Introduction
Osteoblasts, which are amongst the most important
cells in the bone development and remodeling process, are
responsible for the synthesis, secretion and mineralization of bone
matrix and have an important effect in bone development,
maturation, repair and remodeling (1,2).
Mesenchymal stem cells (MSCs) first differentiated into
osteoprogenitor cells, then into preosteoblasts, osteoblasts, and
finally into osteocytes (3).
G protein-coupled receptor kinase interacting
protein 1 (GIT1) is widely present in mammals and birds and exists
mainly in the focal adhesion and cytoplasmic complex structure of
cells (4). It plays an important
role in cell growth and migration through interaction with a
variety of proteins, including Rac1, p21-activated kinase,
phospholipase C and paxillin (5). A
previous study has indicated that GIT1 can be expressed in both
osteoblasts and osteoclasts (6),
suggesting that GIT1 may be involved in the bone metabolic process.
Cell spreading and Boyden chamber assays have shown that GIT1
(Y321F) mutant inhibits the motility and migration of osteoblasts
induced by platelet-derived growth factor (PDGF), and it has been
demonstrated that phosphorylation of GIT1 tyrosine 321 is required
for association with focal adhesion kinase at focal adhesions and
for PDGF-activated migration of osteoblasts (7). Therefore, GIT1 possesses a vital
function in osteoblasts.
MicroRNA (miRNA), an endogenous non-coding
single-stranded small RNA, exists extensively in the biological
genome. It is able to pair with and bind to 3′ untranslated regions
(3′UTRs) of the targeted mRNA incompletely to degrade mRNA or
repress the translation of mRNA, and is involved in regulating more
than half of all gene expression (8). In addition, miRNA is an important
post-transcriptional regulatory factor and plays an extensive and
important role in cell proliferation, differentiation, apoptosis,
tissues development, oncogenesis and other physiological processes
(9). Studies have shown that miRNAs
also have a significant regulatory effect on osteoblastic
differentiation and bone development (10,11). In
the osteoblast differentiation process, miRNA closely associated
with this process, known as ‘osteomiRs’, are present, which can
regulate the differentiation process via many pathways (11,12). For
example, in the late stage of osteoblast differentiation, which is
the bone matrix deposition and mineralization phase, miR-29a and
miR-29c inhibit the expression of the marker gene osteonectin so as
to regulate the bone remodeling process (13).
Nevertheless, studies concerning miRNAs targeting
GIT1 are rare and, to the best of our knowledge, no studies have
involved miRNAs and GIT1 in osteoblasts. Given the important role
of GIT1, the present study aimed to investigate the regulatory
relationship and the interaction between GTI1 and relevant miRNA in
osteoblasts.
Materials and methods
Cell culture and osteoblastic
differentiation
Bone marrow-derived human MSCs (HMSCs) and human
osteoblasts were purchased from ScienCell Research Laboratories,
Inc. (Carlsbad, CA, USA). HMSCs were cultured in mesenchymal stem
cell medium (MSCM; cat. no. 7501; ScienCell Research Laboratories,
Inc.), and osteoblasts were cultured in osteoblast medium (ObM;
cat. no. 4601; ScienCell Research Laboratories, Inc.). Osteoblastic
differentiation was induced by changing to media containing 10% FBS
supplemented with 100 ng/ml bone morphogenetic protein 2 (BMP-2;
R&D Systems, Inc., Minneapolis, MN, USA).
Overexpression vector construction and
transfection
TRIzol (Thermo Fisher Scientific, Inc., Waltham, MA,
USA) was used to extract RNA from osteoblasts according to the
instructions of the manufacturer. Following this, the RNA was
subjected to reverse transcription-polymerase chain reaction
(RT-PCR) to amplify the coding region of GIT1 using an Advantage RT
for PCR kit (Takara Biotechnology Co., Ltd., Dalian, China). The
product was digested with KpnI and EcoRI (Takara
Biotechnology Co., Ltd.), and cloned into pcDNA3.1 vectors (Thermo
Fisher Scientific, Inc.), sequenced and verified in a 3730xl DNA
Analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc.). The
primers used for GIT1 amplification are shown in Table I. Transfection of human osteoblasts
was carried out using Lipofectamine 2000, following the
instructions of the manufacturer (Thermo Fisher Scientific, Inc.,)
in 6-well cell culture plates when the cell confluence reached
~70%. The concentration of GIT1 overexpression vector for
transfection was 4 µg/well, and that of human GIT1 small
interfering RNA (siRNA; sc-35,477; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA), miR-125a-3p mimics and inhibitors (Shanghai
GenePharma Co., Ltd., Shanghai, China) was 50 nM/well. For
osteoblastic differentiation, the medium was replaced 4 h after
transfection with fresh medium containing 10% FBS and 100 ng/ml
BMP-2.
 | Table I.Sequences of primers. |
Table I.
Sequences of primers.
| Primer set | Sense (5′-3′) | Antisense
(5′-3′) | Product size
(bp) |
|---|
| GIT1 CDS
amplification | GGGGTACCGCCACCATGTCCCGAAAGGGGCCG | CGGAATTCTCACTGCTTCTTCTCTCGGGTG |
|
| Quantitative PCR |
|
|
|
|
GAPDH |
GGTATCGTGGAAGGACTC |
GTAGAGGCAGGGATGATG | 128 |
| GIT1 |
ATGTATGAACCTGGCTCTG |
TGAATAGATGGCGTCGTC | 114 |
|
Runx2 |
CAAGGACAGAGTCAGATTAC |
GTGGTAGAGTGGATGGAC | 119 |
|
Osterix |
TGCTTGAGGAGGAAGTTC |
CTTTGCCCAGAGTTGTTG | 111 |
| GIT1 3′UTR
amplification |
CCGCTCGAGCCTCTCTCCCCACACCCTCA |
ATAAGAATGCGGCCGCTAACA
GCTCATGGTCACTTCTTTAT |
|
RT-quantitative PCR (qPCR)
Total RNA was isolated from cultured cell samples
using TRIzol according to the manufacturer's instructions, and a
mirVana miRNA Isolation kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.) was used to purify the miRNAs. Expression levels
were measured by RT-qPCR. First, cDNA was synthesized by RT using
random and oligo-dT primers or specific primers for miRNA-125a-3p
and the GoScript Reverse Transcription System (Promega Corporation,
Shanghai, China). Thermal cycling was performed as follows: one
cycle of 95°C for 5 min and 40 cycles of 95°C for 30 sec, 55°C for
30 sec and 72°C for 30 sec. qPCR was conducted using the GoTaq qPCR
Master mix (Promega Corporation) with an ABI PRISM® 7500
Sequence Detection system (Applied Biosystems) following the
manufacturer's instructions. The reaction was performed at one
cycle of 95°C for 2 min, followed by 40 cycles of 95°C for 15 sec
and 60°C for 32 sec. For measurement of miRNA expression, specific
primers for miRNA-125a-3p and U6 were used. The primer sequences
are shown in Table I. Three
independent experiments were conducted for each sample. Data were
analyzed using the 2−ΔΔCq method (14).
Western blot analysis
Total cellular proteins were extracted by incubating
cells in radioimmunoprecipitation assay (RIPA) buffer (sc-24948;
Santa Cruz Biotechnology, Inc.), separating the proteins (50 µg per
lane) by 10% SDS-PAGE and transferring them to nitrocellulose
membranes (Bio-Rad Laboratories, Inc., Shanghai, China). The
membranes were blocked with 5% non-fat milk for 1 h and then
incubated with GIT1 (cat. no. 2919; rabbit anti-human polyclonal
antibody; 1:1,000; Cell Signaling Technology, Inc., Beverly, MA,
USA) or GAPDH (14C10) rabbit monoclonal antibody (cat. no. 2118;
rabbit anti-human monoclonal antibody; 1:1,000; Cell Signaling
Technology, Inc.) in 5% non-fat milk overnight at 4°C.
Immunoreactive proteins were visualized using horseradish
peroxidase (HRP)-conjugated secondary antibodies (cat. no. 7074;
anti-rabbit IgG, HRP-linked antibody; 1:7,000–8,000 Cell Signaling
Technology, Inc.) and enhanced chemiluminescence reagents (Pierce;
Thermo Fisher Scientific, Inc.). Images were analyzed using
Image-Pro Plus 6.0 software (Media Cybernetics, Inc., Rockville,
MD, USA). Each band was scanned with background correction, and
values were averaged and expressed as the mean ± standard deviation
(SD).
Dual luciferase assay
GIT1 3′UTR was cloned into the dual-luciferase
reporter vector psiCHECK-2 (Promega Corporation), and the seed
region of the miR-125a-3p binding site in the GIT1 3′UTR was
mutated using a QuikChange II Site-Directed Mutagenesis kit
(Stratagene; Agilent Technologies, Inc., Santa Clara, CA, USA). For
luciferase assay, HEK293 cells (American Type Culture Collection,
Manassas, VA, USA) were transfected at 80% confluency in 24-well
dishes with psiCHECK-2 (with GIT1 3′UTR or GIT1 3′UTR mutation),
and miR-125a-3p mimics by Lipofectamine 2,000. Cells were analyzed
at 24 h post-transfection. Firefly and Renilla luciferase
activities were quantified in lysates using the Dual Luciferase
Reporter Assay kit (Promega Corporation) on a Glomax 20/20
luminometer (Promega Corporation) according to the manufacturer's
recommendations. Luciferase readings were corrected for background
and firefly luciferase values were normalized to Renilla to
control for transfection efficiency. All assays were performed in
triplicate in three independent experiments.
Immunoprecipitation
HEK293 cells were lysed in RIPA buffer containing
protease inhibitors (Roche Diagnostics, Basel, Switzerland) and
centrifuged at 10,000 × g for 10 min at 4°C. For
immunoprecipitation, the lysates were incubated with monoclonal
anti-argonaute 2 (Ago2) antibody (1:50; cat. no. 2897; Cell
Signaling Technology, Inc.) overnight at 4°C. Subsequently, 5 ml
hybridoma was coupled to 80 µl Protein-G-Sepharose (GE Healthcare
Life Sciences, Chalfont, UK). Beads were subsequently incubated
with 10 ml HEK293 lysate for 5 h under constant rotation at 4°C.
After incubation, the beads were washed three times with washing
buffer. Finally, the beads were washed once with PBS.
Co-precipitated RNA was extracted using phenol:chloroform:isoamyl
alcohol (25:24:1; cat. no. 15593-031; Invitrogen; Thermo Fisher
Scientific, Inc.). The RNA pellet was used for RT-qPCR.
5-Bromo-2´-deoxyuridine (BrdU) cell
proliferation assay
BrdU assay was used to investigate the effects of
miR-125a-3p and GIT1 on cell proliferation. Briefly, cultured cells
were seeded into a 6-well plate and incubated for 24 h.
Transfections with miR-125a-3p mimics and inhibitors, plasmids and
siRNAs were then carried out using Lipofectamine 2000. A BrdU Cell
Proliferation Assay kit (cat. no. 6813; Cell Signaling Technology,
Inc.) was used for the quantification of cell proliferation
according to the protocol provided by the manufacturer. Cell
proliferation was expressed as a mean percentage of that of the
control (set at 100%).
Alkaline phosphatase (ALP)
measurement
Cells were washed twice and then lysed on ice for 30
min in lysis buffer composed of Mammalian Protein Extraction
Reagent and Protease Inhibitor Cocktail (CW Biotech Co., Beijing,
China). In the following step, lysates were collected and
centrifuged at 10,000 × g for 10 min, and the supernatants
were transferred into a new tube for the detection of ALP activity.
The activity of ALP was measured using a commercially available
Alkaline Phosphatase Assay kit (Nanjing Jiancheng Bioengineering
Institute, Nanjing, China). Supernatants were also analyzed for
protein concentration using a bicinchoninic acid (BCA) assay kit
(Beyotime Institute of Biotechnology, Haimen, China), and ALP
activity was normalized for total protein concentration. The
absorbance values at 490 and 570 nm, respectively for ALP and BCA
analysis, were detected with a Multifunctional Microplate Reader
(Bio-Rad Laboratories, Inc.).
Enzyme-linked immunosorbent assay
(ELISA)
Osteocalcin (OC) secretions from culture
supernatants were assessed using an ELISA kit (cat. no. KAQ1381;
Invitrogen). A 25-µl portion of each sample was added to the
appropriate wells, and then 100 µl working anti-OST-HRP conjugate
was added to all wells and the plates were incubated for 2 h at
room temperature. Solution was thoroughly aspirated or decanted
from the wells and discarded, and the wells were then washed 3
times. Chromogen solution (100 µl) to each well, and the plate was
incubated for 30 min at room temperature in the dark. Stop solution
(100 µl) was added to each well, and the absorbance of each well
was read at 450 nm.
Statistical analysis
The experiments were carried out at least in
triplicate and results were expressed as mean ± SD. SPSS
statistical package (SPSS 17.0 for Windows; SPSS, Inc., Chicago,
IL, USA) was used for statistical analysis. Differences were
analyzed by non-parametric statistical analysis (Mann-Whitney U
tests) between control and treated groups, and P<0.05 was
considered to indicate a statistically significant difference.
Results
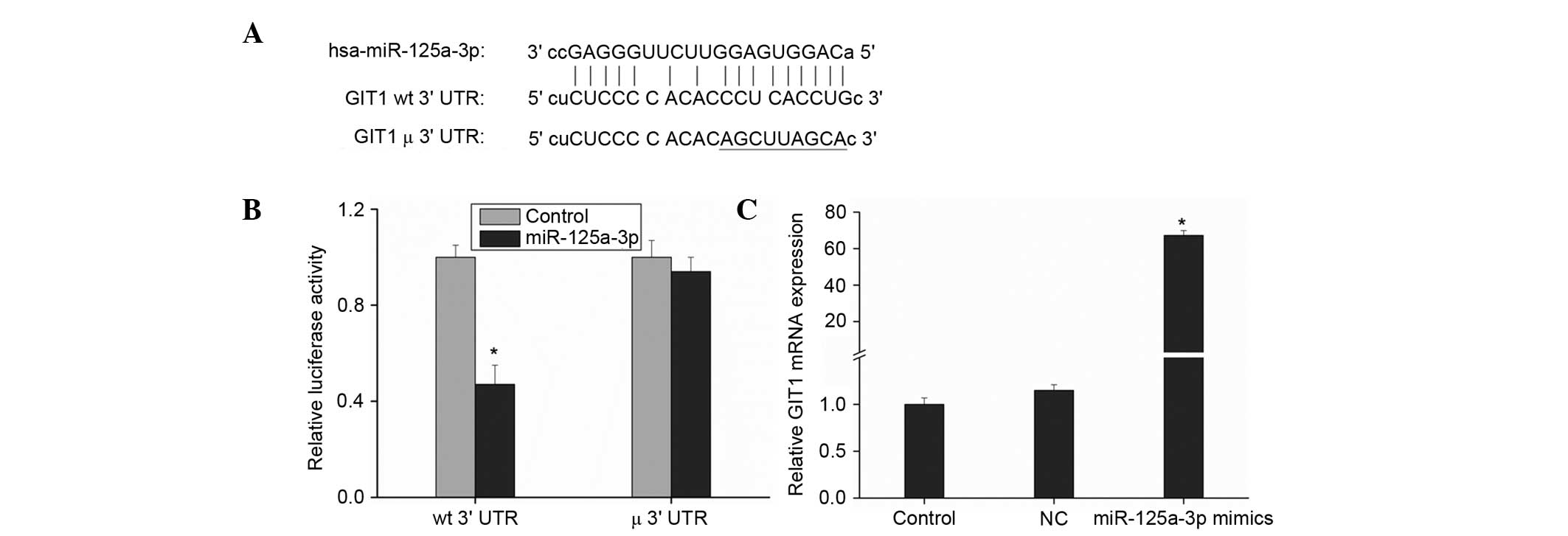
Prediction and verification of
candidate miRNAs of GIT1
The software applications miRanda, TargetScanHuman,
miRBase and miRWalk were used to predict miRNAs targeting GIT1, and
the results revealed that miR-125a-3p had good matching sites with
the GIT1 3′UTR (Fig. 1A). miR-125b,
another member of the miR-125 family, was considered to be
associated with osteoblast differentiation on the basis of a
previous study (15). It has been
indicated to achieve a regulatory effect by inhibiting the
expression of the cell proliferation-associated ErbB2 gene to
suppress stromal cell line ST2 proliferation and then directly
affect osteoblast differentiation (15,16).
Therefore, the regulatory relationship between miR-125a-3p and GIT1
in osteoblasts is worthy of further investigation.
First, a dual-luciferase reporter assay was used to
verify whether there are binding sites of miR-125a-3p in GIT1. The
software applications miRanda (microrna.org/microrna/getGeneForm.do), TargetScanHuman
(targetscan.org/vert_71/), miRBase
(mirbase.org/) and miRWalk (zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/)
predicted that GIT1 has miR-125a-3p binding sites. GIT1 3′UTR was
amplified (see Table I for primers)
and cloned into the dual-luciferase reporter vector psiCHECK-2 and
the potential target sites of miR-125a-3p were mutated (Fig. 1A). These two plasmids were then
transfected into HEK293 cells together with miR-125a-3p mimics,
respectively, in order to analyze the change of luciferase
expression. Cells transfected with blank reporter vector were taken
as the control. The cells transfected with the vector containing
GIT1 3′UTR clones showed a reduction of ~50% in luciferase
expression level, with a statistically significant difference,
(P<0.01 vs. control) while those transfected with the vector
containing mutant GIT1 3′UTR clones had a non-significant change in
the luciferase expression level (Fig.
1B). This illustrates that miR-125a-3p did have significant
binding sites in the GIT1 3′UTR, suggesting that GIT1 is a target
gene of miR-125a-3p.
Ago2 is a core component of the RNA-induced
silencing complex, associating with both miRNAs and their mRNA
targets (17). Thus, immunopurifying
Ago2 under the appropriate conditions might retain associated
miRNAs and mRNAs, allowing miRNA targets to be identified.
Therefore, monoclonal anti-Ago2 antibody was immobilized on
Protein-G-Sepharose beads and incubated with HEK293 cell lysates;
then the co-immunoprecipitated Ago-bound RNAs were extracted and
subjected to RT-qPCR to detect GIT1 mRNA expression. The results
shown in Fig. 1C indicate that GIT1
mRNA expression increased significantly following transfection with
miR-125a-3p mimics. This further illustrates that miR-125a-3p can
targetedly regulate GIT1.
Association between the expression of
miR-125a-3p and GIT1 in osteoblast differentiation
miR-125b is associated with osteoblast
differentiation (15), but it is
unclear whether miR-125a-3p is also associated with osteoblast
differentiation. Thus, further investigation of this is necessary.
After adding BMP-2 to HMSCs for induction of differentiation, the
changes of miR-125a-3p and GIT1 expression were detected. According
to the results, when HMSCs were not induced to differentiate, as
the confluence tended to 100%, the expression of miR-125a-3p
increased gradually (Fig. 2A).
However, while HMSCs were differentiated to osteoblasts, the
expression of miR-125a-3p also increased over time, but the levels
of increase were significantly suppressed (P<0.01; Fig. 2A), which indicated that miR-125a-3p
expression was reduced as osteoblast differentiation proceeded.
Moreover, the expression level of GIT1 was not significantly
changed when HMSCs were in normal culture; however, when BMP-2 was
added to induce osteoblast differentiation, GIT1 expression
gradually increased as the induction of differentiation proceeded
(Fig. 2B). From the aforementioned
results, in the osteoblastic differentiation process, miR-125a-3p
expression was negatively correlated with GIT1 expression, and
upregulation of GIT1 expression may be caused by the inhibition of
miR-125a-3p expression.
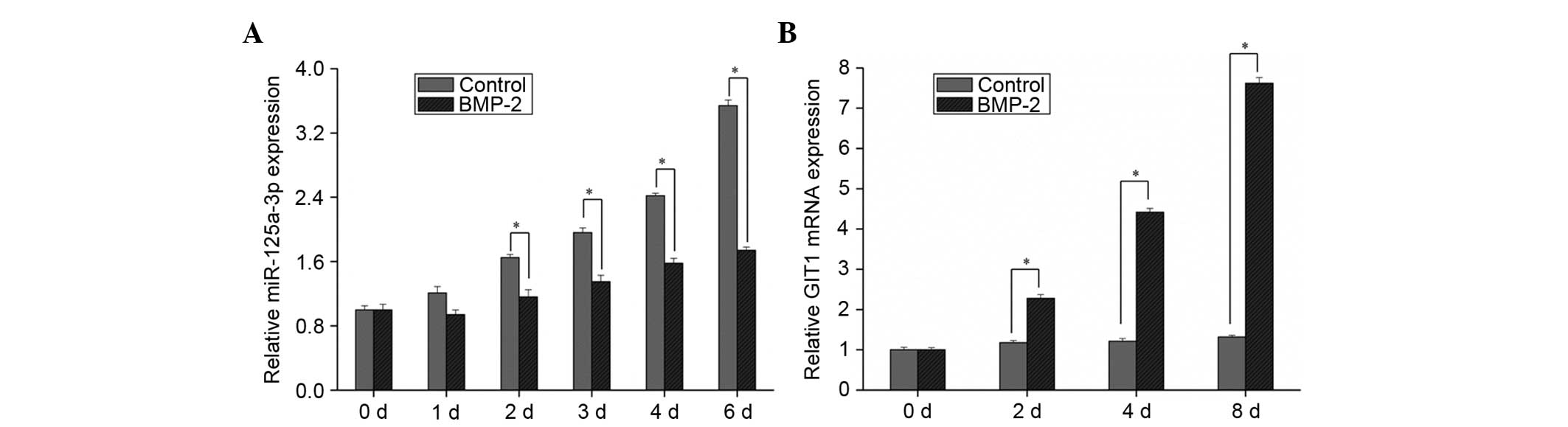 | Figure 2.Relative expression of miR-125a-3p and
GIT1 during osteoblastic differentiation. Human bone-marrow derived
mesenchymal stem cells were harvested after 1, 2, 3, 4, 6 and 8
days after BMP-2 addition, and the expression levels of (A)
miR-125a-3p and (B) GIT1 were measured. Results are presented as
the mean plus standard deviation. *P<0.01 as indicated. Control,
cells without BMP-2; BMP-2, cells treated with BMP-2; miR,
microRNA; GIT1, G protein-coupled receptor kinase interacting
protein 1; BMP, bone morphogenetic protein; d, days. |
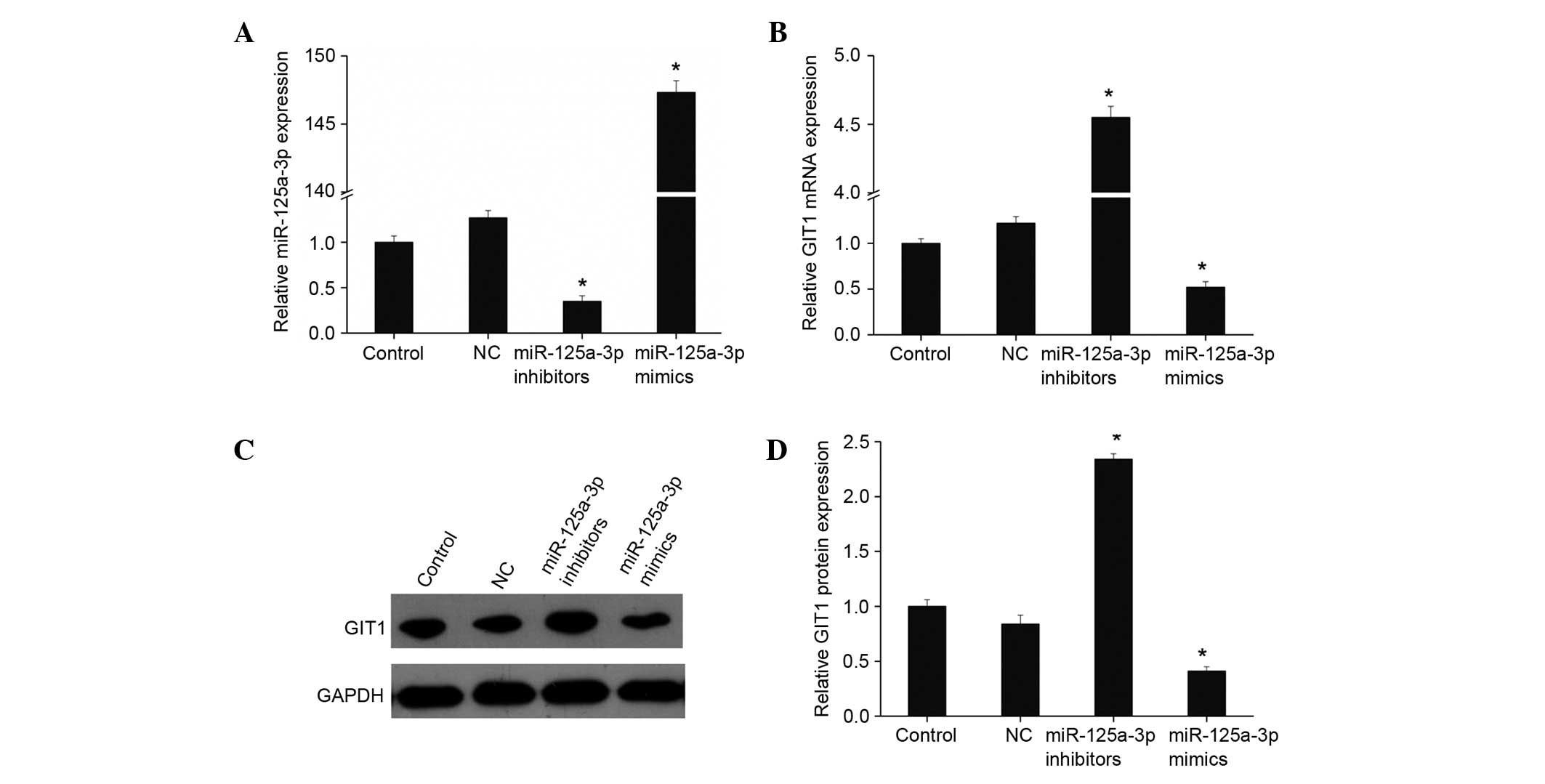
miR-125a-3p downregulates GIT1
expression in osteoblasts
On the basis of the aforementioned results, it may
be speculated that miR-125a-3p regulates GIT1. miR-125a-3p mimics
and inhibitors were transfected into human osteoblasts, and
fluorescence qPCR was used to detect the expression of miR-125a-3p
and GIT1. As shown in Fig. 3A, when
mimics were transfected, the expression level of miR-125a-3p was
significantly upregulated, while the expression of miR-125a-3p was
significantly inhibited following the addition of miR-125a-3p
inhibitors (both P<0.01). The expression of GIT1 was
downregulated with upregulation of miR-miR-125a-3p expression, as
shown by fluorescence qPCR and western blotting (Fig. 3B-D). By contrast, GIT1 expression
increased markedly when miR-125a-3p expression was inhibited. This
suggests that miR-125a-3p targetedly regulates the expression of
GIT1 in osteoblasts.
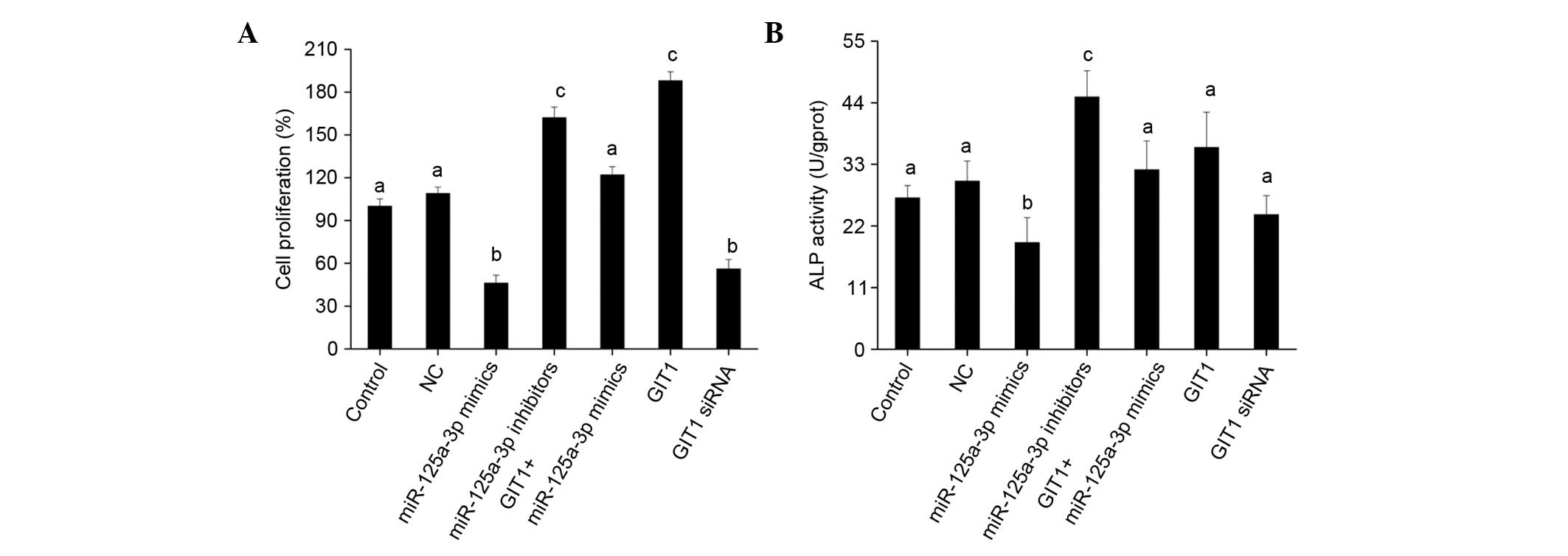
miR-125a-3p affects osteoblast
proliferation and differentiation by regulating GIT1
expression
The impact of miR-125a-3p and GIT1 on osteoblast
proliferation and differentiation was investigated. The
proliferation of osteoblasts transfected with miR-125a-3p mimics
declined significantly compared with the control while the
proliferation of those transfected with miR-125a-3p inhibitors
increased (both P<0.01; Fig. 4A).
This suggests that miR-125a-3p suppresses osteoblast proliferation.
The proliferation of osteoblasts transfected with GIT1
overexpression vector was increased markedly, while their
proliferation was inhibited following transfection with GIT1 siRNA
to reduce GIT1 expression (Fig. 4A),
which indicates that GIT1 promotes osteoblast proliferation. In
addition, following co-transfection with miR-125a-3p mimics and
GIT1 overexpression vector, upregulation of GIT1 expression blocked
the proliferation-suppressing effect of miR-125a-3p, and the
osteoblast proliferation was increased; a significant difference
was found compared with the miR-125a-3p mimics transfection group
(Fig. 4A). This indicates that
miR-125a-3p can targetedly regulate GIT1 expression to inhibit
osteoblast proliferation.
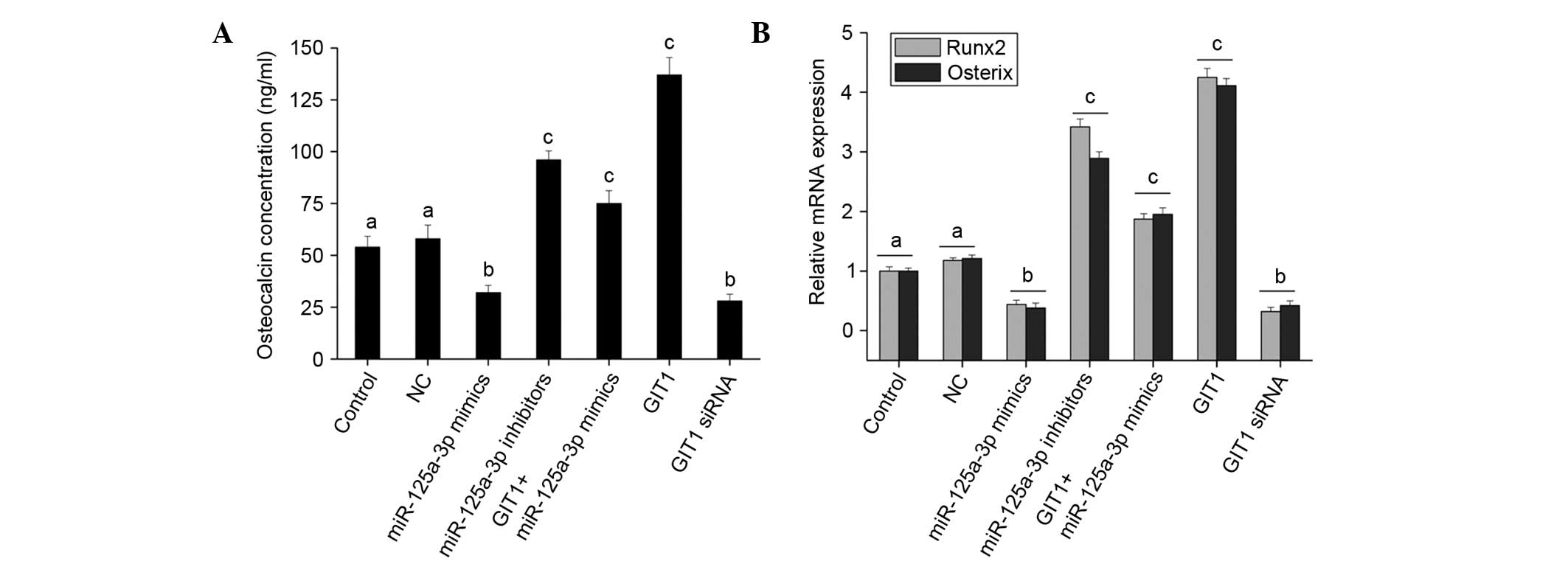
Further analysis of osteoblast differentiation
indicates that the upregulation of miR-125a-3p expression reduced
ALP activity (Fig. 4B), OC secretion
(Fig. 5A) and the expression of
Runx2 and osterix (Fig. 5B). That
is, miR-125a-3p had an inhibitory effect on osteoblast
differentiation, and downregulation of its expression promoted
osteoblast differentiation. GIT1 promoted osteoblast
differentiation, although not very strongly (Figs. 4B and 5). Following co-transfection with
miR-125a-3p mimics and GIT1 overexpression vector, the inhibitory
effect of miR-125a-3p on osteoblast differentiation was weakened
(Figs. 4B and 5). Hence, in osteoblasts, it may be
concluded that miR-125a-3p affects the proliferation and
differentiation of osteoblasts by regulating the expression of
GIT1.
Discussion
Osteoblasts play an important role in bone
formation, development, remodeling and repair processes, and
various growth factors may affect osteoblast proliferation,
differentiation and extracellular matrix synthesis (18).
In recent years, the relationship between miRNAs and
bone formation has attracted increasing attention. Notably, the
knockout of Dicer and Drosha leads to generation failure of all
miRNAs in cells, thus causing a loss of differentiation capacity of
bone marrow stroma stem cells into osteoblasts and adipocytes
(19). This suggests that an
appropriate expression level of miRNAs is essential for the
osteoblastic differentiation of stem cells. In addition, numerous
studies have shown that GIT1 plays a significant role in the growth
and migration of osteoblasts (6–7,20). Consequently, investigation of the
effect of GIT1 and relevant miRNAs in osteoblasts is worthwhile.
However, studies of miRNAs targeting GIT1 are rare at present, with
only a few relevant reports in tumors (21,22).
In the present study, prediction of miRNAs targeting
GIT1 indicated that there is good matching site for miR-125a-3p in
the GIT1 3′UTR. A dual-luciferase reporter assay indicated that
GIT1 is one of the target genes of miR-125a-3p. Therefore, in
osteoblasts, miR-125a-3p may targetedly regulate GIT1 to play its
role. Investigation of the BMP-2-induced differentiation process
from HMSCs to osteoblasts indicates that miR-125a-3p expression
increases gradually, but the level of increase is significantly
suppressed compared with that of non-differentiating HMSCs,
indicating that the inhibitory effect of miR-125a-3p expression is
increasingly evident osteoblast differentiation proceeds. These
results are similar to those of miR-125b reported by Mizuno et
al (14), which indicates that
miR-125a-3p may have a similar effect to miR-125b. miR-125b is the
first reported miRNA to be associated with osteoblast
differentiation (15). It was also
found that the expression level of miR-125b decreased in the
induction process of ST2 cells to osteoblasts, and after the
introduction of miR-125b the expression level of ALP was lowered in
the osteoblastic induction process of ST2 cells; by contrast, after
adding miR-125b inhibitors the expression of ALP was enhanced
(15). This indicates that miR-125b
negatively regulates the osteoblastic differentiation process.
In the present study, as induced differentiation
proceeded, the expression level of GIT1 increased significantly.
Therefore, in the osteoblastic differentiation process, miR-125a-3p
expression was negatively correlated with GIT1 expression; thus, an
increase of GIT1 expression may be caused by suppressed miR-125a-3p
expression. When miR-125a-3p mimics and inhibitors were each
transfected into human osteoblasts, the findings were consistent
with the speculation above. Specifically, when the miR-125a-3p
expression level was changed, the expression level of GIT1 showed
the opposite effect. This illustrates that miR-125a-3p targetedly
regulates the expression of GIT1 in osteoblasts, and one of the
pathways by which miR-125a-3p exerts its effects may involve the
regulation of GIT1.
The impact of miR-125a-3p and GIT1 on osteoblastic
proliferation and differentiation was determined in the present
study, and the results indicated that miR-125a-3p targetedly
regulates GIT1 expression to inhibit osteoblastic proliferation and
differentiation. Feng et al (23) showed that miRNA-125a inhibited cell
growth by targeting glypican-4 in the human embryonic kidney cell
line 293T (HEK293T). Ninio-Many et al (24) found that miRNA-125a-3p reduced cell
proliferation and migration by targeting Fyn in HEK293T cells. The
study of Svensson et al (25)
showed that inhibition of miRNA-125a promoted human endothelial
cell proliferation and viability through an antiapoptotic
mechanism. In addition, Guo et al (26) suggested that miRNA-125a repressed
cell growth by targeting HuR in breast cancer. All the
aforementioned findings are consistent with the conclusions of the
present study, which both indicate a proliferation inhibiting
effect of miR-125a-3p. In the study of Huang et al (27), the authors concluded that miR-125b
was a key regulatory factor of osteoblastic differentiation that
acted by directly targeting core binding factor β and indirectly
acting on Runx2 at an early stage osteoblastic differentiation. Guo
et al (28) reported that
miR-125a plays a biological function in osteoclastogenesis through
a novel TRAF6/NFATc1/miR-125a regulatory feedback loop, and that
miR-125a was markedly downregulated during the macrophage colony
stimulating factor- and receptor activator of nuclear factor-κB
ligand-induced osteoclastogenesis of circulating CD14+
peripheral blood mononuclear cells (PBMCs). Overexpression of
miR-125a in CD14+ PBMCs inhibited osteoclastogenesis,
whereas the inhibition of miR-125a promoted osteoclastogenesis.
Therefore, miR-125a-3p inhibits osteoblast proliferation and
differentiation, and one of the pathways involves the regulation of
GIT1.
In summary, miR-125a-3p affects osteoblast
proliferation and differentiation through the regulation of GIT1.
This finding increases our understanding of the physiological and
pathological roles of miRNAs in osteoblastic differentiation and
maturation processes, and should provide a theoretical basis for
research into bone physiology and pathology. This may assist in the
diagnosis and targeted treatment of metabolic bone disease and
associated drug development.
Acknowledgements
This study was supported by Health Bureau of Wuxi
City Foundation for Youths (grant no. Q201407).
References
|
1
|
Lemaire V, Tobin FL, Greller LD, Cho CR
and Suva LJ: Modeling the interactions between osteoblast and
osteoclast activities in bone remodeling. J Theor Biol.
229:293–309. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Alford AI, Kozloff KM and Hankenson KD:
Extracellular matrix networks in bone remodeling. Int J Biochem
Cell Biol. 65:20–31. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Arpornmaeklong P, Brown SE, Wang Z and
Krebsbach PH: Phenotypic characterization, osteoblastic
differentiation and bone regeneration capacity of human embryonic
stem cell-derived mesenchymal stem cells. Stem Cells Dev.
18:955–968. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schmalzigaug R, Phee H, Davidson CE, Weiss
A and Premont RT: Differential expression of the ARF GAP genes GIT1
and GIT2 in mouse tissues. J Histochem Cytochem. 55:1039–1048.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Manabe R, Kovalenko M, Webb DJ and Horwitz
AR: GIT1 functions in a motile, multi-molecular signaling complex
that regulates protrusive activity and cell migration. J Cell Sci.
115:1497–1510. 2002.PubMed/NCBI
|
|
6
|
Menon P, Yin G, Smolock EM, Zuscik MJ, Yan
C and Berk BC: GPCR kinase 2 interacting protein 1 (GIT1) regulates
osteoclast function and bone mass. J Cell Physiol. 225:777–785.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ren Y, Yu L, Fan J, Rui Z, Hua Z, Zhang Z,
Zhang N and Yin G: Phosphorylation of GIT1 tyrosine 321 is required
for association with FAK at focal adhesions and for PDGF-activated
migration of osteoblasts. Mol Cell Biochem. 365:109–118. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hassan MQ, Gordon JA, Beloti MM, Croce CM,
van Wijnen AJ, Stein JL, Stein GS and Lian JB: A network connecting
Runx2, SATB2 and the miR-23a~27a~24-2 cluster regulates the
osteoblast differentiation program. Proc Natl Acad Sci USA.
107:19879–8435. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hu R, Li H, Liu W, Yang L, Tan YF and Luo
XH: Targeting miRNAs in osteoblast differentiation and bone
formation. Expert Opin Ther Targets. 14:1109–1120. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kapinas K and Delany AM: MicroRNA
biogenesis and regulation of bone remodeling. Arthritis Res Ther.
13:2202011. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kapinas K, Kessler CB and Delany AM:
miR-29 suppression of osteonectin in osteoblasts: Regulation during
differentiation and by canonical Wnt signaling. J Cell Biochem.
108:216–224. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mizuno Y, Yagi K, Tokuzawa Y,
Kanesaki-Yatsuka Y, Suda T, Katagiri T, Fukuda T, Maruyama M, Okuda
A, Amemiya T, et al: miR-125b inhibits osteoblastic differentiation
by down-regulation of cell proliferation. Biochem Biophys Res
Commun. 368:267–272. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Scott GK, Goga A, Bhaumik D, Berger CE,
Sullivan CS and Benz CC: Coordinate suppression of ERBB2 and ERBB3
by enforced expression of micro-RNA miR-125a or miR-125b. J Biol
Chem. 282:1479–1486. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Meister G, Landthaler M, Patkaniowska A,
Dorsett Y, Teng G and Tuschl T: Human Argonaute2 mediates RNA
cleavage targeted by miRNAs and siRNAs. Mol Cell. 15:185–197. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chau JF, Leong WF and Li B: Signaling
pathways governing osteoblast proliferation, differentiation and
function. Histol Histopathol. 24:1593–1606. 2009.PubMed/NCBI
|
|
19
|
Oskowitz AZ, Lu J, Penfornis P, Ylostalo
J, McBride J, Flemington EK, Prockop DJ and Pochampally R: Human
multipotent stromal cells from bone marrow and microRNA: Regulation
of differentiation and leukemia inhibitory factor expression. Proc
Natl Acad Sci USA. 105:18372–7. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu Y, Zhang Y, Yin Q, Xia H and Wang J:
Platelet-derived growth factor promotes osteoblast proliferation by
activating G-protein-coupled receptor kinase interactor-1. Mol Med
Rep. 10:1349–54. 2014.PubMed/NCBI
|
|
21
|
Huang WC, Chan SH, Jang TH, Chang JW, Ko
YC, Yen TC, Chiang SL, Chiang WF, Shieh TY, Liao CT, et al:
miRNA-491-5p and GIT1 serve as modulators and biomarkers for oral
squamous cell carcinoma invasion and metastasis. Cancer Res.
74:751–64. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chan SH, Huang WC, Chang JW, Chang KJ, Kuo
WH, Wang MY, Lin KY, Uen YH, Hou MF, Lin CM, et al: MicroRNA-149
targets GIT1 to suppress integrin signaling and breast cancer
metastasis. Oncogene. 33:4496–507. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Feng C, Li J, Ruan J and Ding K:
MicroRNA-125a inhibits cell growth by targeting glypican-4.
Glycoconj J. 29:503–11. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ninio-Many L, Grossman H, Shomron N,
Chuderland D and Shalgi R: microRNA-125a-3p reduces cell
proliferation and migration by targeting Fyn. J Cell Sci.
126:2867–176. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Svensson D, Gidlöf O, Turczyńska KM,
Erlinge D, Albinsson S and Nilsson BO: Inhibition of microRNA-125a
promotes human endothelial cell proliferation and viability through
an antiapoptotic mechanism. J Vasc Res. 51:239–245. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guo X, Wu Y and Hartley RS: MicroRNA-125a
represses cell growth by targeting HuR in breast cancer. RNA Biol.
6:575–583. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang K, Fu J, Zhou W, Li W, Dong S, Yu S,
Hu Z, Wang H and Xie Z: MicroRNA-125b regulates osteogenic
differentiation of mesenchymal stem cells by targeting Cbfβ in
vitro. Biochimie. 102:47–55. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Guo LJ, Liao L, Yang L, Li Y and Jiang TJ:
MiR-125a TNF receptor-associated factor 6 to inhibit
osteoclastogenesis. Exp Cell Res. 321:142–152. 2014. View Article : Google Scholar : PubMed/NCBI
|