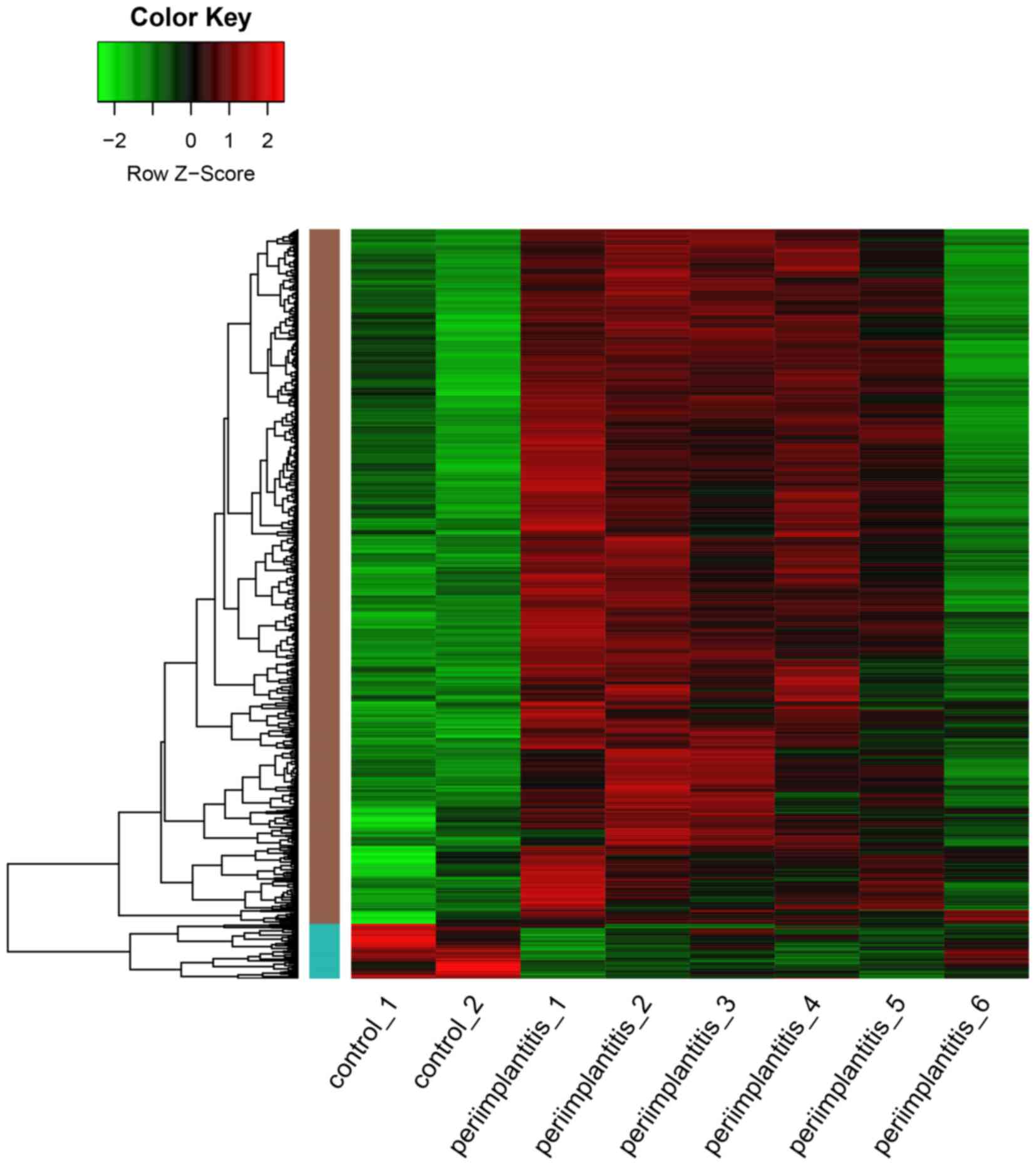
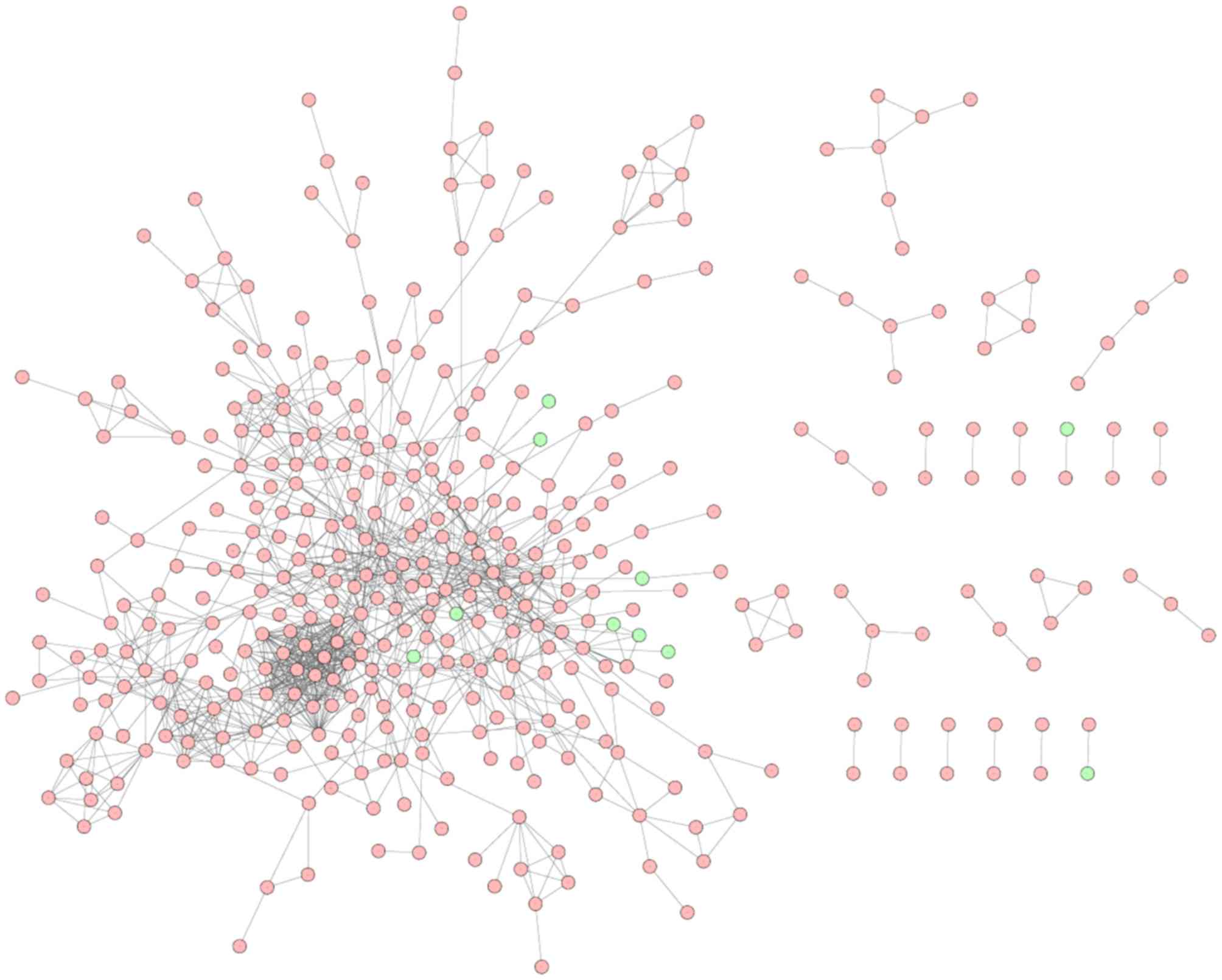
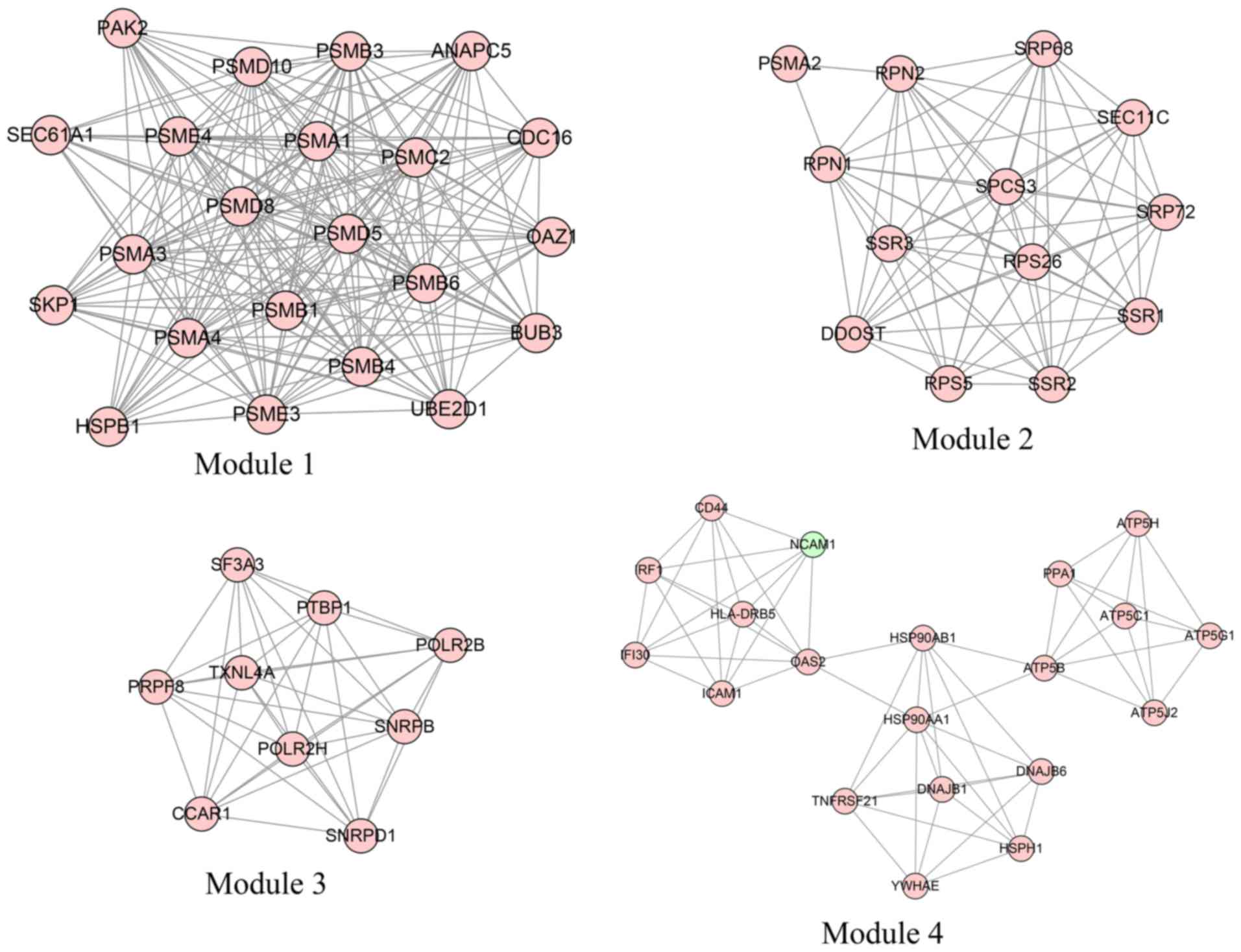
|
1
|
Schminke B, Vom Orde F, Gruber R,
Schliephake H, Bürgers R and Miosge N: The pathology of bone tissue
during peri-implantitis. J Dent Res. 94:354–361. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mombelli A: Microbiology and antimicrobial
therapy of peri-implantitis. Periodontol 2000. 28:177–189. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Berglundh T, Persson L and Klinge B: A
systematic review of the incidence of biological and technical
complications in implant dentistry reported in prospective
longitudinal studies of at least 5 years. J Clin Periodontol.
29:(Suppl 3). 197–212, 232–233. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Becker ST, Beck-Broichsitter BE, Graetz C,
Dörfer CE, Wiltfang J and Häsler R: Peri-implantitis versus
periodontitis: Functional differences indicated by transcriptome
profiling. Clin Implant Dent Relat Res. 16:401–411. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Theocharis AD, Seidel C, Borset M, Dobra
K, Baykov V, Labropoulou V, Kanakis I, Dalas E, Karamanos NK,
Sundan A and Hjerpe A: Serglycin constitutively secreted by myeloma
plasma cells is a potent inhibitor of bone mineralization in vitro.
J Biol Chem. 281:35116–35128. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rakic M, Struillou X, Petkovic-Curcin A,
Matic S, Canullo L, Sanz M and Vojvodic D: Estimation of bone loss
biomarkers as a diagnostic tool for peri-implantitis. J
Periodontol. 85:1566–1574. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bordin S, Flemmig TF and Verardi S: Role
of fibroblast populations in peri-implantitis. Int J Oral
Maxillofac Implants. 24:197–204. 2009.PubMed/NCBI
|
|
8
|
Irshad M, Scheres N, Anssari Moin D,
Crielaard W, Loos BG, Wismeijer D and Laine ML: Cytokine and matrix
metalloproteinase expression in fibroblasts from peri-implantitis
lesions in response to viable Porphyromonas gingivalis. J
Periodontal Res. 48:647–656. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Casado PL, Canullo L, de Almeida Filardy
A, Granjeiro JM, Barboza EP and Leite Duarte ME: Interleukins 1β
and 10 expressions in the periimplant crevicular fluid from
patients with untreated periimplant disease. Implant Dent.
22:143–150. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Park SY, Kim KH, Gwak EH, Rhee SH, Lee JC,
Shin SY, Koo KT, Lee YM and Seol YJ: Ex vivo bone morphogenetic
protein 2 gene delivery using periodontal ligament stem cells for
enhanced re-osseointegration in the regenerative treatment of
peri-implantitis. J Biomed Mater Res A. 103:38–47. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Carvalho BS and Irizarry RA: A framework
for oligonucleotide microarray preprocessing. Bioinformatics.
26:2363–2367. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene Ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Altermann E and Klaenhammer TR:
PathwayVoyager: pathway mapping using the Kyoto Encyclopedia of
Genes and Genomes (KEGG) database. BMC Genomics. 6:602005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
da Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2008. View Article : Google Scholar
|
|
17
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jeong H, Mason SP, Barabási AL and Oltvai
ZN: Lethality and centrality in protein networks. Nature.
411:41–42. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Estrada E and Rodríguez-Velázquez JA:
Subgraph centrality in complex networks. Phys Rev E Stat Nonlin
Soft Matter Phys. 71:0561032005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Goh KI, Oh E, Kahng B and Kim D:
Betweenness centrality correlation in social networks. Phys Rev E
Stat Nonlin Soft Matter Phys. 67:0171012003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Csardi G and Nepusz T: The igraph software
package for complex network research. Inter Journal Complex
Systems. 1695:1–9. 2006.
|
|
23
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bucci M, Roviezzo F, Cicala C, Sessa WC
and Cirino G: Geldanamycin, an inhibitor of heat shock protein 90
(Hsp90) mediated signal transduction has anti-inflammatory effects
and interacts with glucocorticoid receptor in vivo. Br J Pharmacol.
131:13–16. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wallin RP, Lundqvist A, Moré SH, von Bonin
A, Kiessling R and Ljunggren HG: Heat-shock proteins as activators
of the innate immune system. Trends Immunol. 23:130–135. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Qin S, Ni M, Wang X, Maurier-Mahé F,
Shurland DL and Rodrigues GA: Inhibition of RPE cell sterile
inflammatory responses and endotoxin-induced uveitis by a
cell-impermeable HSP90 inhibitor. Exp Eye Res. 93:889–897. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rice JW, Veal JM, Fadden RP, Barabasz AF,
Partridge JM, Barta TE, Dubois LG, Huang KH, Mabbett SR, Silinski
MA, et al: Small molecule inhibitors of Hsp90 potently affect
inflammatory disease pathways and exhibit activity in models of
rheumatoid arthritis. Arthritis Rheum. 58:3765–3775. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
De Nardo D, Masendycz P, Ho S, Cross M,
Fleetwood AJ, Reynolds EC, Hamilton JA and Scholz GM: A central
role for the Hsp90· Cdc37 molecular chaperone module in
interleukin-1 receptor-associated-kinase-dependent signaling by
Toll-like receptors. J Biol Chem. 280:9813–9822. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shimp SK III, Parson CD, Regna NL, Thomas
AN, Chafin CB, Reilly CM and Rylander Nichole M: HSP90 inhibition
by 17-DMAG reduces inflammation in J774 macrophages through
suppression of Akt and nuclear factor-κB pathways. Inflamm Res.
61:521–533. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hultin M, Gustafsson A, Hallström H,
Johansson LÅ, Ekfeldt A and Klinge B: Microbiological findings and
host response in patients with peri-implantitis. Clin Oral Implants
Res. 13:349–358. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tokoro Y, Yamamoto T and Hara K: IL-1 beta
mRNA as the predominant inflammatory cytokine transcript:
Correlation with inflammatory cell infiltration into human gingiva.
J Oral Pathol Med. 25:225–231. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Panagakos FS, Aboyoussef H, Dondero R and
Jandinski JJ: Detection and measurement of inflammatory cytokines
in implant crevicular fluid: A pilot study. Int J Oral Maxillofac
Implants. 11:794–799. 1996.PubMed/NCBI
|
|
33
|
Aboyoussef H, Carter C, Jandinski JJ and
Panagakos FS: Detection of prostaglandin E2 and matrix
metalloproteinases in implant crevicular fluid. Int J Oral
Maxillofac Implants. 13:689–696. 1998.PubMed/NCBI
|
|
34
|
Lee SS, Jang JH, Kim KS, Yoo YJ, Kim YS
and Lee SK: Failure of bone regeneration after demineralized bone
matrix allograft in human maxillary sinus floor elevation. Basic
Appl Pathol. 2:125–130. 2009. View Article : Google Scholar
|
|
35
|
Bullon P, Fioroni M, Goteri G, Rubini C
and Battino M: Immunohistochemical analysis of soft tissues in
implants with healthy and peri-implantitis condition, and
aggressive periodontitis. Clin Oral Implants Res. 15:553–559. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Borsani E, Salgarello S, Stacchiotti A,
Mensi M, Boninsegna R, Ricci F, Zanotti L, Rezzani R, Sapelli P,
Bianchi R and Rodella LF: Altered immunolocalization of heat-shock
proteins in human peri-implant gingiva. Acta Histochem.
109:221–227. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Borm ME, van Bodegraven AA, Mulder CJ,
Kraal G and Bouma G: A NFKB1 promoter polymorphism is involved in
susceptibility to ulcerative colitis. Int J Immunogenet.
32:401–405. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cochran DL: Inflammation and bone loss in
periodontal disease. J Periodontol. 79:(8 Suppl). 1569–1576. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Theoleyre S, Wittrant Y, Tat SK, Fortun Y,
Redini F and Heymann D: The molecular triad OPG/RANK/RANKL:
Involvement in the orchestration of pathophysiological bone
remodeling. Cytokine Growth Factor Rev. 15:457–475. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rakić M, Nikolić-Jakoba N, Struillout X,
Petković-Curcin A, Stamatović N, Matić S, Janković S, Aleksić Z,
Vasilić D, Leković V and Vojvodić D: Receptor activator of nuclear
factor kappa B (RANK) as a determinant of peri-implantitis.
Vojnosanit Pregl. 70:346–351. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Rakic M, Lekovic V, Nikolic-Jakoba N,
Vojvodic D, Petkovic-Curcin A and Sanz M: Bone loss biomarkers
associated with peri-implantitis. A cross-sectional study. Clin
Oral Implants Res. 24:1110–1116. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Karban AS, Okazaki T, Panhuysen CI,
Gallegos T, Potter JJ, Bailey-Wilson JE, Silverberg MS, Duerr RH,
Cho JH, Gregersen PK, et al: Functional annotation of a novel NFKB1
promoter polymorphism that increases risk for ulcerative colitis.
Hum Mol Genet. 13:35–45. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zou YF, Wang F, Feng XL, Tao JH, Zhu JM,
Pan FM and Su H: Association of NFKB1-94ins/delATTG promoter
polymorphism with susceptibility to autoimmune and inflammatory
diseases: A meta-analysis. Tissue antigens. 77:9–17. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li H, Gao L, Shen Z, Li CY, Li K, Li M, Lv
YJ, Li CX, Gao TW and Liu YF: Association study of NFKB1 and SUMO4
polymorphisms in Chinese patients with psoriasis vulgaris. Arch
Dermatol Res. 300:425–433. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kosoy R and Concannon P: Functional
variants in SUMO4, TAB2, and NFkappaB and the risk of type 1
diabetes. Genes Immun. 6:231–235. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Elliott PJ, Zollner TM and Boehncke WH:
Proteasome inhibition: A new anti-inflammatory strategy. J Mol Med
(Berl). 81:235–245. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Marfella R, D'Amico M, Di Filippo C, Baldi
A, Siniscalchi M, Sasso FC, Portoghese M, Carbonara O, Crescenzi B,
Sangiuolo P, et al: Increased activity of the ubiquitin-proteasome
system in patients with symptomatic carotid disease is associated
with enhanced inflammation and may destabilize the atherosclerotic
plaque: Effects of rosiglitazone treatment. J Am Coll Cardiol.
47:2444–2455. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Goldberg AL and Rock K: Not just research
tools-proteasome inhibitors offer therapeutic promise. Nat Med.
8:338–340. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lecker SH, Goldberg AL and Mitch WE:
Protein degradation by the ubiquitin-proteasome pathway in normal
and disease states. J Am Soc Nephrol. 17:1807–1819. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ciechanover A: The ubiquitin proteolytic
system and pathogenesis of human diseases: A novel platform for
mechanism-based drug targeting. Biochem Soc Trans. 31:474–481.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang J and Maldonado MA: The
ubiquitin-proteasome system and its role in inflammatory and
autoimmune diseases. Cell Mol Immunol. 3:255–261. 2006.PubMed/NCBI
|
|
52
|
Nothwang HG, Tamura T, Tanaka K and
Ichihara A: Sequence analyses and inter-species comparisons of
three novel human proteasomal subunits, HsN3, HsC7-I and HsC10-II,
confine potential proteolytic active-site residues. Biochim Biophys
Acta. 1219:361–368. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Gallastegui N and Groll M: The 26S
proteasome: Assembly and function of a destructive machine. Trends
Biochem Sci. 35:634–642. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Le Tallec B, Barrault MB, Courbeyrette R,
Guérois R, Marsolier-Kergoat MC and Peyroche A: 20S proteasome
assembly is orchestrated by two distinct pairs of chaperones in
yeast and in mammals. Mol Cell. 27:660–674. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Murata S, Yashiroda H and Tanaka K:
Molecular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol.
10:104–115. 2009. View Article : Google Scholar : PubMed/NCBI
|