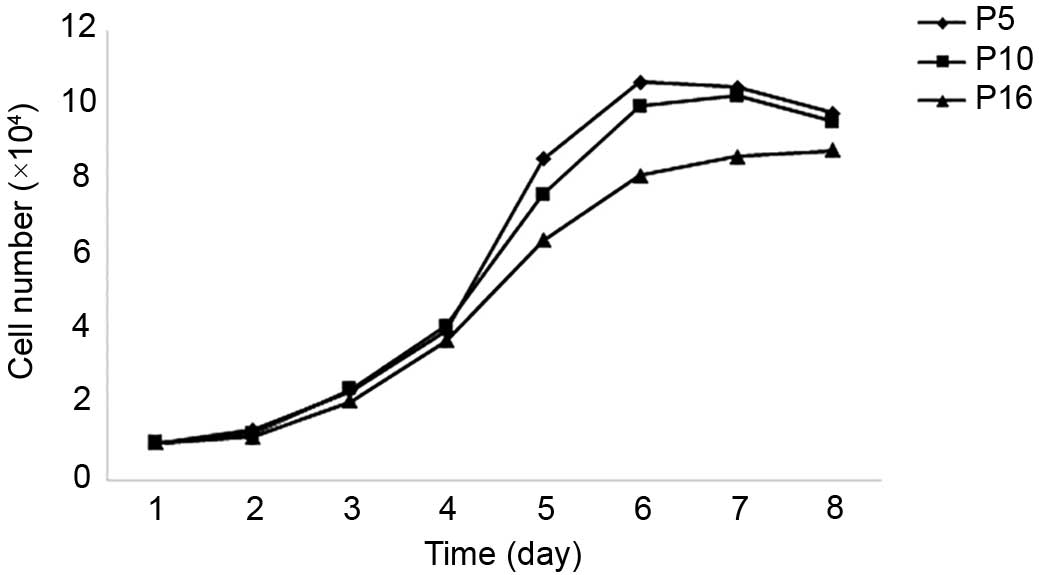
The growth curve of MMSCs from three different
passages appeared as typically sigmoidal, which consisted of a
latent phase, a logarithmic phase and a plateau phase (Fig. 2). The PDTs were ~33.32, 35.33 and
47.04 h at P5, P10 and P16, respectively.
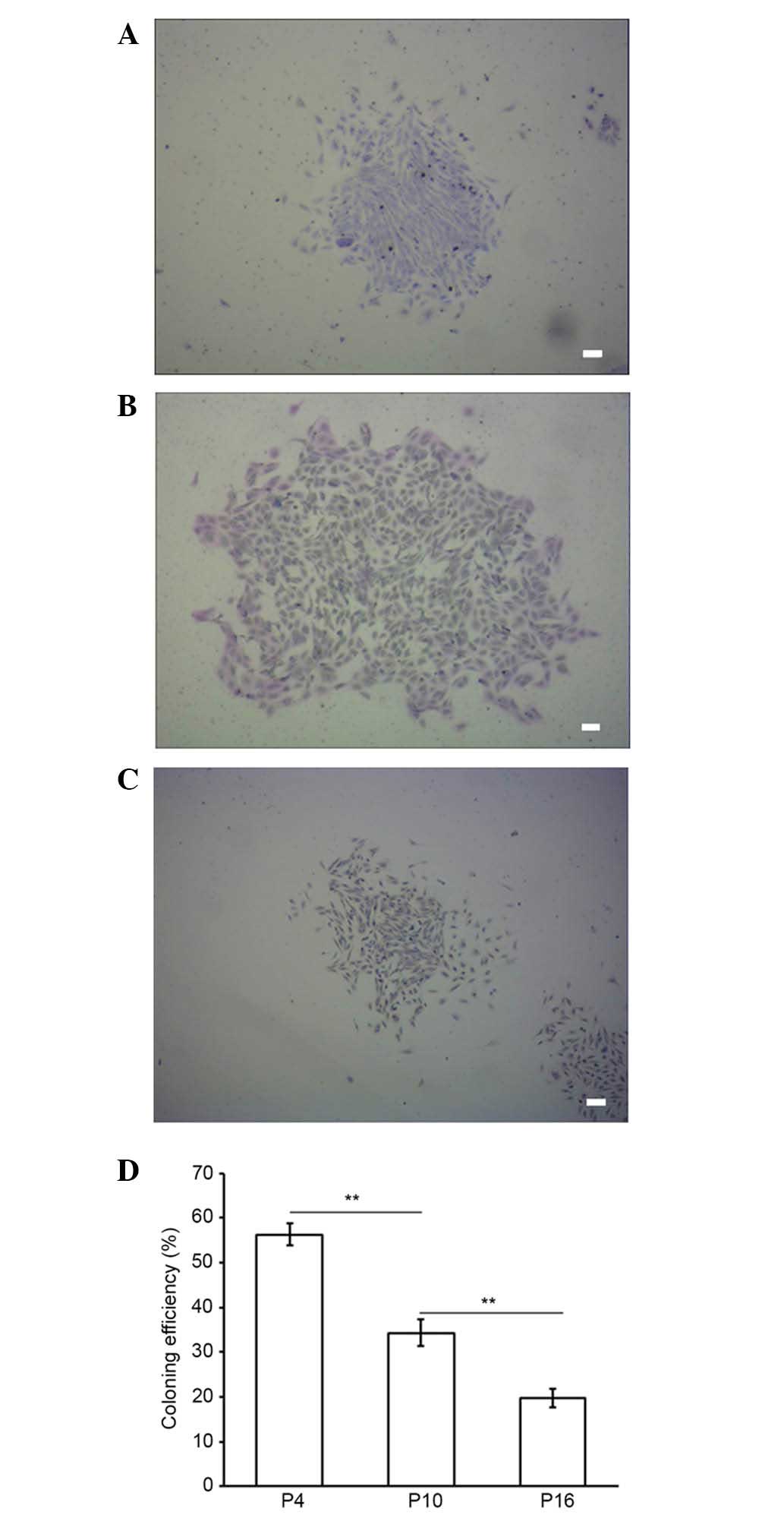
Colony formation of MMSCs from three different
passages was evaluated by staining with Giemsa after two weeks. The
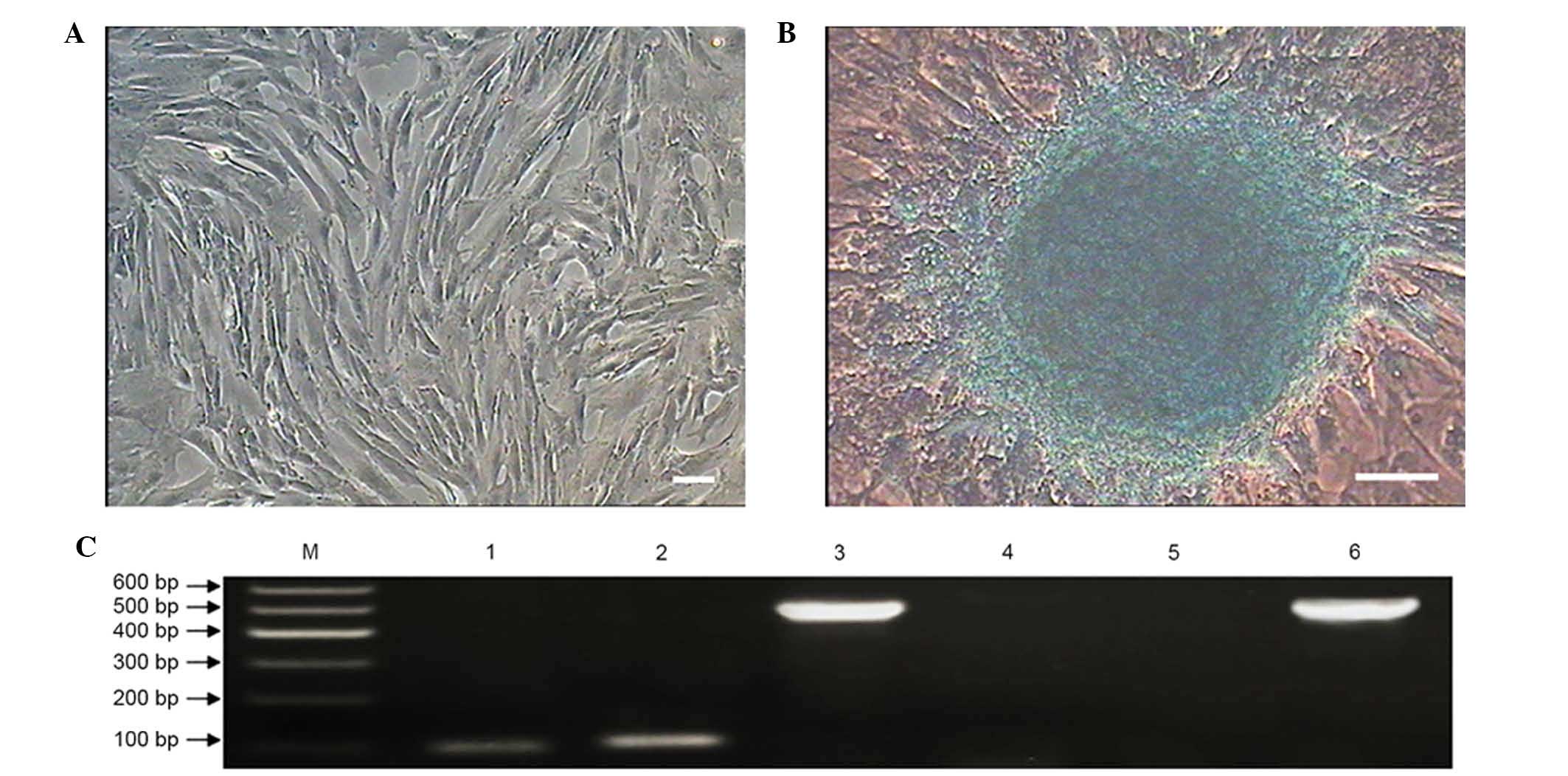
colony-forming efficiency rates of the MMSCs at P4, 10 and 16 were
56.33±2.52, 34.33±3.06 and 19.67±2.08%, respectively (Fig. 3). P4 MMSC proliferation capability
was significantly higher than P10 and P16 (P<0.01), and P10 was
also significantly higher than P16 (P<0.01; Fig. 3D). These results suggest that the
proliferation capacity of the cultured MMSCs declined significantly
in a passage-dependent manner (Fig.
3).
Characterization of MMSCs
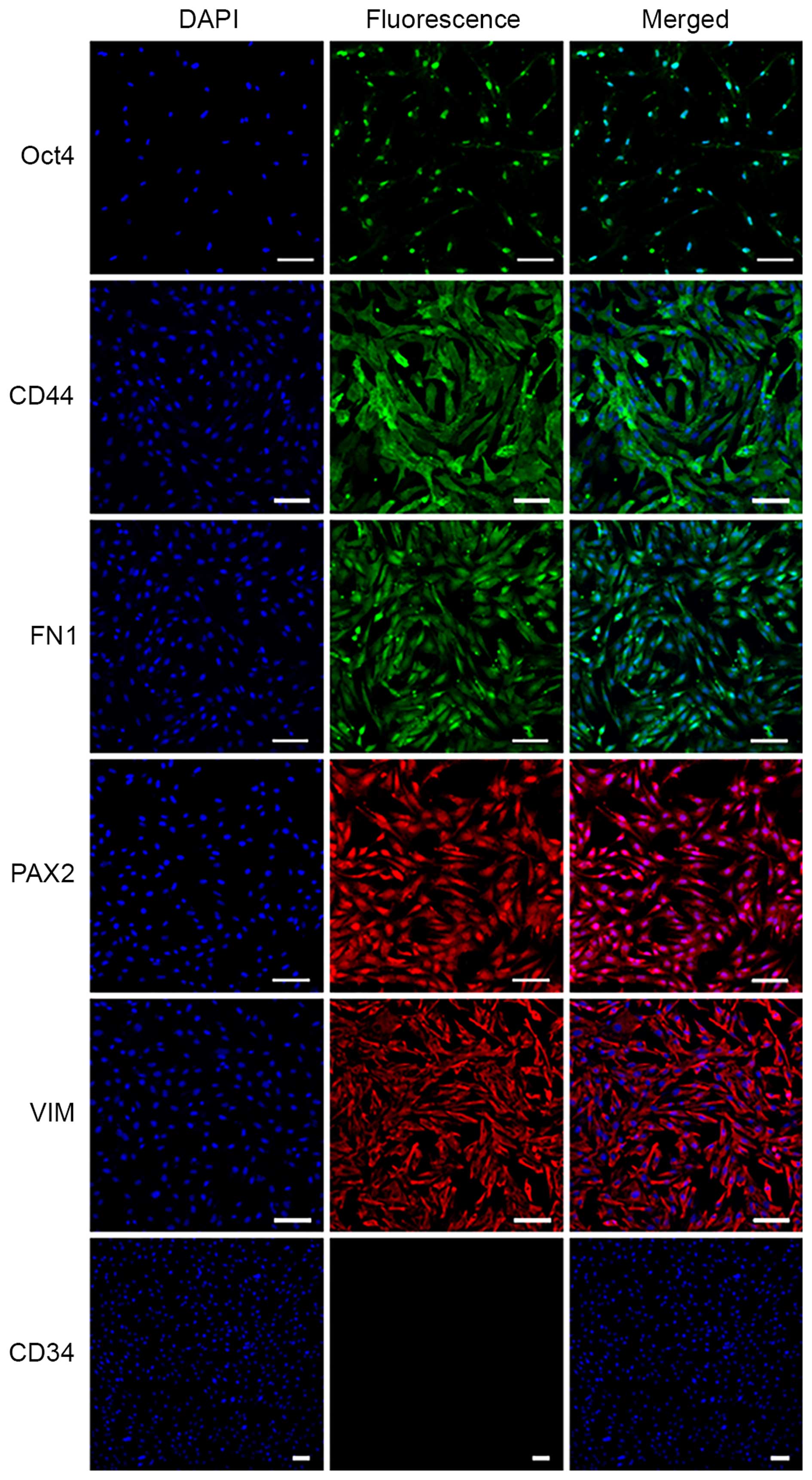
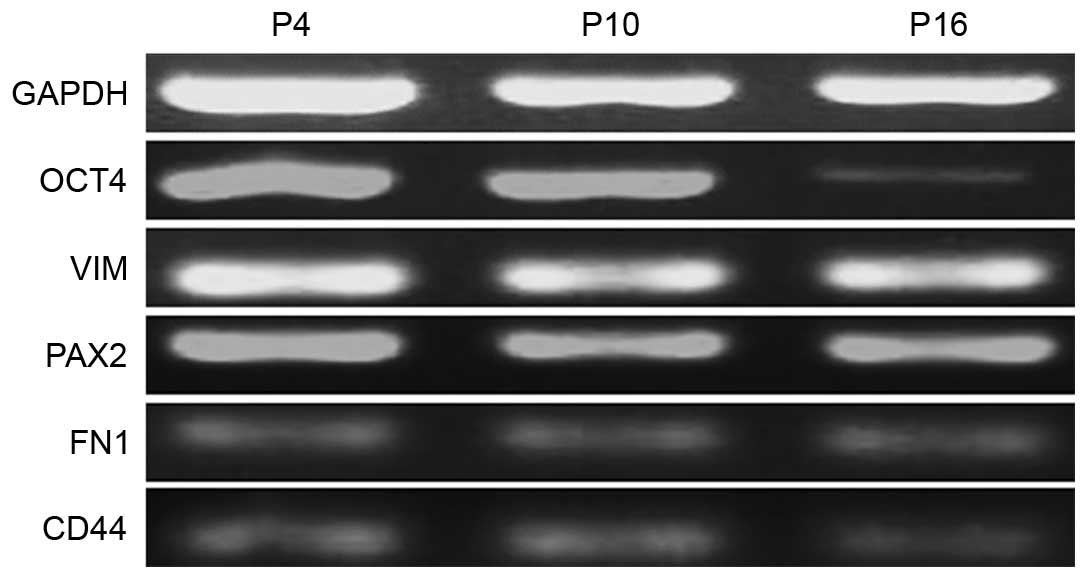
Markers of MMSCs
The specific markers of MMSCs were detected by
immunofluorescence and RT-PCR analyses. The results of
immunofluorescence showed that the MMSCs expressed Oct-4, VIM,
CD44, FN1 and PAX2, but was negative for CD34 expression (Fig. 4). Similarly, the results of RT-PCR
showed that MMSCs positively expressed Oct-4, CD73, CD44, PAX2 and
VIM (Fig. 5).
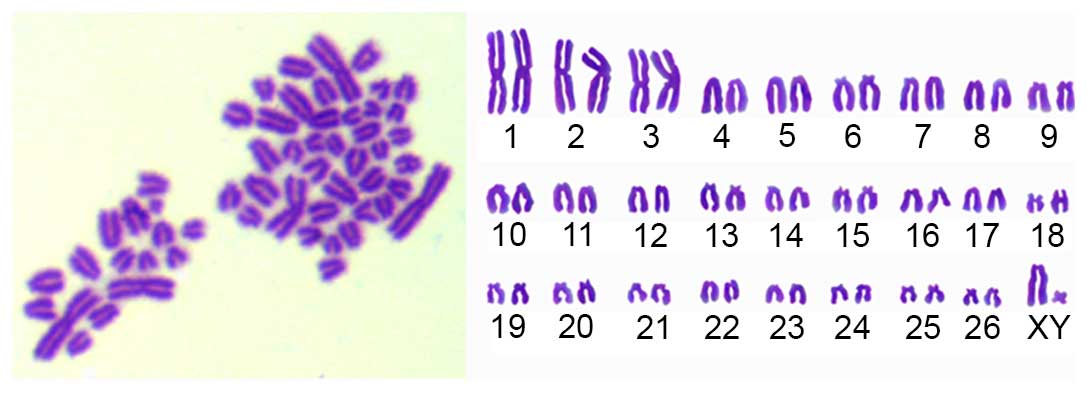
Karyotype analysis
The sheep MMSCs were diploid (2n=54). The sex
chromosome type is XY (Fig. 6). In
this study, the MMSCs chromosome numbers were calculated in 100
spreads per passage of P1-P20 to count the diploid rates. The
results showed diploid cells accounted for 95%, implying that the
cultured cells possessed of genetic stability.
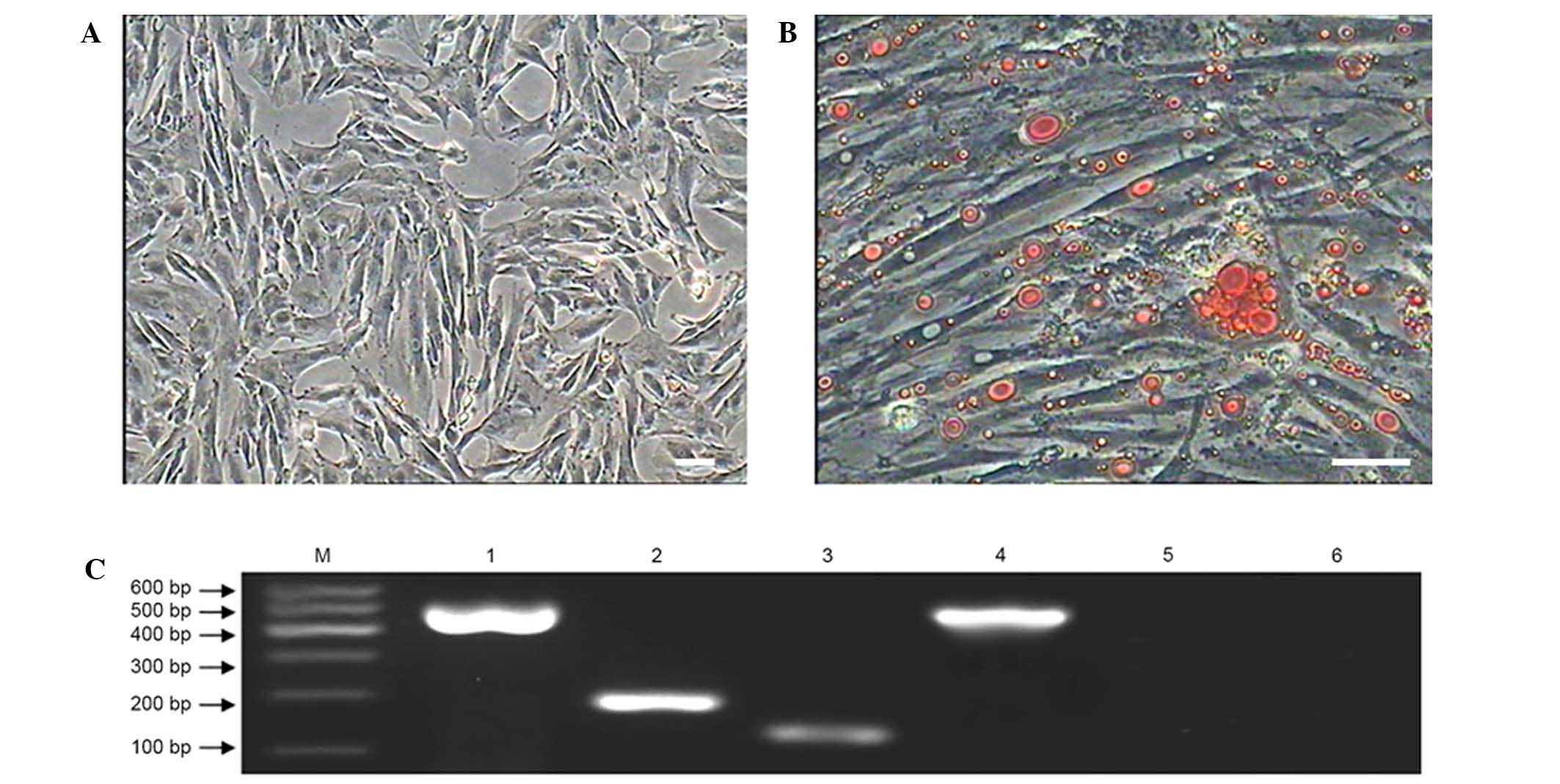
Adipogenic differentiation of MMSCs
When the MMSCs were cultured in adipogenic inducing
medium, the morphology of cells changed slowly from a shuttle shape
to an oblate shape and the cytoplasm contained numerous tiny lipid
droplets after 7 days (Fig. 7). With
the induction time extended to 12 days, the tiny lipid droplets
aggregated to form an increased number of larger droplets (Fig. 7B). Adipogenic differentiation of
MMSCs was detected by Oil Red O staining (3). The MMSCs cultured in complete medium as
a control group were negative for Oil Red O staining (Fig. 7A).
RT-PCR results showed that after incubation with
adipogenic inducing medium for 12 days the cells expressed the
adipocyte-specific genes peroxisome proliferator-activated
receptor-gamma and lipoprotein lipase, while the cells in the
control group were negative for expression of these genes (Fig. 7C).
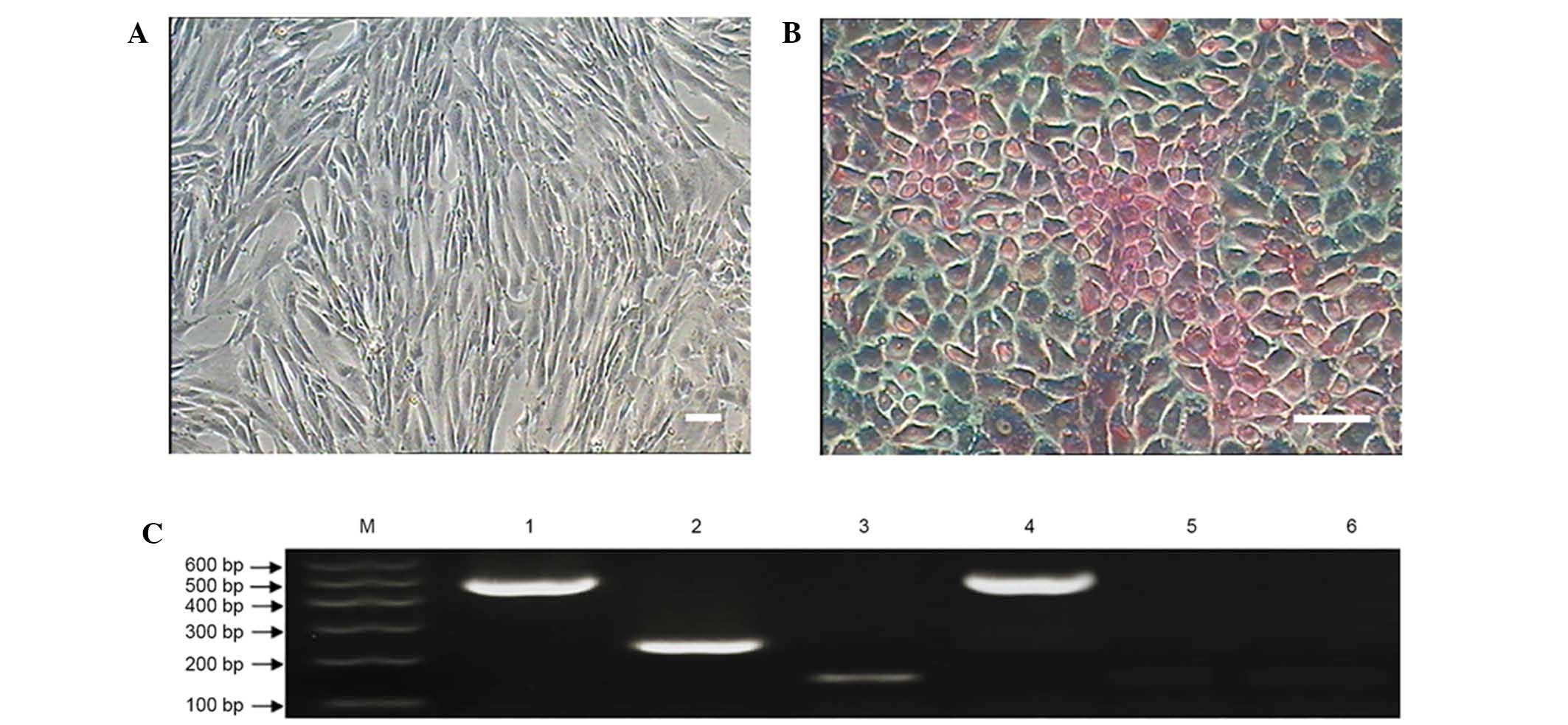
Hepatocellular differentiation of MMSCs
After incubating the MMSCs in hepatocellular medium
for 7 days, the cells displayed obvious morphological changes, with
a small population of small round-shaped cells observed (Fig. 8). After 14 days of differentiation,
the cells showed a rising/piled morphology with light nuclei and
dark cytoplasm that were stained with PAS (Fig. 8B) (23). The control group cells cultured in
complete medium showed no such changes (Fig. 8A).
To evaluate the characterization of the MMSC-derived
hepatocytes, two typical markers of hepatic cell gene expression
were examined at the mRNA level. The RT-PCR results showed that the
induced cells expressed typical markers of hepatic cells, such as
alpha-fetoprotein and albumin (Fig.
8C), while the control group cells cultured in complete medium
expressed no hepatic markers.
Chondrogenic differentiation of MMSCs
The sheep MMSCs were cultured in chondrogenic
medium, and growth slowed. The MMSCs showed obvious morphological
changes and formed small colonies after culture in chondrogenic
medium for 12 days (Fig. 9). After
20 days of differentiation, Alcian blue staining revealed an
increased number and size of colonies (Fig. 9B) (24). The control group cells cultured in
complete medium showed no such effects (Fig. 9A).
RT-PCR was performed to analyze the chondrogenic
differentiation of MMSCs. The results showed cartilage-specific
genes sex determining region Y-box 9 and collagen type II were
optimistic expressed in the induced group, but negatively expressed
in the control group (Fig. 9C).
Discussion
In the present study, we successfully isolated MMSCs
from the kidneys of six-week-old sheep fetuses. In addition, we
observed the MMSCs isolated from six- and 10-week-old fetuses with
significant differences in cell viability (data not shown). MMSCs
were isolated from the six- and 10-week-old fetus kidneys, and the
cell adherence of six-week-old MMSCs was more efficient than the
10-week-old cells, which demonstrated that younger animals were
more suitable for separating the MMSCs.
Previous studies have demonstrated that MMSCs
express the stem cell marker Oct-4, which usually is associated
with stem cell self-renewal and maintaining pluripotency (25,26). In
addition, MMSCs were shown to differentiate in vitro into
adipocytes, hepatocytes and chondrocytes.
We detected the protein expression of VIM, CD44,
PAX2 and FN1 in MMSCs using immunofluorescence and mRNA expression
of VIM, CD44, PAX2 and CD73 using RT-PCR. VIM is a type III
intermediate filament protein that is expressed in mesenchymal
cells (27). CD44, a receptor for
hyaluronic acid and a protein coding gene, encodes a cell-surface
glycoprotein involved in cell adhesion, cell-cell interactions and
migration such as collagens, osteopontin and matrix
metalloproteinases (28). PAX2, a
protein coding gene, expressed in fetal kidneys, throughout the
branching ureteric bud Wolffian and Mullerian ducts, has an
essential role in urogenital tract development (29). Transcription factor of PAX2 that may
have a key function in kidney cell differentiation (29). In addition, it plays a critical role
in the branching ureteric the development of renal epithelium,
dysregulated during early stages of nephrogenesis (29). FN1 encodes fibronectin, a
glycoprotein present in extracellular matrix and in a dimeric or
multimeric form at the cell surface (30). FN is involved in cell adhesion and
migration processes. CD73 (Ecto-5′-nucleotidase) is a
multifunctional ectoenzyme that metabolizes adenosine
5′-monophosphate into adenosine (31). CD73 is also a
glycosyl-phosphatidylinositol-conjugated, membrane-bound
glycoprotein. CD73 is a signaling molecule and has been shown to
participate in purine salvaging and purinergic cascades that lead
to cell metabolism (31). This
pathway has been shown to be important in immunological signaling
and is involved in signal transduction and cell adhesion (31). In summary, the specific markers of
MSCs and kidney were shown to be positively expressed in MMSCs by
immunofluorescence and RT-PCR analyses.
Twice diploid karyotype and the stably
characteristic chromosome number, shape and structure are the
prerequisite for the cell growth and functioning (32). Therefore, karyotype analysis is a
simple and practical method for distinguishing normal cells from
variants. Sheep have 27 pairs of chromosomes, with sex chromosomes
X and Y (33). The sheep MMSCs
cultured in the present study were all normal diploids. In our
experiment, we found that the sheep MMSCs were diploid (2n=54,
XY).
The multipotency of stem cells is an important
prerequisite for autologous cell therapy (34). As they are conducive to mass
preparation, MMSCs can serve as ideal experimental cells for tissue
engineering research (8). The
developmental repertoire of tissue stem cells in vivo is
subject to the control of gene-encoded transcription factors and
extracellular signals (35);
however, the differentiation mechanism remains unclear in
vitro.
In the present study, we induced sheep MMSCs to
differentiate into adipocytes, hepatocytes and chondrocytes, under
different induction conditions. The various inducing factors can
affect the direction of differentiation of the MMSCs, which
originate from mesoblastoma and can be differentiated into endoderm
and ectoderm cell types in vitro. Therefore, the
multi-potentiality and convenient procurement of the MMSCs cultured
herein suggest that they may offer a potential option for tissue
engineering and cellular transplantation therapy (10).
Despite the multi-lineage differentiation of the
sheep MMSCs being successful in vitro, the technical
difficulties and safety concerns related to using the cells for
tissue recovery in vivo remain unsolved. Therefore,
additional studies are needed with regard to using these cells for
future research and therapy.
In conclusion, MMSCs were isolated from the kidney
of six-week-old sheep fetuses, and underwent analyses of their
self-renewal ability and differentiation potential in vitro.
These results suggest the potential utility of the sheep MMSCs as a
source of stem cells for regenerative medical therapies.
Acknowledgements
The present study was supported by the Agricultural
Science and Technology Innovation Program (cxgc-ias-01), the
National Natural Science Foundation of China (grant nos. 31201765
and 31272403; 31472064), the China Postdoctoral Science Foundation
(grant no. 2015M571182) and the National Infrastructure of Animal
Germplasm Resources (2014).
References
|
1
|
Bai C, Hou L, Ma Y, Chen L, Zhang M and
Guan W: Isolation and characterization of mesenchymal stem cells
from chicken bone marrow. Cell Tissue Bank. 14:437–451. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gao Y, Zhu Z, Zhao Y, Hua J, Ma Y and Guan
W: Multilineage potential research of bovine amniotic fluid
mesenchymal stem cells. Int J Mol Sci. 15:3698–3710. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li X, Gao Y, Hua J, Bian Y, Mu R, Guan W
and Ma Y: Research potential of multi-lineage chicken amniotic
mesenchymal stem cells. Biotech Histochem. 89:172–180. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Naghdi M, Tiraihi T, Namin SA and
Arabkheradmand J: Transdifferentiation of bone marrow stromal cells
into cholinergic neuronal phenotype: A potential source for cell
therapy in spinal cord injury. Cytotherapy. 11:137–152. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Drost AC, Weng S, Feil G, Schäfer J,
Baumann S, Kanz L, Sievert KD, Stenzl A and Möhle R: In vitro
myogenic differentiation of human bone marrow-derived mesenchymal
stem cells as a potential treatment for urethral sphincter muscle
repair. Ann N Y Acad Sci. 1176:135–143. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tamama K, Sen CK and Wells A:
Differentiation of bone marrow mesenchymal stem cells into the
smooth muscle lineage by blocking ERK/MAPK signaling pathway. Stem
Cells Dev. 17:897–908. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shafiee A, Kabiri M, Ahmadbeigi N, Yazdani
SO, Mojtahed M, Amanpour S and Soleimani M: Nasal septum-derived
multipotent progenitors: A potent source for stem cell-based
regenerative medicine. Stem Cells Dev. 20:2077–2091. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bai C, Li X, Hou L, Zhang M, Guan W and Ma
Y: Biological characterization of chicken mesenchymal
stem/progenitor cells from umbilical cord Wharton's jelly. Mol Cell
Biochem. 376:95–102. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gao Y, Bai C, Xiong H, Li Q, Shan Z, Huang
L, Ma Y and Guan W: Isolation and characterization of chicken
dermis-derived mesenchymal stem/progenitor cells. Biomed Res Int.
2013:6262582013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Osafune K: Cell therapy for kidney injury:
Different options and mechanisms-kidney progenitor cells. Nephron
Exp Nephrol. 126:642014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu L, Chen D, Yi ZW, Liu XH, Wu XC, Dang
XQ, He QN, He XJ and Mo SH: Nephroprotective effects of subcapsular
transplantation of metanephric mesenchymal cells on
gentamicin-induced acute tubular necrosis in rats. World J Pediatr.
8:156–163. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Oliver JA, Barasch J, Yang J, Herzlinger D
and Al-Awqati Q: Metanephric mesenchyme contains embryonic renal
stem cells. Am J Physiol Renal Physiol. 283:F799–F809. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Al-Awqati Q and Oliver JA: Stem cells in
the kidney. Kidney Int. 61:387–395. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang Y, Jiang H, Bai Y, Liang J, Zhao A
and Dou L: Research progress of bone marrow-derived mesenchymal
stem cells in the treatment of chronic kidney disease. Hainan Med
J. 27:968–972. 2016.
|
|
15
|
Xiangyu Z, Guangyuan Z, Zhongliang C,
Deming Y, Tao D, Guanqun J, Shuai M, Guohua L, Mujun L and Yingjian
Z: Microvesicles derived from human Wharton's Jelly mesenchymal
stromal cells ameliorate renal ischemia-reperfusion injury in rats
by suppressing CX3CL1. Stem Cell Res Ther. 5:402014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yeo RWY, Lai RC, Tan KH and Lin SK:
Exosome: A novel and safer therapeutic refinement of mesenchymal
stem cell. Exosomes Microvesicles. 1:72013.
|
|
17
|
Dressler GR: Advances in early kidney
specification, development and patterning. Development.
136:3863–3874. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
McManus LM and Mitchell RN: Pathobiology
of Human Disease. 1st. Elsevier Science Publishing Co., Inc.;
Amsterdam: 2014, View Article : Google Scholar
|
|
19
|
Barber C, Garnham L, Lovell S, Camus H and
Persaud M: Galvanising the role of learning disability nursing. Br
J Nurs. 17:S32008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Baran SW and Ware CB: Cryopreservation of
rhesus macaque embryonic stem cells. Stem Cells Dev. 16:339–344.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sun CC, Su Pang JH, Cheng CY, Cheng HF,
Lee YS, Ku WC, Hsiao CH, Chen JK and Yang CM: Interleukin-1
receptor antagonist (IL-1RA) prevents apoptosis in ex vivo
expansion of human limbal epithelial cells cultivated on human
amniotic membrane. Stem Cells. 24:2130–2139. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kawarai S, Hashizaki K, Kitao S, Nagano S,
Madarame H, Neo S, Ishikawa T, Furuichi M, Hisasue M, Tsuchiya R,
et al: Establishment and characterization of primary canine
hepatocellular carcinoma cell lines producing alpha-fetoprotein.
Vet Immunol Immunopathol. 113:30–36. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang Z, Sun B, Zhao X, Shao B, An J, Gu Q,
Wang Y, Dong X, Zhang Y and Qiu Z: Erythropoietin and
erythropoietin receptor in hepatocellular carcinoma: Correlation
with vasculogenic mimicry and poor prognosis. Int J Clin Exp
Pathol. 8:4033–4043. 2015.PubMed/NCBI
|
|
24
|
Karystinou A, Roelofs AJ, Neve A,
Cantatore FP, Wackerhage H and De Bari C: Yes-associated protein
(YAP) is a negative regulator of chondrogenesis in mesenchymal stem
cells. Arthritis Res Ther. 17:1472015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Babaie Y, Herwig R, Greber B, Brink TC,
Wruck W, Groth D, Lehrach H, Burdon T and Adjaye J: Analysis of
Oct4-dependent transcriptional networks regulating self-renewal and
pluripotency in human embryonic stem cells. Stem Cells. 25:500–510.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee J, Kim HK, Rho JY, Han YM and Kim J:
The human OCT-4 isoforms differ in their ability to confer
self-renewal. J Biol Chem. 281:33554–33565. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Challa AA and Stefanovic B: A novel role
of vimentin filaments: Binding and stabilization of collagen mRNAs.
Mol Cell Biol. 31:3773–3789. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Vikesaa J, Hansen TV, Jønson L, Borup R,
Wewer UM, Christiansen J and Nielsen FC: RNA-binding IMPs promote
cell adhesion and invadopodia formation. Embo J. 25:1456–1468.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Barua M, Stellacci E, Stella L, Weins A,
Genovese G, Muto V, Caputo V, Toka HR, Charoonratana VT, Tartaglia
M and Pollak MR: Mutations in PAX2 associate with adult-onset FSGS.
J Am Soc Nephrol. 25:1942–1953. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Purushothaman A, Bandari SK, Liu J, Mobley
JA, Brown EE and Sanderson RD: Fibronectin on the surface of
myeloma cell-derived exosomes mediates exosome-cell interactions. J
Biol Chem. 291:1652–1663. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Garavaglia S, Bruzzone S, Cassani C,
Canella L, Allegrone G, Sturla L, Mannino E, Millo E, De Flora A
and Rizzi M: The high-resolution crystal structure of periplasmic
Haemophilus influenzae NAD nucleotidase reveals a novel enzymatic
function of human CD73 related to NAD metabolism. Biochem J.
441:131–141. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ji M, Guan W, Gao Y, Li L, Bai C, Ma Y and
Li C: Cultivation and biological characterization of chicken
primordial germ cells. Braz Arch Biol Technol. Feb 26–2016.(Epub
ahead of print). View Article : Google Scholar
|
|
33
|
Pauciullo A, Perucatti A, Cosenza G,
Iannuzzi A, Incarnato D, Genualdo V, Di Berardino D and Iannuzzi L:
Sequential cross-species chromosome painting among river buffalo,
cattle, sheep and goat: A useful tool for chromosome abnormalities
diagnosis within the family Bovidae. PloS One. 9:e1102972014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nagoshi N, Shibata S, Kubota Y, Nakamura
M, Nagai Y, Satoh E, Morikawa S, Okada Y, Mabuchi Y, Katoh H, et
al: Ontogeny and multipotency of neural crest-derived stem cells in
mouse bone marrow, dorsal root ganglia, and whisker pad. Cell Stem
Cell. 2:392–403. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Guilak F, Cohen DM, Estes BT, Gimble JM,
Liedtke W and Chen CS: Control of stem cell fate by physical
interactions with the extracellular matrix. Cell Stem Cell.
5:17–26. 2009. View Article : Google Scholar : PubMed/NCBI
|