Introduction
Heart disease-related deaths are the highest in most
societies and congenital heart diseases have childhood origins,
necessitating their early diagnosis and treatment. In fact,
congenital heart diseases account for approximately 40% of prenatal
deaths and over 20% of mortality in the first few months after
birth (1). Congenital heart disease
affects approximately 1% of all newborns and is the causative
factor for more deaths within the first year of life as compared to
all other genetic defects (2). A
complete cure of congenital heart defect during early childhood is
rare, but advances in treatment approaches have increased life
expectancy and led to an expansion of adult population with
clinical manifestation of congenital heart defects in up to 90% of
the children born with congenital heart diseases (3). During development, at the embryonic
stage itself, formation of the functional heart is of utmost
importance and the process of cardiac tissue formation is a highly
complex multi-cell lineage differentiation process. The
uninterrupted contractile function of heart from embryonic stage
till the end of life is critical for survival of the organism. The
most common congenital defects are seen in heart development that
lead to a wide variety of abnormalities such as, arrhythmias,
cardiomyopathies, heart failure, and malformed valves all leading
to sudden death (4). Development of
heart and cardiac function is regulated by multiple transcription
factors, signaling proteins and complexes that orchestrate cardiac
morphogenesis and myogenesis and control cardiac contractility
(5). Several complex genetic
pathways are linked to heart development and pathology and a clear
understanding of these players at molecular level is essential in
improving the prognosis of patients with cardiac abnormalities
(6). Regulation of cardiac gene
expression involves multiple independent enhancers that play a
critical role in maintaining a restricted and specific pattern of
gene expression in the heart. It is now recognized that cardiac
transcriptional pathways are intimately regulated by microRNAs
(miRNAs) (7). miRNAs are a class of
small, regulatory RNAs, approximately 22 nucleotides in length at
mature stage and are evolutionarily conserved and also coded by
specific genes. These miRNAs normally function to suppress
expression of genes they target by inhibiting translation and/or
promoting degradation of target protein-coding mRNA by base pairing
(8,9). Considering that the miRNAs play their
role post-transcriptionally by adding another layer of regulation
of cardiac gene expression, they are proposed to act as ‘rheostats’
and ‘switches’ for controlling various aspects of cardiac
development, function, and their dysregulation can lead to cardiac
abnormalities including hypertrophy, arrhythmia and ischemia
(10,11). Since their discovery 25 years ago,
>1,400 miRNAs have been identified in mammals (12) and are proposed to regulate the
expression of >50% of all protein coding genes (13). It has been reported that the
expression of several miRNAs change in diseased hearts, emphasizing
their role in cardiac disease (14).
These findings are corroborated by experimental studies on animals
addressing specific roles of different miRNAs (15–17).
Role of miRNAs in normal heart development
and function
Genomic sequences coding for miRNAs are located in
the intergenic, intronic, and exonic regions in chromosomes.
Intergenic miRNAs are produced from their own transcriptional units
whereas intronic and exonic miRNAs are transcribed and expressed
along their host genes. A subset of intronic miRNAs are transcribed
in the opposite orientation of the host genes and these miRNAs have
their own cis-regulatory elements for expression control
(18). Biogenesis of miRNAs starts
with transcription of the corresponding gene in the nucleus to
generate primary miRNA, which is further processed to produce ~70
nucleotide long precursor miRNA, by a complex of Drosha and
DiGeorge syndrome critical region 8 (DGCR8). Precursor miRNA exits
the nucleus facilitated by exportin 5 in the nuclear membrane
(19), and is cut by Dicer, a type
of RNAse III endonuclease, to give rise to miRNA duplex, in the
cytosol. A single arm of the resulting ~22-nucleotide duplex is
taken selectively into the RNA-induced silencing complex (RISC),
whereas the other stem arm is presumably degraded. The miRNA-loaded
RISC targets the mRNAs, by complementary base pairing the target
sequence(s) within the 3′ untranslated region. A perfect or
near-perfect complementary base pairing between RISC-bound miRNA
and targeted mRNA leads to rapid degradation of the targeted mRNA.
However, in most cases, animal miRNAs are only imperfectly
complementary to their targeted mRNAs, resulting in suppressed
translation from that mRNA or sequestration of the targeted mRNA to
cytoplasmic P-bodies (20). The
significance of miRNAs in development in general and specifically
for cardiac development was realized in gene deletion experiments
in mice and zebrafish, where Dicer gene is deleted. These animals
suffered arrested development from gastrulation stage in
association with almost total lack of miRNAs (21). Similarly animals with mutated Dicer
showed abnormal somitogenesis and heart development (22). Tissue specific deletion of Dicer in
mouse heart, employing Cre-Lox system, under the control of the
postnatally expressed α-myosin heavy chain promoter, led to
deranged expression of cardiac contractile proteins and significant
sarcomere disarray, in association with greatly decreased cardiac
function. These hearts rapidly developed dilated cardiomyopathy and
heart failure after birth (23). The
abnormal heart function seen in Dicer mutant mice closely resembles
the human dilated cardiomyopathy and heart failure and in fact,
failing human hearts are shown to have low levels of Dicer protein,
suggesting an important role for miRNAs in dilated cardiomyopathies
and heart failure in patients (23).
On the other hand, deletion of Dicer during mouse heart
development, using Cre-recombinase under the control of Nkx2.5
promoter, led to embryonic lethality with defective heart
morphogenesis (15) and this
indicated the differential role of miRNAs during and after the
development of the heart.
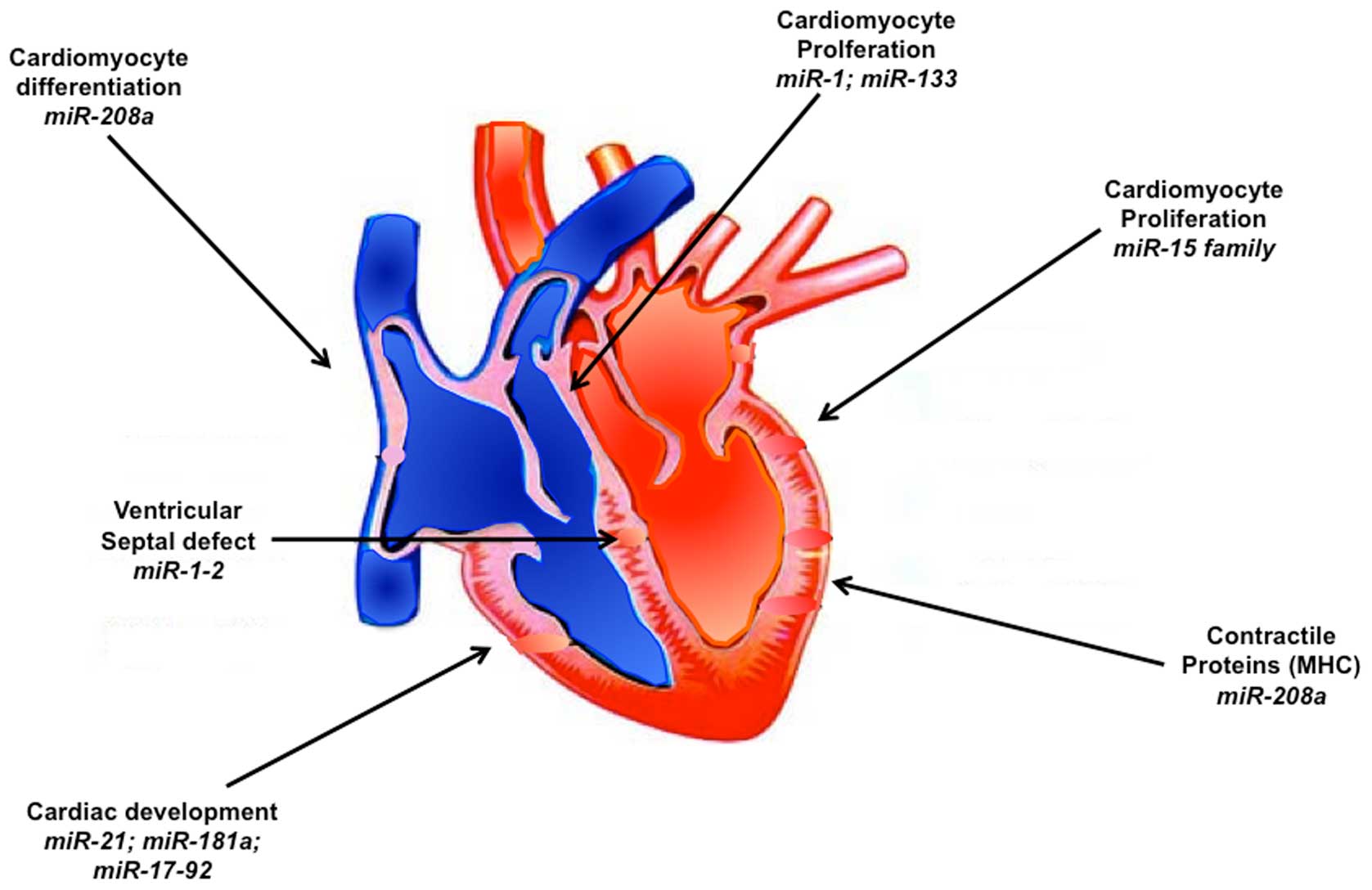
Studies on individual miRNAs expressed in heart
revealed important and critical contribution of many of these
miRNAs during heart development (Fig.
1). Thus miR-1 and miR-133 are experimentally verified to play
a role in the cardiac development. These miRNAs are produced from
the same polycistronic transcripts, encoded by two separate genes.
In mice miR-1-1 and miR-133a-2 are clustered on chromosome 2,
whereas miR-1-2 and miR-133a-1 are clustered on chromosome 18
(24). Expression of these miRNAs is
regulated by muscle transcriptional networks, consisting of serum
response factor and myocardin for cardiac muscle expression and
MyoD and myocyte enhancer factor 2 for skeletal muscle expression
(25). Even though miR-1-1 and
miR-1-2 are encoded by separate genes, they have identical
nucleotide sequence and thus appear to target the same mRNAs.
However, miR-1-2 null mice in utero display pericardial
edema, which is consistent with embryonic myocardial dysfunction
and it appears that miR-1-2 has non-redundant roles with miR-1-1 in
the heart, even though these two miRNAs have overlapping expression
patterns and sequences (24). There
appears to be a fine balance of the effects of these miRNAs as
their excess activity or loss of function can be detrimental to the
development and function of heart. Thus, miR-1 overexpression in
the embryonic heart blocks expansion of ventricular myocardium by
inhibiting cardiomyocyte proliferation (25) and also injection of Xenopus embryos
with miR-1 arrests the cardiac development (24). On the other hand, targeted deletion
of miR-1-2 was found to cause ~50% embryonic lethality in mice
because of ventricular septal defects, with the remaining surviving
mice with the deletion facing mortality at later stage due to
conduction system defects (15).
Understanding how exactly the miRNAs influence heart
development is critical. Although miR-1 can potentially target
several genes in the heart, one important validated target is Hand2
cardiac transcription factor. Thus, deletion of Hand2 leads to
similar ventricular myocyte developmental problems (26) as miR-1 over-production, which also
reduces expression of Hand2 (25).
Expression of miR-133a-1/miR-1-2 and miR-133a-2/miR-1-1 is found
throughout the ventricular myocardium and also in interventricular
septum from embryonic stage E8.5 until adulthood (27). Even though deletion of either
miR-133a-1 or miR-133a-2 has no obvious deleterious effects on the
heart, loss of both the miRs leads to ventricular septal defects
and chamber dilatation resulting in late embryonic and neonatal
lethality (28). Among the other
miRNAs, miR-196a, which is found in fetal human heart is known to
regulate HOXB8-Shh signaling, that is essential for cardiac
septation, outflow tract morphogenesis as well as valve formation
(29).
Congenital cardiac defects and altered
miRNAs
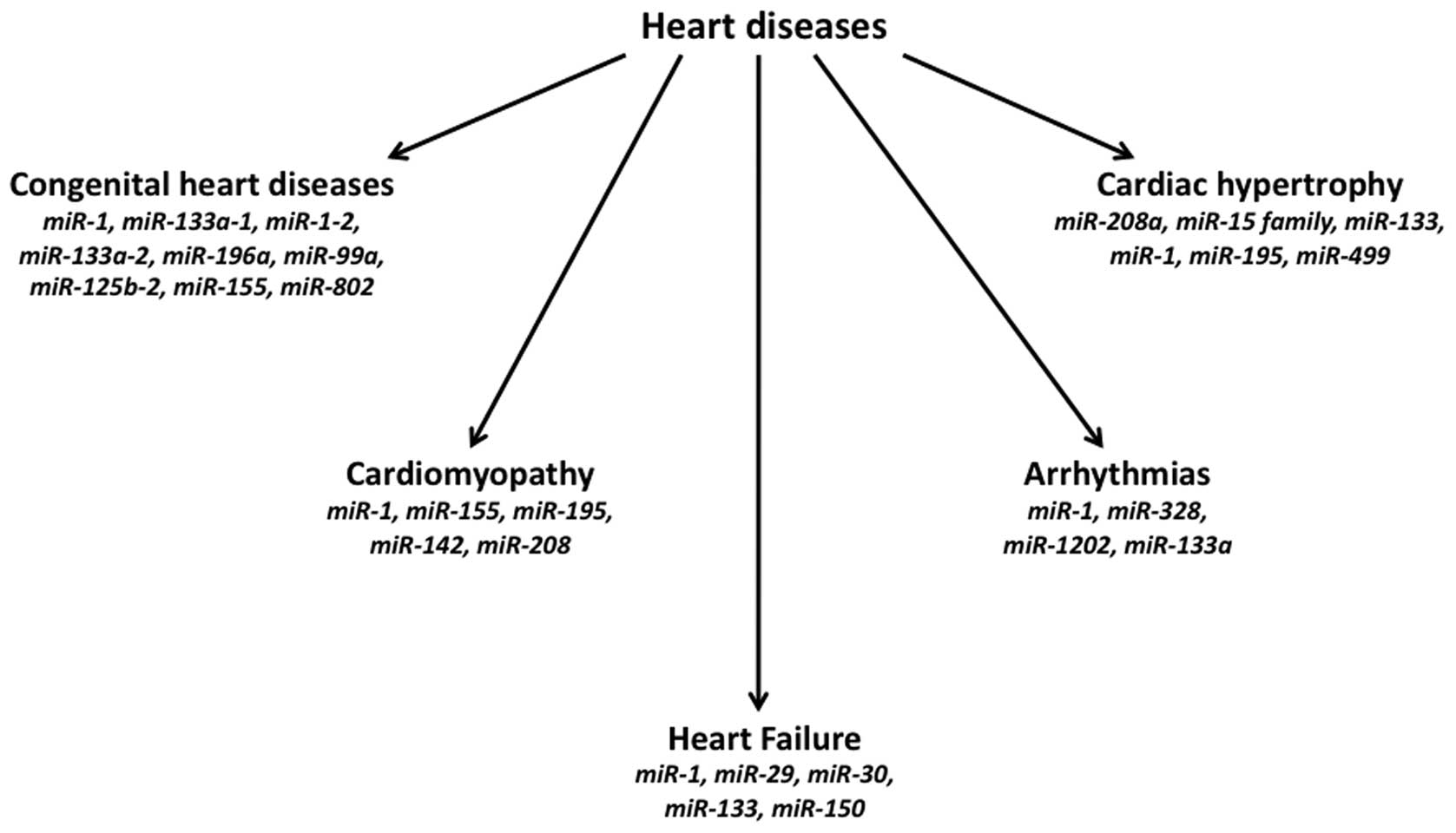
Several congenital cardiac defects have been found
to be associated with altered expression of different miRNAs
(Fig. 2). The hearts of patients
with the most common genetic defect leading to cardiac
abnormalities, the trisomy 21 (Down syndrome), contain an extra
Hsa21 chromosome, and were found to overexpress 5 miRs: miR-99a,
let-7c, miR-125b-2, miR-155, and miR-802, present on human
chromosome 21 (30). It has been
observed that ~61 miRNAs, which target the networks of cardiac
development, show altered expression in the right ventricular
myocardium of children with non-syndromic tetralogy of Fallot
(31). In another condition of human
fetal single ventricle malformation, 38 miRNAs were found to be
downregulated and 10 miRNAs showed upregulation in cardiac tissues,
as compared to normal control cardiac tissue (32). Patients suffering from DG syndrome,
which is caused by a deletion of the DGCR8 on chromosome 22
(22q11.2), develop congenital heart disease. This deletion causes
loss of a component of the RISC, thereby leading to impaired miRNA
expression, which can potentially contribute to congenital heart
defects (33). As this syndrome
results from haploinsufficiency of the 22q11.2 locus, the
possibility that disturbances in miRNA expression potentially
contribute to the gene dosage sensitivity of this disease. It is
interesting to note that while many miRNAs can be perturbed with
minimal effects on phenotype under normal conditions, the same
miRNA disturbances can have profound impact on phenotype under
stress conditions (10). The
apparent minimal effects of miRNAs under non-stress conditions as
compared to their specific involvement during remodeling responses
of diseased tissues make miRNAs attractive therapeutic targets for
inactivating disease-inducing miRNAs with little or no off-target
effects on normal non-stress tissues. Specific expression patterns
of miRNAs have been observed in several cardiac disorders,
including hypertrophy, heart failure, ischemic cardiomyopathy,
post-myocardial infarction (MI) remodelling (14,34–36).
Cardiac injury following acute MI is known to increase the
circulating levels of certain myocardial-derived miRNAs, such as
miR-1, miR-133, miR-499 and miR-208, and it has been proposed that
the specifically increased miRNAs can be useful as both diagnostic
as well as prognostic biomarkers (37,38).
Importance of miRNAs in cardiac remodeling
and cardiac hypertrophy
Heart being a highly sensitive tissue to various
stresses and external pathological stimuli, responds by undergoing
extensive cardiac remodeling, which is essentially cardiac
hypertrophy (39). In cardiac
hypertrophy there is an increase in the size of cardiomyocyte
and/or myofibrillar volume without any change in cardiomyocyte
number. This elevated cell size and volume is needed to maintain
normal cardiac output under stress conditions. There is a
reactivation of fetal cardiac genes during cardiac hypertrophy,
indicating that the cardiac genes involved in heart development
before birth are redeployed during cardiac hypertrophic growth. It
has been recognized that expression of several miRNAs is both up-
and down-regulated (Fig. 2) in
experimental models of cardiac hypertrophy and also in samples from
failing human hearts (35,40). It has been demonstrated that miR-195
expression is elevated during cardiac hypertrophy in human hearts
and in mouse hypertrophic hearts and that miR-195 alone is
sufficient to induce hypertrophic growth in cultured rat
cardiomyocytes (40).
During heart development and in diseased hearts,
miR-208a and miR-208b are differentially expressed coinciding with
the expression of their host genes, Myh6 and Myh7, respectively.
These miRNAs regulate the host gene switch during development and
under stress conditions in a feedback manner (16,41).
Gain-of-function studies showed that cardiac specific
overexpression of miR-208a alone is able to induce cardiac
hypertrophy and cardiac conduction defects (41). Expression of the Myh7/miR-208b is
regulated by miR-208a, which also acts upstream of Myh7b/miR-499 in
adult hearts. Another important target of miR-208a is thyroid
hormone receptor-associated protein 1, a key component of the
thyroid hormone signal pathway and an important player in cardiac
hypertrophy (16,41). While miR-195, miR-214 or miR-21 are
upregulated during cardiac hypertrophy, the expression of miR-1,
miR-133a, miR-93 and miR-181 is downregulated (17,42).
Repression of miR-133a is found to be sufficient to lead to cardiac
hypertrophy both in vivo and in vitro, probably
through the upregulation of RhoA, Cdc42, and NELFA/Whsc2 (17). On the other hand, knockout of both
miR-133a-1 and miR-133a-2, leads to partial embryonic lethality
with the remaining living mutant mice showing cardiomyopathy but
not cardiac hypertrophy (28),
indicating different experimental approaches may lead to different
and some times discrepant results. It has been shown that
deregulation of miR-499, which controls the expression of
sarcomeric genes, contributes to the pathogenesis of cardiac
hypertrophy (43).
miRNAs as therapeutic targets and
biomarkers
Many of the properties of miRNAs make them
clinically relevant because miRNA expression is altered in diseased
hearts, making miRNAs as potential biomarkers for the diagnosis of
cardiovascular disease. Another important feature of miRNAs is
their smaller size, which makes their in vivo delivery
feasible. Besides, considering that single miRNAs target multiple
mRNAs coding for different proteins of a common pathway, it becomes
easier to efficiently control a given pathway or biological
function with a single miRNA. However, exploitation of this
property also calls for caution to avoid potential off-target
activities of the chosen miRNAs. It has been proposed that miR-1 is
a potential biomarker for early diagnosis of acute MI, as
circulating miR-1 levels are markedly increased in these patients
and the circulating miR-1 levels can also be used to differentiate
between acute MI and other cardiac events such as angina pectoris
(44,45), non-acute MI (46), and other cardiovascular diseases
(47). Appearance of miR-499 in the
blood is considered to be an indicator of heart damage following
acute MI. Plasma miR-499 levels are increased several-fold in acute
MI patients but are below detection in other conditions such as
congestive heart failure and acute coronary syndrome and also in
normal controls (48). It is
interesting to note that cardiospecific regenerative capacity of
miR-499 has the ability to promote preferential differentiation of
cardiac stem cells (CSCs) to mature functional cardiomyocytes. In
human CSCs, miR-499 overexpression led to improved regenerative
potential of miR-499-overexpressing cell grafts, increased
myocardial tissue repair, and better restoration of heart mass
(49). Without such miR-499
overexpression, transplants of normal CSC to post-MI myocardium
lead to the generation of cardiomyocyte-like immature cells that
failed to differentiate to mature cardiomyocytes.
Conclusions
Heart disease related deaths are the highest in most
societies and congenital heart diseases account for a significant
part, particularly in infants and children. Increased survival of
the children with congenital heart defects with advances in medical
technologies, led to elevated number of adults with heart diseases.
Various types of miRNAs are implicated in the development of heart
and dysregulation of specific miRNAs is associated with congenital
and other cardiac defects. Normal cardiac function as well as
stress responsive cardiac hypertrophy is controlled among other
factors, by specific miRNAs. Recent studies strongly implicate
certain miRNAs such as miR-499 as useful biomarkers of a given
heart disease. Therapeutic application of miRNAs is also being
envisaged. More work is needed for the effective use of miRNAs
either as diagnostic and prognostic markers and as therapeutic
agents.
References
|
1
|
Trojnarska O, Grajek S, Katarzyński S and
Kramer L: Predictors of mortality in adult patients with congenital
heart disease. Cardiol J. 16:341–347. 2009.PubMed/NCBI
|
|
2
|
Rosamond W, Flegal K, Furie K, Go A,
Greenlund K, Haase N, Hailpern SM, Ho M, Howard V, Kissela B, et
al: American Heart Association Statistics Committee and Stroke
Statistics Subcommittee: Heart disease and stroke statistics - 2008
update: A report from the American Heart Association Statistics
Committee and Stroke Statistics Subcommittee. Circulation.
117:e25–e146. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Khairy P, Ionescu-Ittu R, Mackie AS,
Abrahamowicz M, Pilote L and Marelli AJ: Changing mortality in
congenital heart disease. J Am Coll Cardiol. 56:1149–1157. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bruneau BG: The developmental genetics of
congenital heart disease. Nature. 451:943–948. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Olson EN: Gene regulatory networks in the
evolution and development of the heart. Science. 313:1922–1927.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Olson EN: A decade of discoveries in
cardiac biology. Nat Med. 10:467–474. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang DZ: MicroRNAs in cardiac development
and remodeling. Pediatr Cardiol. 31:357–362. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kiriakidou M, Tan GS, Lamprinaki S, De
Planell-Saguer M, Nelson PT and Mourelatos Z: An mRNA m7G cap
binding-like motif within human Ago2 represses translation. Cell.
129:1141–1151. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Humphreys DT, Westman BJ, Martin DI and
Preiss T: MicroRNAs control translation initiation by inhibiting
eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc
Natl Acad Sci USA. 102:16961–16966. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu N and Olson EN: MicroRNA regulatory
networks in cardiovascular development. Dev Cell. 18:510–525. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cai B, Pan Z and Lu Y: The roles of
microRNAs in heart diseases: A novel important regulator. Curr Med
Chem. 17:407–411. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rota R, Ciarapica R, Giordano A, Miele L
and Locatelli F: MicroRNAs in rhabdomyosarcoma: Pathogenetic
implications and translational potentiality. Mol Cancer.
10:1202011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Thum T, Galuppo P, Wolf C, Fiedler J,
Kneitz S, van Laake LW, Doevendans PA, Mummery CL, Borlak J,
Haverich A, et al: MicroRNAs in the human heart: A clue to fetal
gene reprogramming in heart failure. Circulation. 116:258–267.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao Y, Ransom JF, Li A, Vedantham V, von
Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ and
Srivastava D: Dysregulation of cardiogenesis, cardiac conduction,
and cell cycle in mice lacking miRNA-1-2. Cell. 129:303–317. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
van Rooij E, Sutherland LB, Qi X,
Richardson JA, Hill J and Olson EN: Control of stress-dependent
cardiac growth and gene expression by a microRNA. Science.
316:575–579. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Carè A, Catalucci D, Felicetti F, Bonci D,
Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, et al:
MicroRNA-133 controls cardiac hypertrophy. Nat Med. 13:613–618.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yi R, Qin Y, Macara IG and Cullen BR:
Exportin-5 mediates the nuclear export of pre-microRNAs and short
hairpin RNAs. Genes Dev. 17:3011–3016. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bernstein E, Kim SY, Carmell MA, Murchison
EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV and Hannon
GJ: Dicer is essential for mouse development. Nat Genet.
35:215–217. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wienholds E, Koudijs MJ, van Eeden FJ,
Cuppen E and Plasterk RH: The microRNA-producing enzyme Dicer1 is
essential for zebrafish development. Nat Genet. 35:217–218. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen JF, Murchison EP, Tang R, Callis TE,
Tatsuguchi M, Deng Z, Rojas M, Hammond SM, Schneider MD, Selzman
CH, et al: Targeted deletion of Dicer in the heart leads to dilated
cardiomyopathy and heart failure. Proc Natl Acad Sci USA.
105:2111–2116. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen JF, Mandel EM, Thomson JM, Wu Q,
Callis TE, Hammond SM, Conlon FL and Wang DZ: The role of
microRNA-1 and microRNA-133 in skeletal muscle proliferation and
differentiation. Nat Genet. 38:228–233. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao Y, Samal E and Srivastava D: Serum
response factor regulates a muscle-specific microRNA that targets
Hand2 during cardiogenesis. Nature. 436:214–220. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Srivastava D, Thomas T, Lin Q, Kirby ML,
Brown D and Olson EN: Regulation of cardiac mesodermal and neural
crest development by the bHLH transcription factor, dHAND. Nat
Genet. 16:154–160. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu N, Williams AH, Kim Y, McAnally J,
Bezprozvannaya S, Sutherland LB, Richardson JA, Bassel-Duby R and
Olson EN: An intragenic MEF2-dependent enhancer directs
muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad
Sci USA. 104:20844–20849. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu N, Bezprozvannaya S, Williams AH, Qi
X, Richardson JA, Bassel-Duby R and Olson EN: microRNA-133a
regulates cardiomyocyte proliferation and suppresses smooth muscle
gene expression in the heart. Genes Dev. 22:3242–3254. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Goddeeris MM, Rho S, Petiet A, Davenport
CL, Johnson GA, Meyers EN and Klingensmith J: Intracardiac
septation requires hedgehog-dependent cellular contributions from
outside the heart. Development. 135:1887–1895. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Latronico MV, Catalucci D and Condorelli
G: MicroRNA and cardiac pathologies. Physiol Genomics. 34:239–242.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
O'Brien JE Jr, Kibiryeva N, Zhou XG,
Marshall JA, Lofland GK, Artman M, Chen J and Bittel DC: Noncoding
RNA expression in myocardium from infants with tetralogy of Fallot.
Circ Cardiovasc Genet. 5:279–286. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yu ZB, Han SP, Bai YF, Zhu C, Pan Y and
Guo XR: microRNA expression profiling in fetal single ventricle
malformation identified by deep sequencing. Int J Mol Med.
29:53–60. 2012.PubMed/NCBI
|
|
33
|
Omran A, Elimam D, Webster KA, Shehadeh LA
and Yin F: MicroRNAs: A new piece in the paediatric cardiovascular
disease puzzle. Cardiol Young. 23:642–655. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ikeda S, Kong SW, Lu J, Bisping E, Zhang
H, Allen PD, Golub TR, Pieske B and Pu WT: Altered microRNA
expression in human heart disease. Physiol Genomics. 31:367–373.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tatsuguchi M, Seok HY, Callis TE, Thomson
JM, Chen JF, Newman M, Rojas M, Hammond SM and Wang DZ: Expression
of microRNAs is dynamically regulated during cardiomyocyte
hypertrophy. J Mol Cell Cardiol. 42:1137–1141. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
van Rooij E, Sutherland LB, Thatcher JE,
DiMaio JM, Naseem RH, Marshall WS, Hill JA and Olson EN:
Dysregulation of microRNAs after myocardial infarction reveals a
role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA.
105:13027–13032. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fichtlscherer S, Zeiher AM, Dimmeler S and
Sessa WC: Circulating microRNAs: Biomarkers or mediators of
cardiovascular diseases? Arterioscler Thromb Vasc Biol.
31:2383–2390. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tijsen AJ, Pinto YM and Creemers EE:
Circulating microRNAs as diagnostic biomarkers for cardiovascular
diseases. Am J Physiol Heart Circ Physiol. 303:H1085–H1095. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hunter JJ and Chien KR: Signaling pathways
for cardiac hypertrophy and failure. N Engl J Med. 341:1276–1283.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
van Rooij E, Sutherland LB, Liu N,
Williams AH, McAnally J, Gerard RD, Richardson JA and Olson EN: A
signature pattern of stress-responsive microRNAs that can evoke
cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA.
103:18255–18260. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Callis TE, Pandya K, Seok HY, Tang RH,
Tatsuguchi M, Huang ZP, Chen JF, Deng Z, Gunn B, Shumate J, et al:
MicroRNA-208a is a regulator of cardiac hypertrophy and conduction
in mice. J Clin Invest. 119:2772–2786. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sayed D, Hong C, Chen IY, Lypowy J and
Abdellatif M: MicroRNAs play an essential role in the development
of cardiac hypertrophy. Circ Res. 100:416–424. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Matkovich SJ, Hu Y, Eschenbacher WH, Dorn
LE and Dorn GW II: Direct and indirect involvement of microRNA-499
in clinical and experimental cardiomyopathy. Circ Res. 111:521–531.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li C, Fang Z, Jiang T, Zhang Q, Liu C,
Zhang C and Xiang Y: Serum microRNAs profile from genome-wide
serves as a fingerprint for diagnosis of acute myocardial
infarction and angina pectoris. BMC Med Genomics. 6:162013.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kuwabara Y, Ono K, Horie T, Nishi H, Nagao
K, Kinoshita M, Watanabe S, Baba O, Kojima Y, Shizuta S, et al:
Increased microRNA-1 and microRNA-133a levels in serum of patients
with cardiovascular disease indicate myocardial damage. Circ
Cardiovasc Genet. 4:446–454. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ai X, Curran JW, Shannon TR, Bers DM and
Pogwizd SM: Ca2+/calmodulin-dependent protein kinase
modulates cardiac ryanodine receptor phosphorylation and
sarcoplasmic reticulum Ca2+ leak in heart failure. Circ
Res. 97:1314–1322. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, He
J, Qin YW and Jing Q: Circulating microRNA: A novel potential
biomarker for early diagnosis of acute myocardial infarction in
humans. Eur Heart J. 31:659–666. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang L, Chen X, Su T, Li H, Huang Q, Wu
D, Yang C and Han Z: Circulating miR-499 are novel and sensitive
biomarker of acute myocardial infarction. J Thorac Dis. 7:303–308.
2015.PubMed/NCBI
|
|
49
|
Hosoda T, Zheng H, Cabral-da-Silva M,
Sanada F, Ide-Iwata N, Ogórek B, Ferreira-Martins J, Arranto C,
D'Amario D, del Monte F, et al: Human cardiac stem cell
differentiation is regulated by a mircrine mechanism. Circulation.
123:1287–1296. 2011. View Article : Google Scholar : PubMed/NCBI
|