Introduction
Although carbapenems antibiotics remain the backbone
therapy for severe suspected bacterial infections, resistance to
this antimicrobial treatment has been increasingly reported
(1). Thus, therapeutic options have
become limited. Multidrug-resistance to antibiotics currently
available, in particular in Gram-negative bacteria, has created a
critical global medical challenge (2). Acinetobacter baumannii is
frequently observed as a nosocomial infection, which causes high
mortality, morbidity and hospitalization cost (3). Crude mortality rate and attributable
mortality of the infection were reported to be 52 and 10–35%,
respectively (4).
Multidrug-resistant A. baumannii is considered as a leading
cause of nosocomial infection, particularly in critically ill
patients (5). A. baumannii
has been reported to be resistant to a broad range of antimicrobial
agents, and the tendency for its epidemic spread has subsequently
extended (6). An increasing drug
resistance of A. baumannii to carbapenems has been
demonstrated by the SENTRY Antimicrobial Surveillance Program,
whose objective was to report antimicrobial susceptibility and
pathogen occurrence data for >40,000 episodes of BSI in 72
medical centers representing 22 nations since January 1997
(7). The emergence of multidrug and
pandrug-resistant A. baumannii has caused major threats to
the infection control and treatment plans in clinical practices
(8). According to our knowledge,
drug resistance of A. baumannii is closely related with the
application of antibacterial agents. However, few studies have been
performed to investigate whether a single antibacterial agent was
able to induce pandrug resistance of A. baumannii. In the
present study, drug resistant A. baumannii strains were
generated in vitro, once sensitive strains were induced with
some commonly used antibiotics, such as FEP, SCF, TZP, LEV, AK, IPM
and CIP. Findings from the current study may provide evidence for
the association between the drug resistance of A. baumannii
strains and antibiotics, which may provide some guidance for the
treatment of A. baumannii infection.
Materials and methods
Sample collection and susceptibility
tests
A total of 16 non-repeated A. baumannii
strains were identified from sputum samples collected from patients
at the Second Xiangya Hospital of Central South University
(Changsha, China) between January 2010 and June 2011 (Patient
characteristics are presented in Table
I). Strains were sensitive to penicillins, cephalosporins,
β-lactam antibiotics, carbapenems, fluoroquinolones, and
aminoglycosides. All isolates were assayed for the antibiotic
susceptibility of cefepime, piperacillin/tazobactam, ciprofloxacin,
amikacin, imipenem, cefoperazone/sulbactam and levofloxacin, which
obtained from The National Institutes for Food and Drug Control of
China (Beijing, China), using the Kirby-Bauer test. Mueller-Hinton
(MH) broth and agar were purchased from Guangzhou Detgerm
Microbiology Technology Co., Ltd. (Guangzhou, China). Results were
analyzed using the performance standards for antimicrobial
susceptibility testing established by the Clinical and Laboratory
Standards Institute (CLSI) (9).
Minimum inhibitory concentration (MIC) was measured using the agar
dilution method. Results were analyzed using the performance
standards established by the CLSI in 2009. Pseudomonas
aeruginosa ATCC27853 (Mecconti, Sp. Zo.o., Warsaw, Poland) was
used as a quality control.
 | Table I.The patient characteristics of the 16
Acinetobacter baumannii strains obtained. |
Table I.
The patient characteristics of the 16
Acinetobacter baumannii strains obtained.
| Case | Gender | Age, years | Sample | Department | Main diagnosis |
|---|
| 1 | Male | 43 | sputum | Department of
hematology | M3 acute myeloid
leukemia; Lung infection |
| 2 | Male | 80 | sputum | Department of
senile disease | Acute exacerbation
of chronic obstructive pulmonary disease |
| 3 | Male | 73 | sputum | Department of
nephrology | Nephrotic syndrome;
Lung infection |
| 4 | Male | 80 | sputum | Department of
senile disease | Acute exacerbation
of chronic obstructive pulmonary disease |
| 5 | Male | 88 | sputum | Department of
senile disease | Acute exacerbation
of chronic obstructive pulmonary disease |
| 6 | Male | 78 | sputum | Department of
senile disease | Interstitial lung
disease; Lung infection |
| 7 | Male | 52 | sputum | Department of
respiratory medicine | Severe
pneumonia |
| 8 | Male | 39 | sputum | ICU | Lung infection
after renal transplantation |
| 9 | Male | 77 | sputum | Department of
senile disease | Type 2 respiratory
failure and Lung infection |
| 10 | Male | 44 | sputum | Department of
hematology | Multiple myeloma;
Lung infection |
| 11 | Male | 56 | sputum | Department of
nephrology | Diabetic
nephropathy; Lung infection |
| 12 | Male | 60 | sputum | Department of
senile disease | Acute exacerbation
of chronic bronchitis |
| 13 | Male | 88 | sputum | Department of
senile disease | Acute exacerbation
of chronic obstructive pulmonary disease |
| 14 | Male | 57 | sputum | Department of
respiratory medicine | Community-acquired
pneumonia |
| 15 | Male | 70 | sputum | Department of
neurology | Cerebral
hemorrhage; Lung infection |
| 16 | Male | 33 | sputum | Department of
infectious diseases | Lung abscess |
Induction of drug-resistant strains
using multi-step selection
A total of 16 strains of A. baumannii that
were sensitive to FEP, cefoperazone-sulbactam (SCF), tazobactam
(TZP), levofloxacin (LEV), amikacin (AK), imipenem (IPM), and
ciprofloxacin (CIP) antibiotics were used for the induction of drug
resistance in the strains. Bacteria were suspended in sterilized
isotonic saline solution to a turbidity of 0.5 McFarland. The
concentration of the suspension was modulated to 109
CFU/ml. Subsequently, 100 µl suspension was inoculated onto MH agar
plates with the seven antibiotics outlined (antibiotic
concentration, 1/4 of the MIC). Following inoculation at 37°C for
24 h, bacteria selected from single colonies were inoculated onto
the MH agar plates at the same concentration as the antibiotics.
Following inoculation for five generations, the resultant
suspensions were inoculated onto the MH agar plates with a doubled
concentration of antibiotics (1/2 of the MIC). The induction
sequence of the drugs was FEP, SCF, TZP, IPM, AK, CIP and LEV.
Sensitivity of the induced strains was determined using the
Kirby-Bauer method according to the CLSI guidelines (9) and MIC detection.
DNA isolation from the A. baumannii
strains
DNA from A. baumannii strains was extracted
using a DNA genome extraction kit (Tiangen Biotech Co., Ltd.,
Beijing, China) prior to the induction of resistance to FEP, SCF,
TZP, IPM, AK, CIP and LEV, according to the manufacturer's
instructions. Extracted DNA was resolved in
tris-ethylenediaminetetraacetic acid buffer, supplemented with
RNase. Purified DNA was aliquoted and stored at −20°C.
Random amplified polymorphic DNA
assay
Random amplified polymorphic DNA (RAPD) assay was
performed using a RAPD analysis kit (GE Healthcare Life Sciences,
Uppsala, Sweden). DNA was amplified by the addition of random
primer AP2 with a sequence of 5′-GTTTCGCTCC-3′ (10). Polymerase chain reaction (PCR)
amplification was performed in a total volume of 20 µl, containing
50 ng DNA template, 2X PCR mix and 2 µl of the primer. Mixtures
were subjected to 45 cycles of amplification (95°C for 45 sec, 33°C
for 45 sec, 72°C for 120 sec for each cycle) with an initial
incubation step at 95°C for 5 min and a final extension step at
72°C for 10 min. Amplified fragments were separated using 1.5%
agarose gel electrophoresis at 150 V/cm for 20 min. Images of the
gels were captured under ultraviolet illumination. Subsequently,
the distribution of the DNA bands obtained before and after drug
induction were compared.
PCR amplification for the
drug-resistant genes
Sequences of β-lactamase genes (OXA-23,
OXA-24, AmpC, TEM-1 and IMP),
fluoroquinolone resistance genes (parC and gyrA),
aminoglycoside resistance genes [aac(3)-I, aac(6′)-I, ant(3′)-I
and aph(3)-Via], 16S rRNA
methylase genes (armA, rmtA and rmtB), and
active efflux gene (adeB) were downloaded from Genbank
(ncbi.nlm.nih.gov/genbank). Specific
primers were designed according to these gene sequences. The
primers (Table II) were synthesized
by Sangon Biotech Co., Ltd., (Shanghai, China). PCR reactions were
performed in a volume of 20 µl containing 2X Taq PCR Master Mix, 10
µmol/l of each primer and 50 ng DNA template. PCR conditions of
each primer are listed in Table
III. The amplification product was electrophoresed on a 1.5%
agarose gel for 20 min with a voltage of 150 V. Following DNA
purification, the DNA samples were sent to Sangon Biotech Co., Ltd.
for sequencing analysis. The partial sequences of parC and
gyrA were compared with that of the NCBI database
respectively. using BLAST analysis (accessible at: https://blast.ncbi.nlm.nih.gov/Blast.cgi).
 | Table II.Primers used in polymerase chain
reaction amplification and the lengths of the resultant
fragments. |
Table II.
Primers used in polymerase chain
reaction amplification and the lengths of the resultant
fragments.
| Gene | Primer | Primer
sequence | Length of product,
bp |
|---|
| TEM-1 | Sense |
TTCGTGTCGCCCTTATTC | 512 |
|
| Anti |
ACGCTCGTCGTTTGGTAT |
|
| IMP | Sense |
CTACCGCAGCAGAGTCTTTG | 587 |
|
| Anti |
AACCAGTTTTGCCTTACCAT |
|
| OXA-23 | Sense |
TGTCATAGTATTCGTCGTT | 453 |
|
| Anti |
TTCCCAAGCGGTAAA |
|
| OXA-24 | Sense |
TTTGCCGATGACCTT | 175 |
|
| Anti |
TAGCTTGCTCCACCC |
|
| AmpC | Sense |
CGACAGCAGGTGGAT | 510 |
|
| Anti |
GGTTAAGGTTGGCATG |
|
| aac(3)-I | Sense |
ACCTACTCCCAACATCAGCC | 158 |
|
| Anti |
ATATAGATCTCACTACGCGC |
|
|
aac(6′)-I | Sense |
TATGAGTGGCTAAATCGA | 395 |
|
| Anti |
CCCGCTTTCTCGTAGCA |
|
| ant(3)-I | Sense |
TGATTTGCTGGTTACGGTGAC | 284 |
|
| Anti |
CGCTATGTTCTCTTGCTTTTG |
|
| aph(3)-VIa | Sense |
ATACAGAGACCACCATACAGT | 234 |
|
| Anti |
GGACAATCAATAATAGCAAT |
|
| armA | Sense |
GGGGTCTTACTATTCTG | 503 |
|
| Anti |
TTCCCTTCTCCTTTC |
|
| rmtA | Sense |
CCTAGCGTCCATCCTTTCCTC | 315 |
|
| Anti |
AGCGATATCCAACACACGATGG |
|
| rmtB | Sense |
ATGAACATCAACGATGCCCTC | 756 |
|
| Anti |
TTATCCATTCTTTTTTATCAAGTATAT |
|
| gyrA | Sense |
GCTGGCTAACGGTAACTC | 305 |
|
| Anti |
GGCTTCAATGGGACTG |
|
| parC | Sense |
CTGAACAGGCTTACTTGAA | 400 |
|
| Anti |
AAGTTATCTTGCCATTCG |
|
| AdeB | Sense |
TACCGGTATTACCTTTGCCGGA | 250 |
|
| Anti |
GTCTTTAAGTGTCGTAAAAGCCAC |
 | Table III.Polymerase chain reaction thermal
cycling conditions. |
Table III.
Polymerase chain reaction thermal
cycling conditions.
| Gene |
Pre-denaturation | Denaturation | Annealing | Extension | Cycles | Final
extension |
|---|
| TEM-1 | 94 (5 min) | 94 (60 sec) | 55 (60 sec) | 72 (50 sec) | 30 | 72 (7 min) |
| IMP | 94 (5 min) | 94 (60 sec) | 55 (60 sec) | 72 (50 sec) | 30 | 72 (7 min) |
| OXA-23 | 94 (5 min) | 94 (30 sec) | 48 (30 sec) | 72 (35 sec) | 30 | 72 (7 min) |
| OXA-24 | 94 (5 min) | 94 (30 sec) | 48 (30 sec) | 72 (35 sec) | 30 | 72 (7 min) |
| AmpC | 94 (5 min) | 94 (30 sec) | 50 (30 sec) | 72 (50 sec) | 30 | 72 (7 min) |
| aac(3)-I | 94 (4 min) | 94 (30 sec) | 55 (30 sec) | 72 (60 sec) | 35 | 72 (7 min) |
|
aac(6′)-I | 94 (4 min) | 94 (30 sec) | 55 (30 sec) | 72 (60 sec) | 35 | 72 (7 min) |
| ant(3)-I | 94 (4 min) | 94 (30 sec) | 55 (30 sec) | 72 (60 sec) | 35 | 72 (7 min) |
| aph(3)-VIa | 93 (2 min) | 93 (20 sec) | 55 (30 sec) | 72 (30 sec) | 30 | 72 (5 min) |
| armA | 94 (5 min) | 94 (30 sec) | 47 (30 sec) | 72 (50 sec) | 30 | 72 (5 min) |
| rmtA | 93 (2 min) | 93 (20 sec) | 55 (30 sec) | 72 (30 sec) | 30 | 72 (5 min) |
| rmtB | 93 (2 min) | 93 (20 sec) | 50 (60 sec) | 72 (60 sec) | 30 | 72 (5 min) |
| gyrA | 94 (4 min) | 94 (30 sec) | 55 (30 sec) | 72 (40 sec) | 30 | 72 (7 min) |
| parC | 94 (4 min) | 94 (30 sec) | 53 (30 sec) | 72 (40 sec) | 30 | 72 (7 min) |
| adeB | 95 (5 min) | 95 (30 sec) | 53 (60 sec) | 72 (90 sec) | 30 | 72 (7 min) |
Reverse transcription and
Semi-quantitative polymerase chain reaction (RT-Semi-qPCR)
assay
Total mRNA of 16 induced strains and sensitive
strains were extracted using a total RNA kit II (Omega Bio-Tek,
Inc. Norcross, GA, USA) according to the manufacturer's protocols.
RT was executed using an ReverTra Ace-α-transcriptase purchased
from Toyobo Co., Ltd., (Osaka, Japan) in a total volume of 25 µl,
following the manufacturer's protocol. PCR was performed in a
volume of 20 µl, comprising 2 µl cDNA. The mRNA expression of
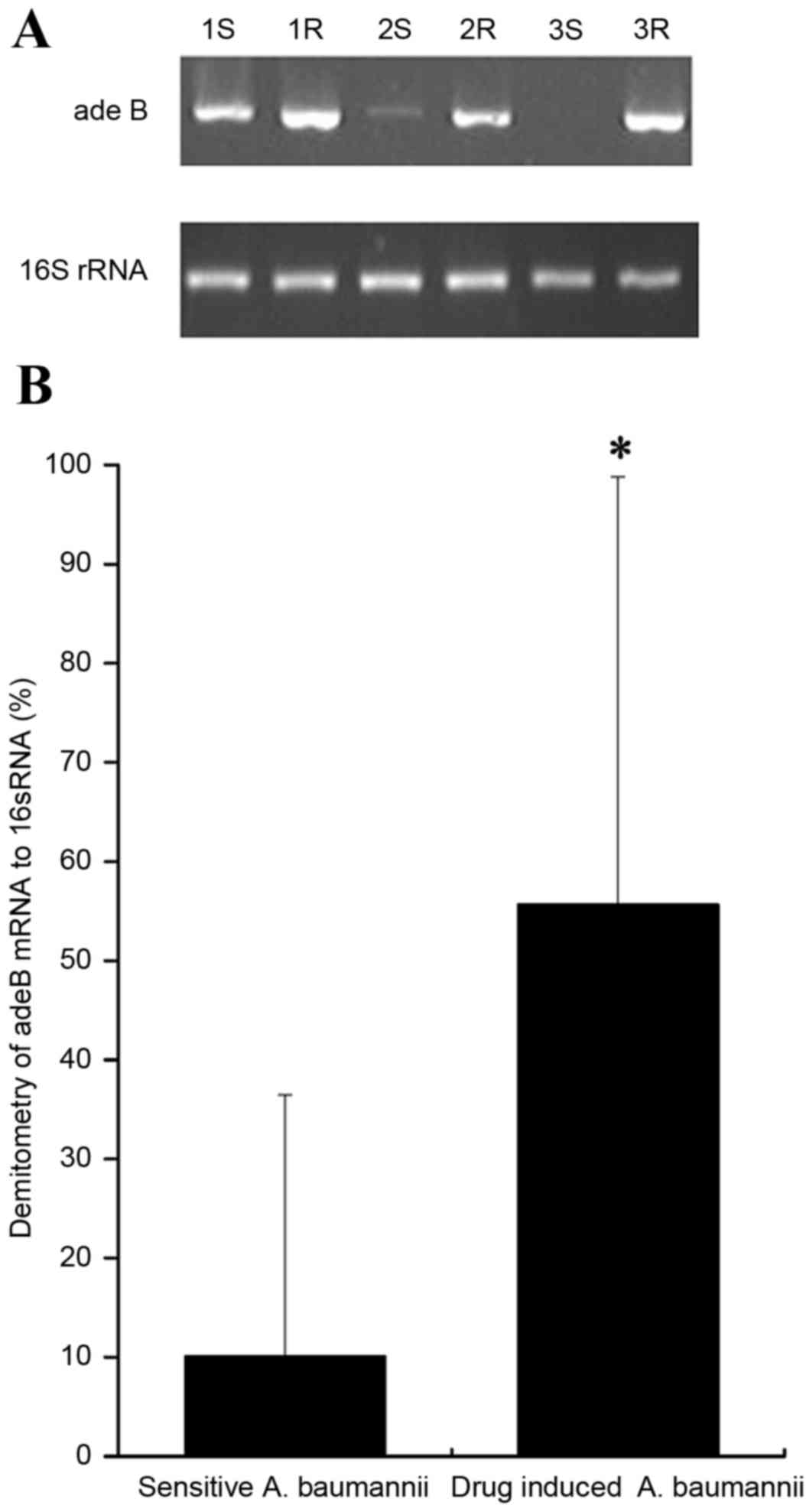
active efflux gene adeB was normalized to 16S rRNA. The
following cycling conditions were used, adeB: 95°C for 5
min, then 35 cycles of 95°C for 30 sec, 53°C for 60 sec and 72°C
for 90 sec, followed by 72°C for 4 min; 16S rRNA: 95°C for 5
min, 35 cycles of 94°C for 30 sec, 55°C for 40 sec and 72°C for 45
sec, followed by 72°C for 4 min. The primers used for 16S rRNA
downloaded from Genbank (accessible at: https://www.ncbi.nlm.nih.gov/genbank/) were forward
5′-GTTATTAGGGAAGAACATATGTG-3′ and reverse
5′-CCACCTTCCTCCGGTTTGTCACC-3′. And the primers used for adeB
were forward 5′-AAAGACTTCAAAGAGCGGACTA-3′ and reverse
5′-ATTGTCACCTTGTGGCAACCCT-3′. The cDNA amplification product was
electrophoresed on a 1.5% agarose gel. Subsequently, the
electrophoretic grayscale were analyzed to assess the product sizes
using Quantity One 4.4.0 software (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) and presented as a mean ± standard
deviation.
Statistical analysis
SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA)
was used for statistical analysis. The rate of induced strains
resistant to each antibiotic and carrying rate of drug-resistant
genes were presented as percentages. Chi-square test was used for
comparing the carrying rate of drug resistance genes. Student's
t-test was performed for the expression of adeB mRNA.
P<0.05 was considered to indicate a statistically significant
difference. The electrophoretic grayscale of adeB was
presented as a mean ± standard deviation. Each experiment was
repeated 3 times.
Results
Source and distribution of the
strains
A total of 16 strains were isolated from sputum
samples obtained from the Department of Senile Disease (43.8%), ICU
(6.25%), Department of Respiratory Medicine (12.5%), Department of
Hematology (12.5%), Department of Nephrology (12.5%), Department of
Infectious Diseases (6.25%) and Department of Neurology (6.2%),
respectively, at the Second Xiangya Hospital of Central South
University.
MIC of the strains following induction
in vitro
Among the 16 strains, 15 strains (93.8%) acquired
drug resistance following in vitro induction. Five strains
were resistant to all the drugs, while 8 strains were resistant to
≥5 drugs, and 2 strains were resistant to <5 drugs. Table IV summarizes the MICs of the 16
strains before and after in vitro induction using FEP, SCF,
TZP, LEV, AK, IPM, and CIP, respectively. The number of strains
resistant to FEP, CIP, LEV, AK, TZP, SCF, and IPM were 13 (81.3%),
12 (75.0%), 11 (68.8%), 11 (68.8%), 9 (56.3%), 8 (50.0%), and
(43.8%), respectively. Minimum drug resistance of the strains was
noted in IPM, followed by SCF and TZP. Following drug induction, a
decrease was observed in the size of the inhibition zone generated
by all the induced strains, and an increase was noted in the MIC to
each drug (Table IV).
 | Table IV.MIC of the 16 strains prior to and
following drug induction. |
Table IV.
MIC of the 16 strains prior to and
following drug induction.
| Strain | FEP | SCF | TZP | LEV | AK | IPM | CIP |
|---|
| 1 | 2/32 | 2/64 | 4/64 | 0.5/8 | 8/64 | 2/16 | 0.5/32 |
| 2 | 2/16 | 2/32 | 4/64 | 0.5/8 | 16/128 | 4/64 | 0.5/32 |
| 3 | 2/16 | 4/64 | 8/64 | 0.5/8 | 8/64 | 0.5/8 | 0.5/32 |
| 4 | 2/32 | 2/64 | 8/64 | 0.5/8 | 8/64 | 8/128 | 0.5/16 |
| 5 | 4/32 | 4/128 | 4/64 | 0.5/8 | 2/16 | 2/16 | 0.25/32 |
| 6 | 2/32 | 4/64 | 4/64 | 0.5/8 | 4/128 | – | 0.5/16 |
| 7 | 4/64 | 4/32 | 8/64 | 1/8 | 8/64 | – | 1/16 |
| 8 | 4/32 | – | 8/64 | 1/8 | 8/64 | – | 0.5/64 |
| 9 | 2/32 | – | – | 0.5/16 | 8/128 | – | 0.5/32 |
| 10 | 2/32 | – | – | 0.5/8 | 8/128 | – | 0.5/16 |
| 11 | 2/64 | 4/64 | 4/64 | – | – | – | – |
| 12 | 4/64 | – | – | – | – | 2/64 | – |
| 13 | 4/64 | – | – | – | 8/64 | 2/64 | – |
| 14 | – | – | – | 0.5/4 | – | – | 0.5/16 |
| 15 | – | – | – | – | – | – | 0.5/16 |
| 16 | – | – | – | – | – | – | – |
Genotyping of AP2
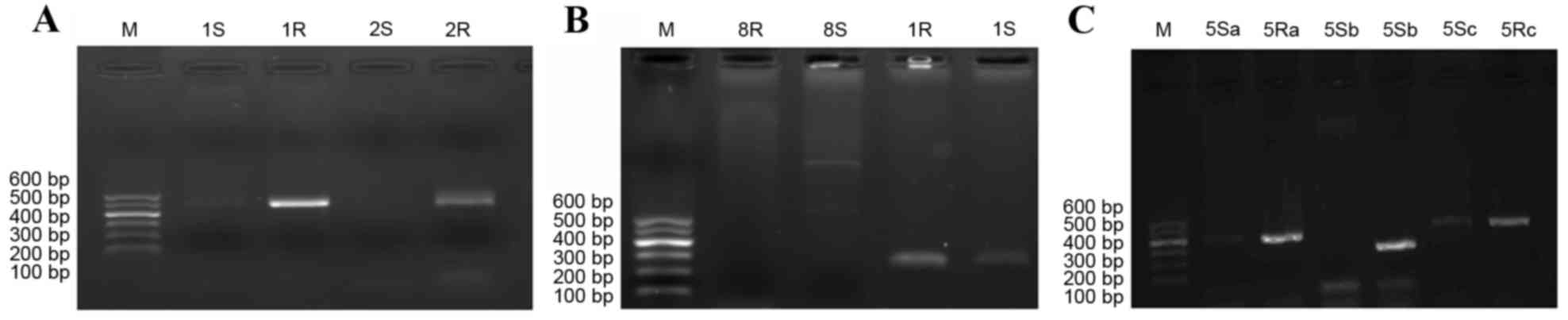
As the RAPD results indicate in Fig. 1, genotyping of the strains varied
depending on the original non-induced strains (16 sensitive
strains) and those obtained after induction in vitro.
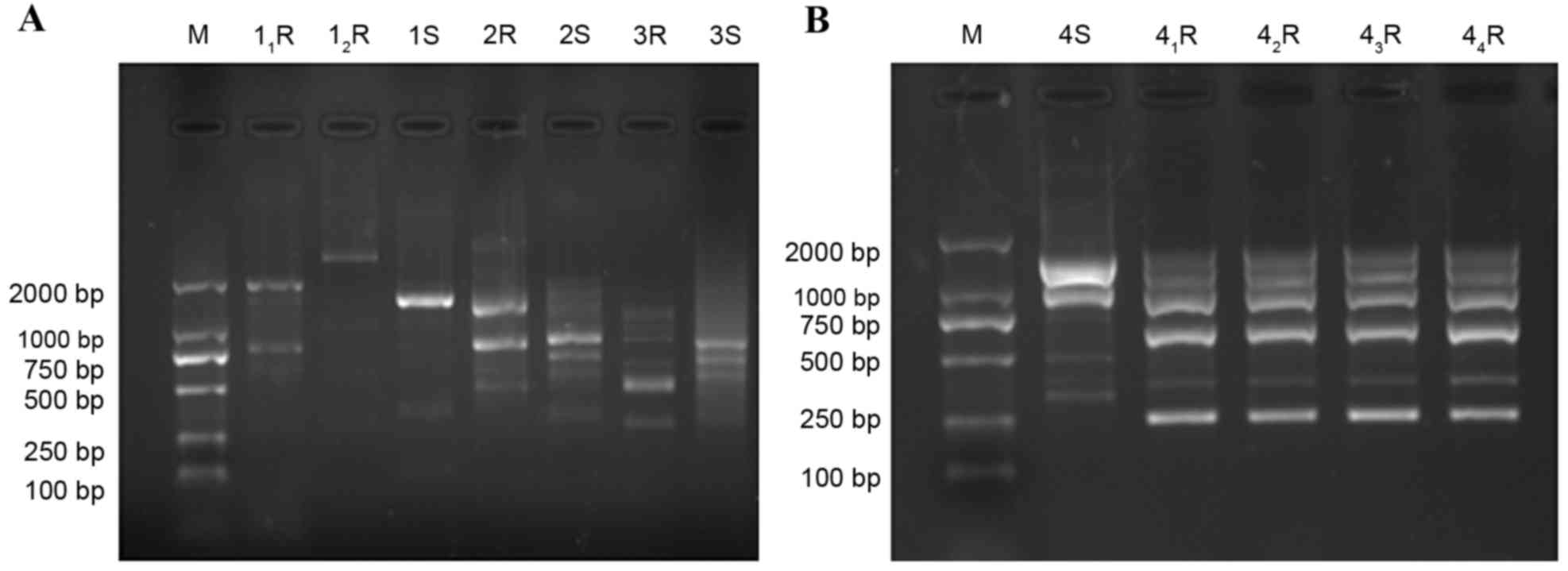 | Figure 1.Genotype of Acinetobacter
baumannii before and after drug induction. (A) 1S, 2S, 3S and
4S, strains prior to drug induction; 11R,
12R, drug-resistant strain number 1; 2R, drug-resistant
strain number 2; 3R, drug-resistant strain number 3 and (B)
41R, 42R, 43R and 44R,
drug-resistant strain number 4. M, marker; S, prior to induction;
R, following induction. |
Identification of the drug resistance
genes
With the exception of four drug resistant genes
(rmtA, IMP, TEM-1, and OXA-24), a
significantly increased positive rate of gene amplification was
noted in the induced strains when compared with drug-susceptible
strains (P<0.05; Fig. 2; Table V). The present study showed that
acquired drug resistance was achieved in A. baumannii
following exposure to low concentrations of antibiotics, in
vitro. Moreover, significant differences were exhibited in the
amplification results of adeB in A. baumannii
following in vitro induction when compared with the results
obtained from the strains without in vitro induction
(χ2=20.257; P<0.05; Table
V).
 | Table V.Carrying rates of drug resistance
gene in sensitive strains and induced strains. |
Table V.
Carrying rates of drug resistance
gene in sensitive strains and induced strains.
| Gene | Carrying rates in
sensitive strains | Carrying rates in
induced strains | χ2 | P-value |
|---|
| aac(3)-I | 12.5 (2/16) | 54.5 (6/11) | – | 0.027 |
| aac(6)-I | 18.8 (3/16) | 63.6 (7/11) | – | 0.024 |
| ant(3)-I | 6.3 (1/16) | 81.8 (9/11) | – | 0.000 |
| aph(3) | 6.3 (1/16) | 54.5 (6/11) | – | 0.009 |
| armA | 12.5 (2/16) | 90.9 (10/11) | – | 0.000 |
| rmtA | 18.8 (3/16) | 45.5 (5/11) | – | 0.144 |
| rmtB | 12.5 (2/16) | 54.5 (6/11) | – | 0.027 |
| IMP | 25.0 (4/16) | 27.0 (10/37) |
0.024 | 0.582 |
| TEM-1 | 68.8 (11/16) | 81.1 (30/37) |
0.970 | 0.261 |
| OXA-24 | 37.5 (6/16) | 43.2 (16/37) |
0.152 | 0.469 |
| OXA-23 | 18.8 (3/16) | 64.9 (24/37) |
9.505 | <0.05 |
| AmpC | 50.0 (8/16) | 91.9 (34/37) | 11.918 | <0.05 |
| gyrA | 100.0 (16/16) | 100.0 (11/11) | – | – |
| parC | 100.0 (16/16) | 100.0 (12/12) | – | – |
| adeB | 18.8 (3/16) | 78.6 (56/71) | 20.257 | <0.05 |
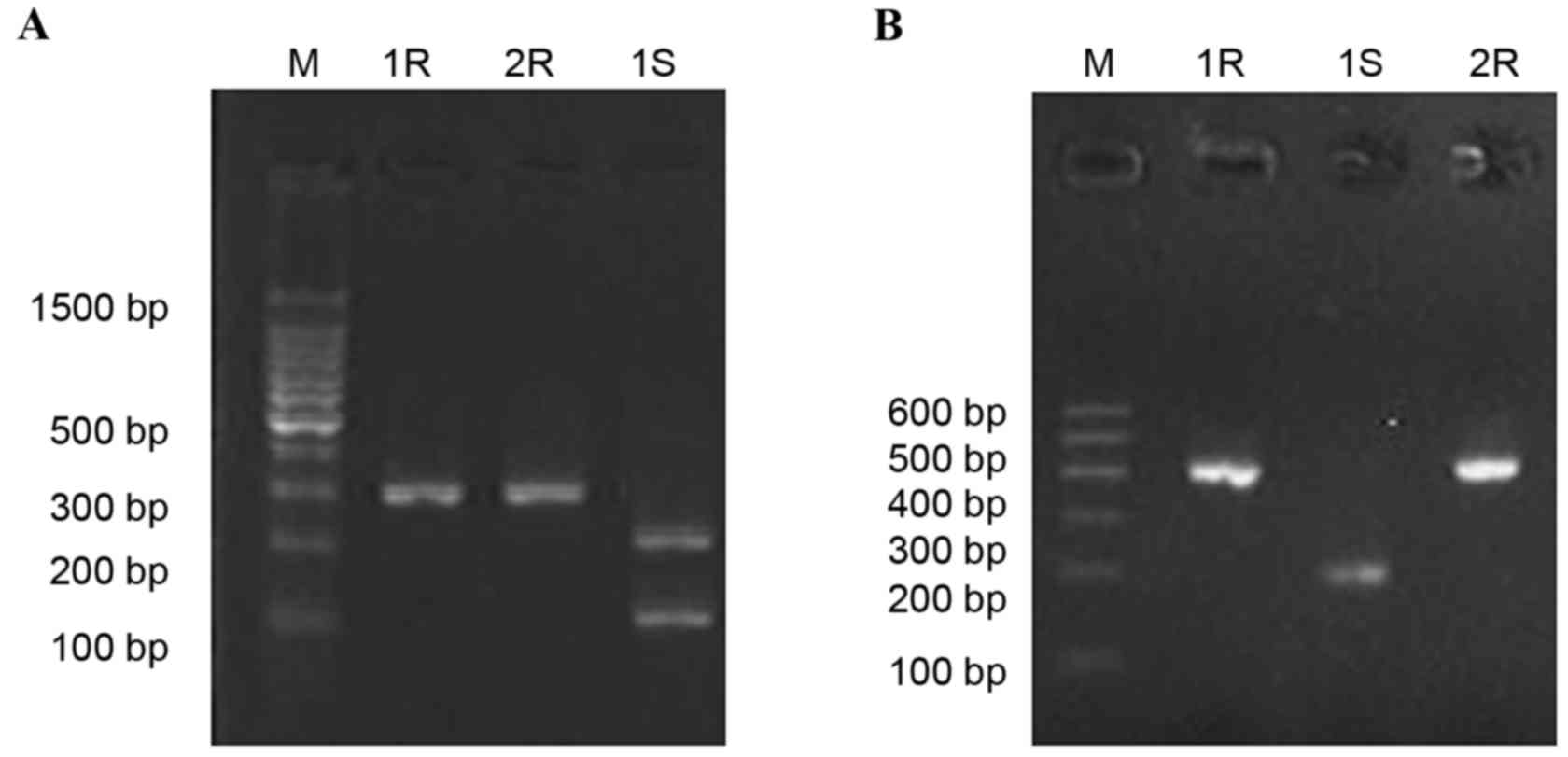
PCR amplification and restriction map
of gyrA and parC
Following drug induction using fluoroquinolone
antibiotics, CIP and LEV, PCR results revealed the amplification of
gyrA and parC before and after induction was
positive. Following digestion using Hinf1, gyrA fragments,
obtained from 10 drug-sensitive strains (10/16; 62.5%) generated
two bands (225 and 80 bp, Fig. 3A),
while the gyrA fragments, obtained from the 11 drug
resistant strains, were not digested by Hinf1. parC
fragments obtained from the drug resistant strains (3/12; 25.0%)
and sensitive strains (11/16; 68.7%) were digested into two bands
(205 and 195 bp, were displayed as one band due to it's similar
molecular weight; Fig. 3B).
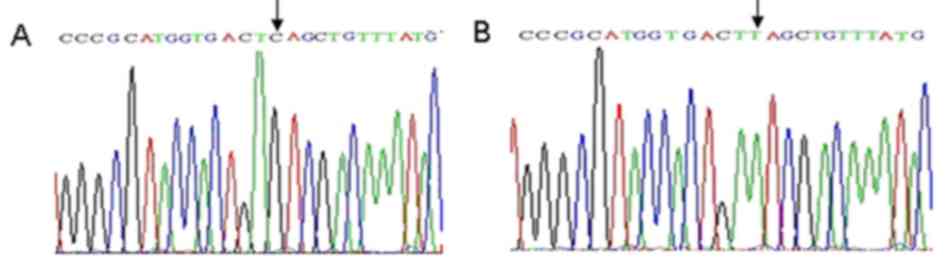
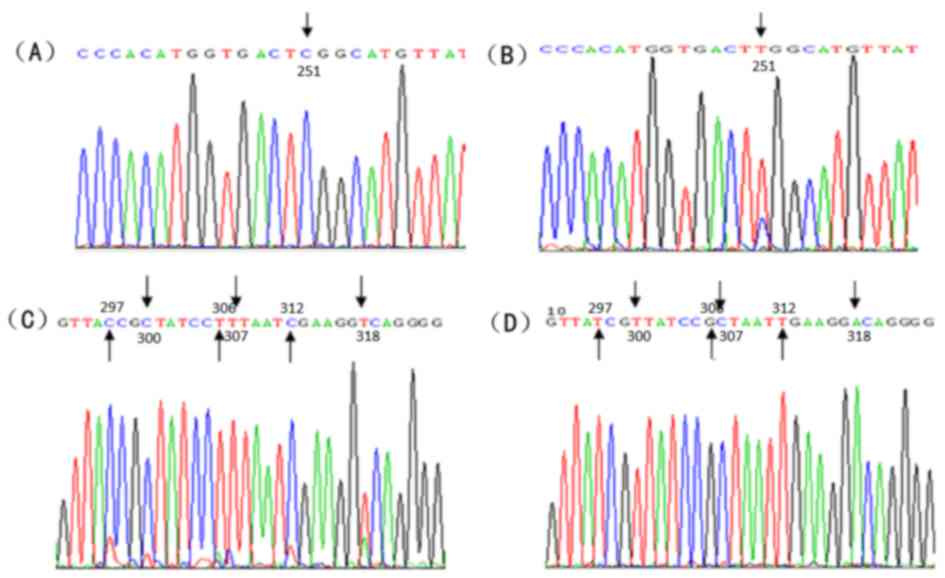
Sequence analyses of gyrA and
parC
Following sequencing, the partial sequences were
compared with that of the NCBI database using BLAST analysis
(https://blast.ncbi.nlm.nih.gov/Blast.cgi). Results
indicated nucleotide (nt) 242 C/T and 275 C/T were in the cutting
site of Hinf1 in gyrA of the strains that underwent drug
induction, which induced the substitution of serine 81 to leucine,
and threonine 92 to methionine (Fig.
4). However, no gene mutation was noted in the cutting site of
the Hinf1. The nt 251 C/T was exhibited in the cutting site of
Hinf1 in parC of the strains that underwent drug induction,
which induced the substitution of serine 84 to leucine (Fig. 5A and B). However, multiple synonymous
mutations were observed, such as nt 297 T/C, nt 300 T/C, nt 306
G/T, nt 307 C/T, nt 312 T/T, and nt 318 A/T (Fig. 5C and D).
Expression of adeB mRNA
Semi-qRT-PCR indicated the lengths of the amplified
fragments for adeB and 16S rRNA were 273 and 750 bp,
respectively. The relative grayscale was calculated according to
the Quantity One 4.4.0 software. Compared with the mRNA expression
levels of the sensitive strains, significant differences were
observed in the adeB mRNA expression of drug-induced strains
(55.69±43.11% vs. 10.08±26.35%; P<0.05; Fig. 6).
Discussion
A. baumannii, which is a gram-negative,
non-fermentative coccobacillus of the family Moraxellaceae, is
considered to be an important cause of ventilator-associated
pneumonia, sepsis, urinary system infection and meningitis
(11–17). Currently, no approved antimicrobial
drugs have been successfully developed to treat A.
baumannii, as it exhibits multidrug resistance. In the present
study, 16 A. baumannii strains were isolated from the
Department of Senile Disease (43.8%), ICU (12.5%), Department of
Respiratory Medicine (12.5%), Department of Hematology (12.5%),
Department of Nephrology (12.5%), and Department of Neurology
(6.2%), respectively, at the Second Xiangya Hospital of Central
South University. The majority of strains were isolated from the
ICU and the Department of Respiratory Medicine in a previous study
(18). We speculated that this
descrepency may be due to the strains selected in this study.
Moreover, the strains were exclusively isolated from sputum
samples, demonstrating that A. baumannii may be an important
cause for respiratory tract infection and was similar to a previous
study using sputum samples from patients with pneumonia as a source
of Acinetobacter baumannii (18). Thus, more attention is required when
monitoring the respirator and the nursing staff to prevent
respiratory tract infections in these Departments.
The mechanism of how A. baumannii develops
resistance towards multiple drugs has been extensively studied
(19–21). The mechanisms underlying resistance
to multiple drugs in A. baumannii have been identified to
include i) the capacity to generate enzymes that may inactivate the
antibacterial agents; ii) changes in the antibacterial-binding
proteins that prevent their action; iii) alternations in the
structure and number of porin proteins that lead to decreased
permeability to antibacterial agents; and iv) the activity of
efflux pumps that further reduce the concentration of antibacterial
agents within the bacterial cell. It has been well-acknowledged
that the long-term exposure to various antibiotics is the major
reason for the formation of multidrug-resistant A. Baumannii
(6). However, few studies have
investigated the mechanism of how drug resistance is established.
With the large-scale application of antibacterial agents,
multidrug-resistant bacteria have been compared with bacteria that
are sensitive to antibacterial agents. To date, various
multidrug-resistant bacteria have been detected. For example,
resistant Pseudomonas aeruginosa and Streptococcus
pyogenes have been induced in vitro, using various
agents such as gentamicin, ciprofloxacin, and azithromycin
(22–26).
In the present study, multi-step selection was used
for the induction of drug-resistant bacteria to avoid the
specificity of the induction. Furthermore, the growth and disperse
pattern of the bacteria in vivo was mimicked in our study.
The initial induction concentration was 1/4 MIC, which revealed
similarity with the low dose clinical medication administered in
our clinical practices (27). A
total of 15 strains (98.3%) resistant to FEP, SCF, TZP, IPM, AK,
CIP and LEV were collected following exposure to increasing
concentrations of the agent. Moreover, sharp increases were noted
in the MIC to these agents post-induction. These results indicated
that significant differences were observed in the drug-resistant
phenotype of A. baumannii in the presence of antibiotics.
Additionally, the drug tolerance of the strains was comparatively
stable. Significant cross resistance was exhibited in the strains
collected in the present study. Our study was consistent with
previous reports (23,28).
The genotype and drug-resistant genes were analyzed
before and after drug induction. The results of the present study
revealed statistical differences in the drug resistance genes and
genotyping. Moreover, a significantly increased positive rate was
observed in the β-lactamase gene in strains subjected to drug
induction, particularly OXA-23 (64.9%) and AmpC
(91.9%). To our knowledge, OXA-23-producing A.
baumannii was resistant to IPM (29–36).
AmpC enzyme was encoded by chromogene, and it's over
expression could induce drug resistance towards penicillin and the
third generation broad-spectrum cephalosporins, through hydrolysis.
We speculated that OXA-23 and AmpC may be associated
with drug resistance towards FEP, SCF, TZP, and IPM as an increased
positive rate was observed in OXA-23 and AmpC. In
A. baumannii, aminoglycosides modifying enzyme genes, such
as AAC(3)-I, AAC(6′)-I
and ANT(3)-I, and 16S
rRNA methylase gene, including armA, rmtA,
rmtB, and qph (3),
were revealed to induce the drug resistance towards the
aminoglycoside antibiotics. This type of drug resistance may be
related with horizontal transmission of the drug-resistant genetic
locus induced by plasmid or transposon. The present findings
indicated that statistical differences were observed in the
carrying rate of armA and rmtB. Based on the present
data, the expression of the aminoglycoside resistance genes was
suggested to be a major cause for the drug resistance to the
aminoglycosides demonstrated herein. In addition, gyrA and
parC were isolated in all strains, and gene mutations of
these genes may be associated with the formation of drug resistance
(29,32). As the efflux pump, which is located
in the outer membrane, has been suggested to have an important role
in multidrug resistance capacity in bacteria (31–33), the
expression of the active efflux gene adeB was determined.
The resistance-nodulation-cell-division-type multidrug efflux pump,
adeb, was associated with aminoglycoside resistance and has
previously been implicated in mediating the level of susceptibility
towards other drugs, such as tetracyclines, chloramphenicol,
erythromycin, trimethoprim, and ethidium bromide (34). In the present study, a significant
increase was noted in the carrying rate of adeB following
in vitro induction; furthermore, significant differences
were exhibited in the expression of adeB in strains that
underwent drug induction when compared with the sensitive strains,
which implied that the efflux pump has a crucial role in the drug
resistance of A. baumannii. Notably, specific drug-resistant
genes were also isolated in A. baumannii obtained from the
clinical practices. However, no drug resistance was noted, which
may be related to the lack of expression and low-level expression
of these genes.
In the present study, notable differences were
observed in the traits of colonies following in vitro
induction. Compared with the colonies formed prior to drug
induction, the profile of colonies were smaller in a pattern of
slow growth. This demonstrated that the growth of the A.
baumannii was notably affected by the antibiotics. Further
studies are required to validate whether the pathogenicity of A.
baumannii increases in the presence of increased drug
resistance to antibiotics.
In conclusion, the present study established a drug
resistant A. baumannii model following in vitro
induction, using multiple-step methods. Furthermore, the active
efflux pump may be involved in the development of drug resistance
in A. baumannii in vitro. This study may provide useful
information for the drug resistance mechanism of the A.
baumannii; however, further studies are necessary to fully
elucidate the drug resistance of A. baumannii in vivo.
Acknowledgments
The present study was supported by the China
National Natural Scientific Foundation (grant no. 81470133), and
the Science and Technology Planning Project of Hunan Province of
China (grant no. 2015JC3035).
References
|
1
|
Bou G, Cerveró G, Domínguez MA, Quereda C
and Martínez-Beltrán J: Characterization of a nosocomial outbreak
caused by a multiresistant Acinetobacter baumannii strain with a
carbapenem-hydrolyzing enzyme: High-level carbapenem resistance in
A. Baumannii is not due solely to the presence of beta-lactamases.
J Clin Microbiol. 38:3299–3305. 2000.PubMed/NCBI
|
|
2
|
Levin AS: Multiresistant Acinetobacter
infections: A role for sulbactam combinations in overcoming an
emerging worldwide problem. Clin Microbiol Infect. 8:144–153. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gulen TA, Guner R, Celikbilek N, Keske S
and Tasyaran M: Clinical importance and cost of bacteremia caused
by nosocomial multi drug resistant acinetobacter baumannii. Int J
Infect Dis. 38:32–35. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shih MJ, Lee NY, Lee HC, Chang CM, Wu CJ,
Chen PL, Ko NY and Ko WC: Risk factors of multidrug resistance in
nosocomial bacteremia due to Acinetobacter baumannii: A case
control study. J Microbiol Immunol Infect. 41:118–123.
2008.PubMed/NCBI
|
|
5
|
Tas T, Kocoglu E, Mengeloglu Z, Bucak O
and Karabörk S: Investigation of in-vitro susceptibility of
multidrug-resistant Acinetobacter baumannii strains isolated from
clinical specimens to tigecycline. Bosn J Basic Med Sci.
13:266–270. 2013.PubMed/NCBI
|
|
6
|
Minandri F, D'Arezzo S, Antunes LC,
Pourcel C, Principe L, Petrosillo N and Visca P: Evidence of
diversity among epidemiologically related carbapenemase-producing
Acinetobacter baumannii strains belonging to international clonal
lineage II. J Clin Microbiol. 50:590–597. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Castanheira M, Wanger A, Kruzel M,
Deshpande LM and Jones RN: Emergence and clonal dissemination of
OXA-24- and OXA-58-Producing Acinetobacter baumannii Strains in
Houston, Texas: Report from the SENTRY Antimicrobial Surveillance
Program. J Clin Microbiol. 46:3179–3180. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lu PL, Siu LK, Chen TC, Ma L, Chiang WG,
Chen YH, Lin SF and Chen TP: Methicillin-resistant Staphylococcus
aureus and Acinetobacter baumannii on computer interface surfaces
of hospital wards and association with clinical isolates. BMC
Infect Dis. 9:1642009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Clinical and Laboratory Standards
Institute, . Performance Standards for Antimicrobial Susceptibility
Testing: Twenty-Second Informational Supplement M100-S22. CLSI.
32:64–65. 2012.
|
|
10
|
Koeleman JG, Stoof J, Biesmans DJ,
Savelkoul PH and Vandenbroucke-Grauls CM: Comparison of amplified
ribosomal DNA restriction analysis, random amplified polymorphic
DNA analysis and amplified fragment length polymorphism
fingerprinting for identification of Acinetobacter genomic species
and typing of Acinetobacter baumannii. J Clin Microbiol.
36:2522–2529. 1998.PubMed/NCBI
|
|
11
|
Hoban DJ, Bouchillon SK and Dowzicky MJ:
Antimicrobial susceptibility of extended-spectrum beta-lactamase
producers and multidrug-resistant Acinetobacter baumannii
throughout the United States and comparative in vitro activity of
tigecycline, a new glycylcycline antimicrobial. Diagn Microbiol
Infect Dis. 57:423–428. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
López-Alvarez B, Martín-Laez R, Fariñas
MC, Paternina-Vidal B, García-Palomo JD and Vázquez-Barquero A:
Multidrug-resistant Acinetobacter baumannii ventriculitis:
Successful treatment with intraventricular colistin. Acta Neurochir
(Wien). 151:1465–1472. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kwa AL, Loh C, Low JG, Kurup A and Tam VH:
Nebulized colistin in the treatment of pneumonia due to
multidrug-resistant Acinetobacter baumannii and Pseudomonas
aeruginosa. Clin Infect Dis. 41:754–757. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ho CM, Ho MW, Chi CY, Lin CD, Lin CW,
Tseng SP, Teng LJ, Chang HY, Chang HL, Chang YF, et al: Repeated
colonization by multi-drug-resistant Acinetobacter calcoaceticus-A.
Baumannii complex and changes in antimicrobial susceptibilities in
surgical intensive care units. Surg Infect (Larchmt). 14:43–48.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Celik IH, Oguz SS, Demirel G, Erdeve O and
Dilmen U: Outcome of ventilator-associated pneumonia due to
multidrug-resistant Acinetobacter baumannii and Pseudomonas
aeruginosa treated with aerosolized colistin in neonates: A
retrospective chart review. Eur J Pediatr. 171:311–316. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Moland ES, Craft DW, Hong SG, Kim SY,
Hachmeister L, Sayed SD and Thomson KS: In vitro activity of
tigecycline against multidrug-resistant Acinetobacter baumannii and
selection of tigecycline-amikacin synergy. Antimicrob Agents
Chemother. 52:2940–2942. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
D'Arezzo S, Capone A, Petrosillo N, Visca
P, GRAB, Ballardini M, Bartolini S, Bordi E, Di Stefano A, Galiè M,
et al: Epidemic multidrug-resistant Acinetobacter baumannii related
to European clonal types I and II in Rome (Italy). Clin Microbiol
Infect. 15:347–357. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Garnacho-Montero J and Amaya-Villar R:
Multiresistant Acinetobacter baumannii infections: Epidemiology and
management. Curr Opin Infect Dis. 23:332–339. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hou C and Yang F: Drug-resistant gene of
blaOXA-23, blaOXA-24, blaOXA-51 and blaOXA-58 in Acinetobacter
baumannii. Int J Clin Exp Med. 8:13859–13863. 2015.PubMed/NCBI
|
|
20
|
Poirel L, Pitout JD and Nordmann P:
Carbapene-mases: Molecular diversity and clinical consequences.
Future Microbio. 2:501–512. 2007. View Article : Google Scholar
|
|
21
|
Sinha M and Srinivasa H: Mechanisms of
resistance to carbapenems in meropenem- resistant Acinetobacter
isolates from clinical samples. Ind J Med Microbiol. 25:121–125.
2008. View Article : Google Scholar
|
|
22
|
Chretien FC and Dubois R: Effect of
nomegestrol acetate on spinability, ferning and mesh dimension of
midcycle cervical mucus. Contraception. 43:55–65. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Radberg G, Nilsson LE and Svensson S:
Development of quinolone-imipenem cross resistance in Pseudomonas
aeruginosa during exposure to ciprofloxacin. Antimicrob Agents
Chemother. 34:2142–2147. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Barclay ML, Begg EJ, Chambers ST and
Peddie BA: The effect of aminoglycoside-induced adaptive resistance
on the antibacterial activity of other antibiotics against
Pseudomonas aeruginosa in vitro. J Antimicrob Chemother.
38:853–858. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Prasad SV, Ballal M and Shivananda PG:
Induction of resistance to fluoroquinolones in clinical and
environmental isolates of Pseudomonas aeruginosa. Indian J Pathol
Microbiol. 50:94–96. 2007.PubMed/NCBI
|
|
26
|
De Vecchi E, Nicola L, Zucchetti E and
Drago L: In vitro induction of resistance by tissue concentrations
of azithromycin, clarithromycin, cefixime and
amoxicillin/clavulanate in clinical isolates of Streptococcus
pyogenes. J Chemother. 18:379–388. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Miller K, O'Neill AJ and Chopra I:
Response of Escherichia coli hypermutators to selection pressure
with antimicrobial agents from different classes. J Antimicrob
Chemother. 49:925–934. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tripodi MF, Durante-Mangoni E, Fortunato
R, Utili R and Zarrilli R: Comparative activities of colistin,
rifampicin, imipenem and sulbactam/ampicillin alone or in
combination against epidemic multidrug-resistant Acinetobacter
baumannii isolates producing OXA-58 carbapenemases. Int J
Antimicrob Agents. 30:537–540. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jeon BC, Jeong SH, Bae IK, Kwon SB, Lee K,
Young D, Lee JH, Song JS and Lee SH: Investigation of a nosocomial
outbreak of imipenem-resistant Acinetobacter baumannii producing
the OXA-23 beta-lactamase in Korea. J Clin Microbiol. 43:2241–2245.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chiu CH, Lee HY, Tseng LY, Chen CL, Chia
JH, Su LH and Liu SY: Mechanisms of resistance to ciprofloxacin,
ampicillin/sulbactam and imipenem in Acinetobacter baumannii
clinical isolates in Taiwan. Int J Antimicrob Agents. 35:382–386.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu YH, Kuo SC, Lee YT, Chang IC, Yang SP,
Chen TL and Fung CP: Amino acid substitutions of quinolone
resistance determining regions in GyrA and ParC associated with
quinolone resistance in Acinetobacter baumannii and Acinetobacter
genomic species 13TU. J Microbiol Immunol Infect. 45:108–112. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wieczorek P, Sacha P, Hauschild T,
Zorawski M, Krawczyk M and Tryniszewska E: Multidrug resistant
Acinetobacter baumannii-the role of AdeABC (RND family) efflux pump
in resistance to antibiotics. Folia Histochem Cytobiol. 46:257–267.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lin L, Ling BD and Li XZ: Distribution of
the multidrug efflux pump genes, adeABC, adeDE and adeIJK, and
class 1 integron genes in multiple-antimicrobial-resistant clinical
isolates of Acinetobacter baumannii-Acinetobacter calcoaceticus
complex. Int J Antimicrob Agents. 33:27–32. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hou PF, Chen XY, Yan GF, Wang YP and Ying
CM: Study of the correlation of imipenem resistance with efflux
pumps AdeABC, AdeIJK, AdeDE and AbeM in clinical isolates of
Acinetobacter baumannii. Chemotherapy. 58:152–158. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Magnet S, Courvalin P and Lambert T:
Resistance-nodulation-cell division-type efflux pump involved in
aminoglycoside resistance in Acinetobacter baumannii strain BM4454.
Antimicrob Agents Chemother. 45:3375–3380. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhou Y, Wu X, Zhang X, Hu Y, Yang X, Yang
Z and Wang M: Genetic Characterization of ST195 and ST365
Carbapenem-Resistant Acinetobacter baumannii Harboring blaOXA-23 in
Guangzhou, China. Microb Drug Resist. 21:386–390. 2015. View Article : Google Scholar : PubMed/NCBI
|