Introduction
The majority of gingival tumors are highly
differentiated squamous cell carcinomas, such as oral squamous cell
carcinoma (OSCC) (1). These tumors
typically grow slowly and present as ulcers (2). In early stages, tumors are able to
infiltrate mandibular alveoli, leading to bone destruction, tooth
mobility and pain (1). Surgical
therapy is recommended for the treatment for OSCC; however, due to
the high degree of malignancy, advanced tumors typically infiltrate
surrounding tissues or migrate distantly (3). Migration, which is the leading cause of
mortality in patients with gingival carcinoma, occurs in >30% of
patients with negative lymph node diagnoses (4). Surgery and chemotherapy are less
effective under these circumstances (1). To reduce tumor recurrence and
metastasis, and improve survival, >70% of patients have
undergone radical treatment which is unnecessary and typically
leads to adverse complications, such as severe facial deformities
and swallowing, speech, neck and shoulder dysfunction (5–7).
Therefore, the identification of accurate diagnostic markers and
novel therapeutic targets of gingival carcinoma is necessary.
At present, the activation of oncogenes and
mutations of tumor suppressor genes are believed to be the primary
mechanisms underlying tumorigenesis (8,9).
Similarly, the interactions and coinciding actions of various
pathways may result in the tumorigenesis and progression of
gingival carcinoma; a process with which various mRNAs and micro
(mi)RNAs are associated (10,11).
Various recent studies have demonstrated that miRNA-214 is
associated with tumorigenesis and progression. For example, Zhang
et al (12) showed that
miRNA-214 may be used as a diagnostic predictor of gastric tumor
and reduce tumor cell proliferation and invasion. Wang et al
(13) demonstrated that miRNA-214
was able to modulate the proliferation and invasion of breast
cancer via tumour protein p53 (p53). Wang et al (13) reported that miRNA-214 had a
suppressive role in bladder cancer via targeting p53 and DNA-damage
regulated protein 1 (14). Wan et
al (15) demonstrated that
miRNA-214 was able to protect cardiomyocytes in follow-up treatment
of myocardial ischemia. Izawa et al (16) reported that miRNA-214 may prevent
liver fibrosis and effectively relieve liver cirrhosis induced by
thioacetamide. These findings suggest that miRNA-214 may be a novel
therapeutic agent for cancer prevention, diagnosis and
treatment.
Protein tyrosine phosphatase gene (PTEN) belongs to
the tumor suppressor gene family. Mutation of PTEN is associated
with the development of various human malignancies, and has key
roles in apoptosis, cell cycle and cell migration (17–23). In
various malignancies, such as prostate cancer (24), brain tumor (6), endometrial cancer (25), glioblastoma (26), melanoma (27), cervical cancer (28), colorectal gland cancer (29), breast cancer (30) and colon cancer (31) PTEN is downregulated. Furthermore,
PTEN is associated with gingival carcinoma and its expression is
decreased in gingival tumor tissues (10). Another recent study has indicated
that PTEN is associated with the occurrence, development and
metastasis of gingival carcinoma (32). However, the up-stream regulation
factor of PTEN in gingival carcinoma remains to be identified.
In the present study, reverse
transcription-quantitative polymerase chain reaction (RT-qPCR),
western blot analysis, gene bioinformatics prediction and ELISA
techniques were performed to evaluate the mRNA and protein
expression of PTEN and miRNA-214 levels in the tumor tissues, blood
and saliva of patients with gingival carcinoma. The association
between PTEN and miRNA-214 was subsequently analyzed.
Materials and methods
Research subjects and selection
criteria
Between January 2013 and March 2015, blood and
saliva samples were harvested from 56 gingival carcinoma patients
in the Affiliated Stomatology Hospital of Kunming Medical
University (Kunming, China). These patients were diagnosed with
gingival carcinoma and chose to receive surgical resection
treatment. As a control, blood and saliva samples were harvested
from 33 patients from the same hospital diagnosed with
non-neoplastic gum disease during the same period. Tumor tissues
and peritumoral tissues were subsequently harvested from the 56
gingival carcinoma patients. The gingival carcinoma patients
included 38 males and 18 females, aged 17–75 years old, with a
median age of 58.6 years. The control patients, included 20 males
and 13 females, aged 25–72 years old, with a median age of 59.5
years. All patients were at the early stages of gingival carcinoma
or non-neoplastic gum disease and were not treated with hormone
therapy, traditional medicine, radiotherapy or chemotherapy prior
to surgery. The gingival carcinoma patients included 30 patients
with gingival carcinoma in the upper gum and 26 patients with
gingival carcinoma in the lower gum, and consisted of one case of
mucoepidermoid carcinoma, two cases of adenoid cystic carcinoma,
two cases of adenocarcinoma, three cases of myoepithelial carcinoma
and 48 cases of squamous cell carcinoma. Prior written and informed
consent were obtained from all patients and the study was approved
by the ethics review board of Affiliated Stomatology Hospital of
Kunming Medical University.
Reagents and instruments
The miRcute miRNA isolation kit (dp501), miRcute
miRNA first strand cDNA synthesis kit (KR201), miRcute miRNA
fluorescent detection kit (FP 401), SuperReal Premix (SYBR Green)
(FP 204) and TIANScript II first strand cDNA synthesis kit (KP 107)
were purchased from Tiangen Biotech Co., Ltd. (Beijing, China). The
PCR-iQ5 system was purchased from Bio-Rad Laboratories, Inc.
(Hercules, CA, USA). The polyclonal rabbit anti-human anti-PTEN
antibody (ab32199), mouse monoclonal anti-β-actin antibody (ab6276)
and secondary goat anti-rabbit antibody (ab6721) were purchased
from Abcam (Cambridge, MA, USA). TRIzol reagent was purchased from
Shanghai Yeasen Biotechnology Co., Ltd. (10606ES60; Shanghai,
China). The bicinchoninic acid protein assay kit was purchased from
Real-Times (Beijing, China) Biotechnology Co., Ltd. (RTP7102;
Beijing, China). The miRNeasy serum/plasma kit for serum RNA
extraction was purchased from Guangzhou Jianlun Biological
Technology Co., Ltd. (JL 217184; Guangzhou, China). The PTEN ELISA
kit was purchased from Wuhan USCN Business Co., Ltd. (sE95822Hu;
Houston, TX, USA). Image Lab 3.0 software was purchased from
Beijing Bio-launching Technologies Co., Ltd. (Beijing, China).
Sample collection
Tumor tissues and peritumoral tissues were preserved
in liquid nitrogen for further analysis. Peripheral blood was
harvested in the morning following overnight fasting and stored at
−20°C following anticoagulation with EDTA.
Prior to saliva collection, research participants
were prohibited from food, water and smoking for 2 h to maintain
oral hygiene. To increase the quantity of collected saliva, the
head of a sterile cotton swab was moistened with 2% citric acid
solution, and placed on the first sidewall of tongue for ~5 sec.
Following saliva collection, the cotton swab would be applied to
the other sidewall. Saliva was collected repeatedly using a sterile
enzyme-free centrifuge tube until a total of 5 ml had been
harvested.
RT-qPCR
The RT-qPCR was carried out according to the
manufacturer's protocol. To detect PTEN, an E.Z.N.A. Total
DNA/RNA/Protein kit (Omega Bio-Tek, Inc., Norcross, GA, USA) and
TRIzol were used according to the manufacturer's protocol to
extract total RNA from samples, and RT was performed to produce
cDNA (TIANScript II cDNA kit; Tiangen Biotech Co., Ltd., Beijing,
China). The total reaction volume was 20 µl: 10 µl SuperReal Premix
(SYBR Green), 0.5 µl upstream primers, 0.5 µl downstream primers, 2
µl cDNA and 7 µl double distilled H2O. Briefly, a 25-µl
RT-qPCR system was performed for 40 cycles according to the
following conditions: An initial denaturation was performed at 95°C
for 3 min, followed by denaturation at 95°C for 12 sec, annealing
at 62°C for 40 sec, and extension at 72°C for 20 sec. The results
were calculated using 2−ΔΔCq method (33). The PTEN/β-actin ratio was calculated.
To detect levels of miRNA-214, RT-qPCR was performed under the same
conditions, using U6 as an internal control. The miRNA-214/U6 ratio
was calculated. Primers sequences are presented in Table I. Each experiment was repeated three
times.
 | Table I.Primers used in reverse
transcription-quantitative polymerase chain reaction. |
Table I.
Primers used in reverse
transcription-quantitative polymerase chain reaction.
| Primer | Primer
sequence |
|---|
| PTEN |
|
|
Forward |
5′-TTGAAGACCATAACCCACCACAG-3′ |
|
Reverse |
5′-CATTACACCAGTTCGTCCCTTTC-3′ |
| β-actin |
|
|
Forward |
5′-AAGATGACCCAGATCATGTTTGAGACC-3′ |
|
Reverse |
5′-GCCAGGTCCAGACGCAGGAT-3′ |
| U6 |
|
|
Forward |
5′-ATTGGAACGATACAGAGAAGATT-3′ |
|
Reverse |
5′-GGAACGCTTCACGAATTTG-3′ |
| miR-214 |
|
|
Forward |
5′-AGCATAATACAGCAGGCACAGAC-3′ |
|
Reverse |
5′-AAAGGTTGTTCTCCACTCTCTCAC-3′ |
Western blot analysis
Total proteins were extracted and separated by 10%
SDS-PAGE and transferred onto a nitrocellulose membrane. Following
blocking with non-fat milk, the membrane was incubated with primary
anti-PTEN (1:500) and anti-β-actin (1:5,000) antibodies at 4°C
overnight. The membrane was subsequently washed with TBST 5 times
for 5 min. Following washing, the membrane was incubated with goat
anti-rabbit secondary antibody (1:3,000) at room temperature for 1
h. The membrane was subsequently developed using enhanced
chemiluminescence plus an enhanced chemiluminescence reagent
(BeyonECL Plus; Beyotime Institute of Biotechnology, Haimen,
China). Image lab 3.0 software was used to analyze results. β-actin
was used as an internal control. The relative expression level of
PTEN was calculated based on the expression of β-actin.
ELISA
Blood samples were centrifuged at 1,000 × g for 10
min at 4°C to separate serum and red blood cells. ELISA was
performed according to the manufacturer's protocol of the PTEN
ELISA kit. Briefly, blank wells and wells for standard sample,
serum and saliva were prepared. In the wells for standard sample,
50-µl standard samples with different concentrations (180, 120, 60,
30, 15, 7.5 and 3.75 pg/ml) were added. In the wells for serum and
saliva, 10 µl serum or saliva and 40 µl dilution solution were
added. Nothing was added to blank wells. Besides blank wells, 100
µl horseradish peroxidase-labeled detection antibody (1:1,000)
mixed with dilution buffer was added to each well. The plate was
sealed and incubated for 1 h at room temperature. Following five
repeated washes with the provided washing buffer, the substrate (50
µl substrate A and 50 µl substrate B) was added to each well and
incubated for 15 min at 37°C. The reaction was terminated by adding
50 µl stopping solution to each well. Optical density was measured
at a wavelength of 450 nm.
Bioinformatics prediction
Bioinformatics prediction is currently the
recommended technique for the study of miRNA function (34). In order to evaluate the regulation
mechanism of PTEN, miRanda (www.microma.org/rnicroma/home.do), TargetScan
(www.targetscan.org), PiTa (www.genie.weizmann.ac.il/pubs/mir07),
RNAhybrid (www.hibiserv.techfak.uni-bielefeld.de/rnahybrid) and
Pictar (www.pictar.mdc-berlin.de) programs were used to
predict the possible regulatory gene of PTEN.
Statistical analysis
Data were processed using SPSS, version 18.0 (SPSS,
Inc., Chicago, IL, USA). Data are presented as the mean ± standard
deviation. The normality of data was tested using the
Kolmogorov-Smirnov test. Comparison among multiple sets of
measurement data was performed using one-way analysis of variance.
When there was homogeneity of variance, least significant
difference and Student-Newman-Keuls tests were performed.
Otherwise, Tamhane's T2 or Dunnett's T3 method was used. P<0.05
was considered to indicate a statistically significant
difference.
Results
Level of PTEN mRNA in samples
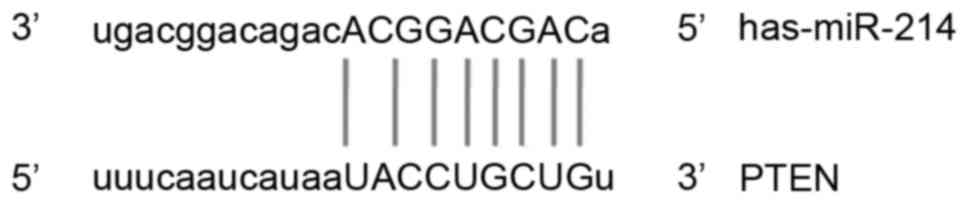
miRNA-214 was identified as one of the genes that
may regulate PTEN (Fig. 1). RT-qPCR
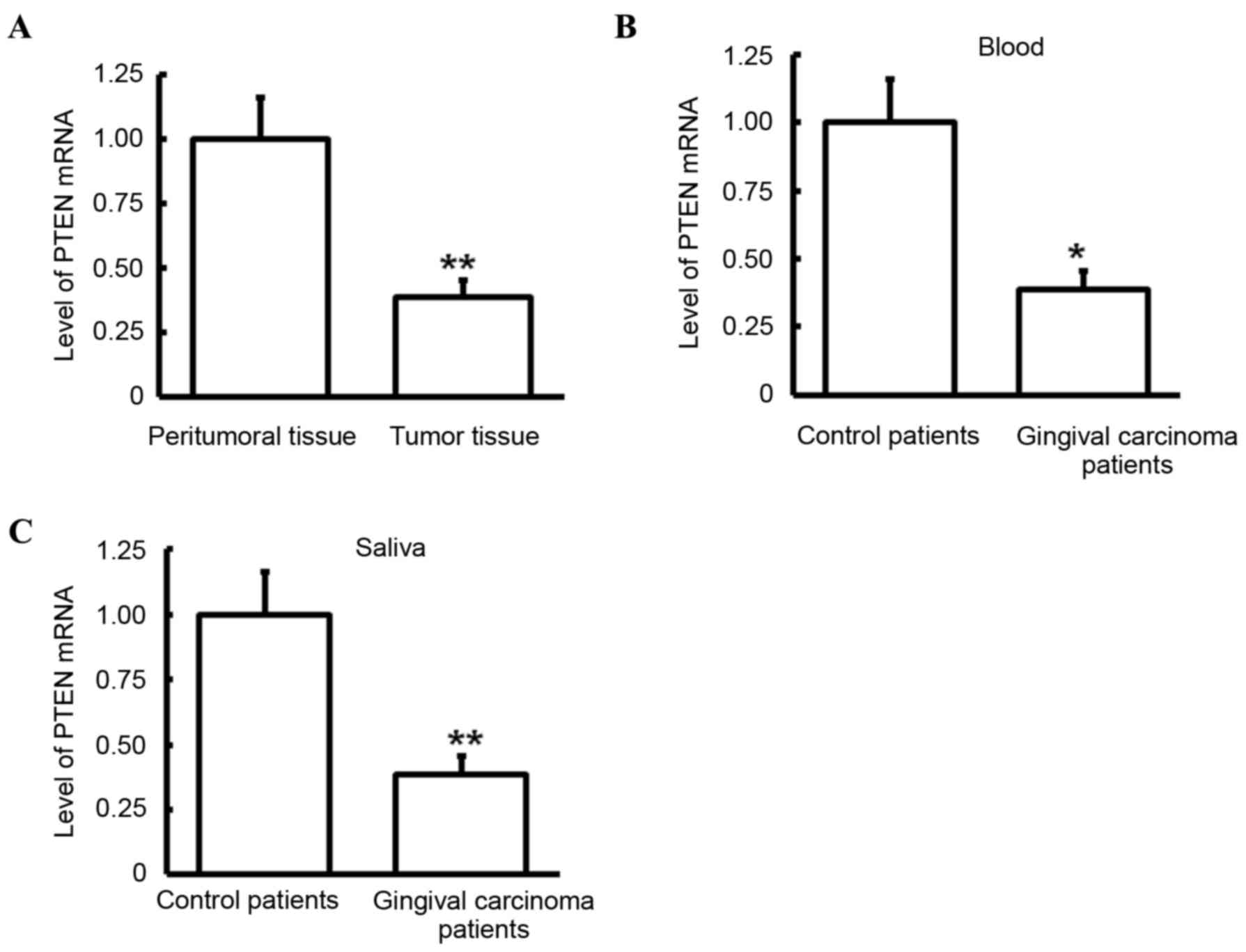
was performed to detect levels of PTEN mRNA in tumor tissues,
peritumoral tissues, blood and saliva. Compared with that in
peritumoral tissues, PTEN mRNA levels in tumor tissues of patients
with gingival carcinoma was significantly lower (P<0.05;
Fig. 2A). Similarly, compared with
those observed in control patients, PTEN mRNA levels in the blood
and saliva of gingival carcinoma patients were significantly
decreased (both P<0.05; Fig. 2B and
C). These results indicate that PTEN may have a role in
gingival carcinoma.
Protein expression level of PTEN in
tumor and peritumoral tissues
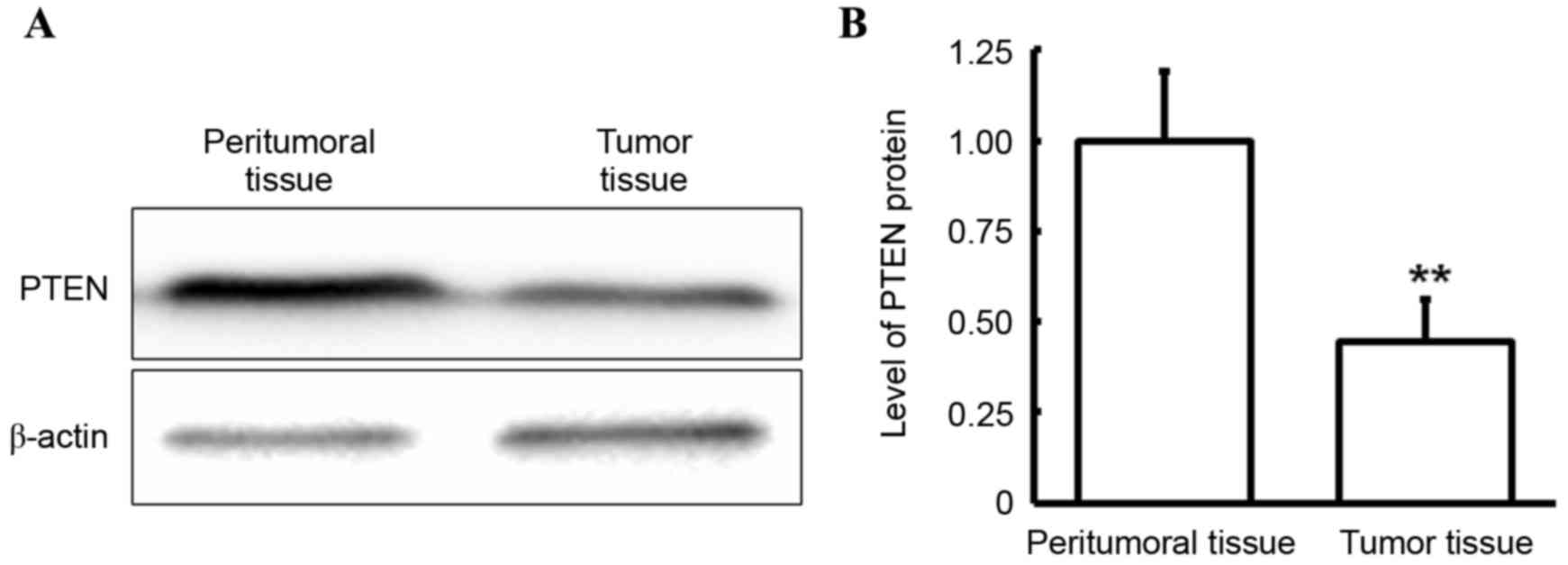
Western blotting was performed to determine the
expression level of PTEN in tumor and peritumoral tissues. As
demonstrated in Fig. 3, in gingival
carcinoma patients, the PTEN protein level in the tumor tissues was
significantly decreased compared with that in peritumoral tissues
(P<0.05). This finding was consistent with that observed in PTEN
mRNA levels, which suggests that PTEN is downregulated at the
transcriptional and protein expression level and that this
downregulation may have a role in gingival carcinoma.
Protein expression level of PTEN in
blood and saliva
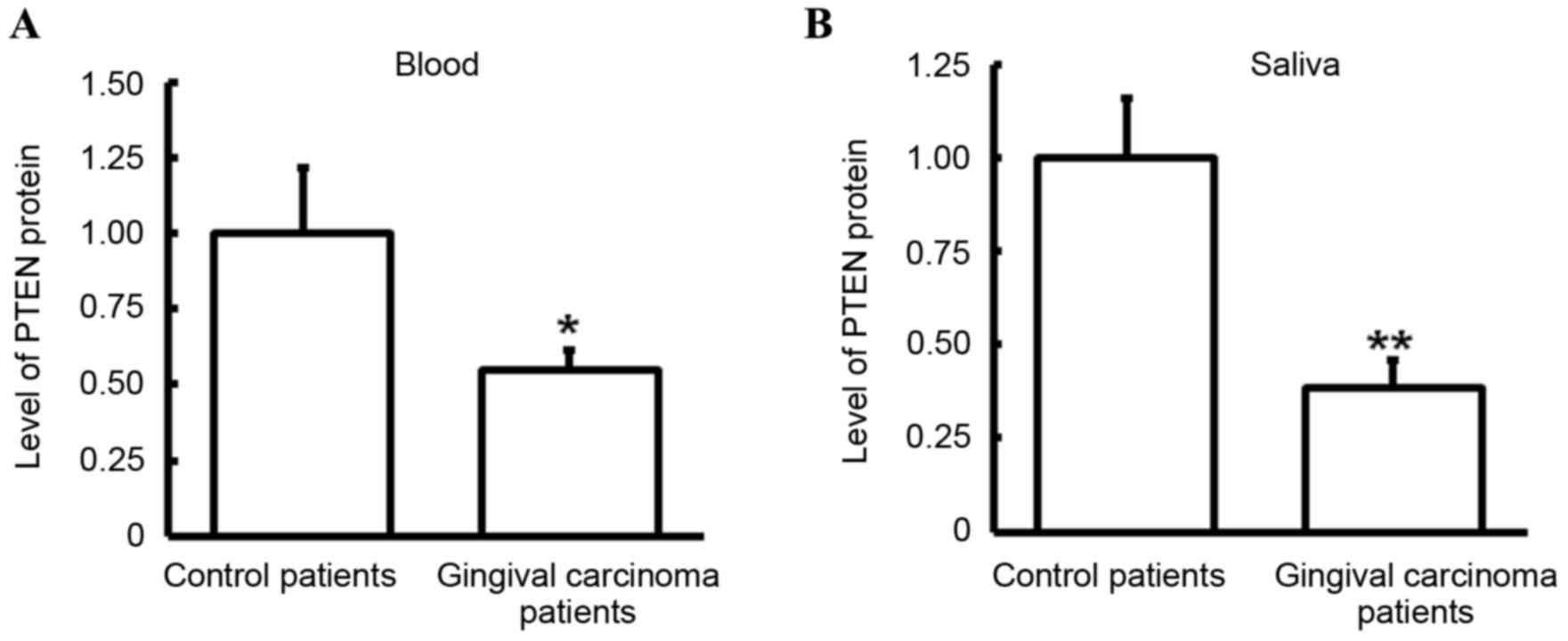
The expression level of PTEN protein in blood and
saliva was detected via ELISA. Compared with control patients, the
expression level of PTEN was significantly decreased in the blood
(P<0.05; Fig. 4A) and saliva
(P<0.01; Fig. 4B) of gingival
carcinoma patients. These results were consistent with those
observed in PTEN mRNA. As blood is a typical channel of gingival
carcinoma metastasis (35), the
decrease of PTEN protein in blood may be associated with gingival
carcinoma metastasis. Furthermore, the downregulation of PTEN
protein level in saliva may be a potential marker of gingival
carcinoma.
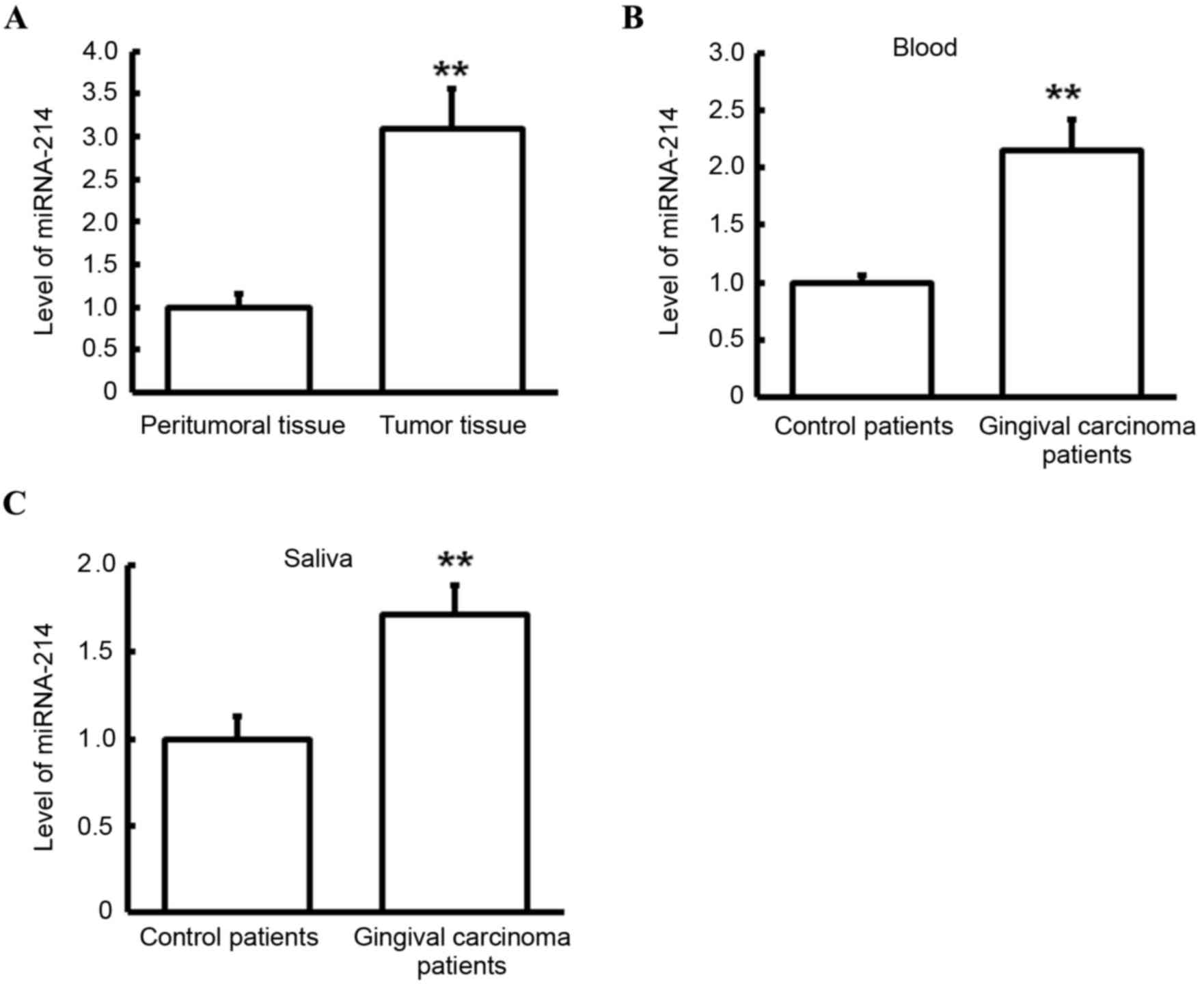
Expression level of miRNA-214 in tumor
tissues, peritumoral tissues, blood and saliva
RT-qPCR was performed to detect expression levels of
miRNA-214 in tumor tissues, peritumoral tissues, blood and saliva.
As presented in Fig. 5A, miRNA-214
level in the tumor tissues of gingival carcinoma patients was
significantly higher than that in peritumoral tissues of gingival
carcinoma patients (P<0.01). Similarly, miRNA-214 levels in the
blood (Fig. 5B) and saliva (Fig. 5C) of gingival carcinoma patients were
significantly increased in comparison with those in control
patients (both P<0.01). In combination with the finding that
miRNA-214 may be a regulatory gene PTEN, these results suggest that
miRNA-214 may have a regulatory effect on gingival carcinoma, and
it may regulate the transcriptional level of PTEN and ultimately
affect the protein level of PTEN.
Discussion
In the present study, the mRNA and protein levels of
PTEN in tumor tissues, peritumoral tissues, blood and saliva of
patients with gingival carcinoma were evaluated. The expression of
miRNA-214, a predicted upstream gene of PTEN, was detected
simultaneously, and the potential regulation of PTEN by miRNA-214
in gingival carcinoma was identified.
Among oral and maxillofacial malignancies, >80%
of tumors are OSCC, and is the sixth most prevalent carcinoma
worldwide (36,37). OSCC typically occurs in the tongue,
gingiva, and cheek (38). It is
clinically characterized by a high degree of malignancy, trend
toward regional lymph node metastasis and poor prognosis (2). The majority of advanced OSCCs exhibit
invasive growth (39), and it is
difficult to surgically remove tumors that have infiltrated
surrounding tissues and migrated distantly, and traditional
chemotherapy has been demonstrated to have a low efficacy in this
scenario (40). The improvement of
early diagnosis and the implementation of individualized treatment
has become necessary to ameliorate the treatment efficacy and
prognosis of OSCC (41). Therefore,
there is a need to clarify the pathogenesis of OSCC with
surrounding tissue infiltration and distant metastasis and to
identify novel gene targets for therapy.
As a tumor suppressor gene, PTEN was discovered in
1997 and is located on chromosome 10 (10q 23.3) (25,42,43). The
downregulation or deletion of PTEN has a role in apoptosis, cell
cycle and cell migration, and is associated with the development of
various human malignancies (44).
Previous research on the effect of PTEN on
tumorigenesis has predominantly focused on endometrial cancer,
glioma, prostate cancer, breast cancer and melanoma (25–31).
However, to the best of our knowledge, the role of PTEN in OSCC has
been rarely reported. Although the mechanism remains to be
elucidated, an abnormal decrease of PTEN has been demonstrated in
the majority of types of tumor (6,25–31). In
the present study, a significant reduction in PTEN expression was
observed in the tumor tissues, blood and saliva of patients with
gingival carcinoma compared with control patients. As tumors may
migrate through blood as well as via regional invasion, the
reduction of PTEN in the blood of patients with gingival carcinoma
suggests that PTEN may have a role in tumor metastasis. It is
possible that a reduction in PTEN reduces apoptosis, whereas
excessive proliferation of cells is associated with increased cell
infiltration and mitigation, leading to tumor metastasis.
Additionally, changes in PTEN levels in the blood and saliva of
patients may be a potential biomarker of gingival carcinoma,
particularly in the early stage of tumorigenesis.
Bioinformatics were applied to predict possible
upstream regulatory genes of PTEN. It has recently been
demonstrated that a class of small, endogenous and non-coding
nucleic acid miRNAs are able to prevent transcription of PTEN mRNA
(45). According to the current
literature, miRNAs have a high level of stability in conventional,
formalin-fixed, paraffin-embedded clinical tissue specimens, which
suggests that they may have potential as diagnostic molecular
markers; however, mRNAs tend to be more susceptible to
environmental impact and are more unstable than miRNAs (46–48).
Furthermore, it has previously been suggested that miRNAs serve
important roles in tumorigenesis via their regulation of protein
coding genes (49,50). The cleavage of PTEN mRNA may lead to
downregulation of PTEN protein. As predicted, miRNA-214 was shown
to be closely associated with PTEN; therefore it may be an upstream
regulatory gene of PTEN. Furthermore, it has previously been
demonstrated that miRNA-214 is able to regulate PTEN in the
processes of tumor invasion and metastasis in human gastric cancer,
breast cancer and cervical cancer (51–53).
In the present study, a significant increase of
miRNA-214 levels was observed in the tumor tissues, blood and
saliva of patients with gingival carcinoma. These results suggest
that PTEN in gingival carcinoma is directly downregulated by
miRNA-214, as the upregulation of miRNA-214 may promote cleavage of
PTEN, thus inhibiting the proapoptotic effect of PTEN protein, the
formation of gingival cysts and ultimately gingival
tumorigenesis.
In conclusion, the balance between miRNA-214 and
PTEN levels may regulate the tumorigenesis and progression of
gingival carcinoma. Additionally, miRNA-214 is more stable than
PTEN in blood and saliva, and may be a more suitable marker for the
early diagnosis of gingival carcinoma.
Acknowledgements
The authors of the present study want to thank Dr
Shenglan Wang from the Oncology Department of the First People's
Hospital of Yunnan Province (Kunming, China).
References
|
1
|
Qiu W: Oral and maxillofacial surgery
(6th). People's Medical Publishing House. Beijing: 274–275.
2008.
|
|
2
|
Girdler NM: Oral ulceration: Benign or
malignant? A diagnostic dilemma. Br Dent J. 168:3861990. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferlito A, Shaha AR and Rinaldo A: The
incidence of lymph node micrometastases in patients pathologically
staged No in cancer of oral cavity and oropharynx. Oral Oncol.
38:3–5. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kademani D: Oral cancer. Mayo Clin Proc.
82:878–887. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Becker MT, Shores CG, Yu KK and Yarbrough
WG: Molecular assay to detect metastatic head and neck squamous
cell carcinoma. Arch Otolaryngol Head Neck Surg. 130:21–27. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Németh Z, Velich N, Bogdan S, Ujpál M,
Szabó G and Suba ZS: The prognostic role of clinical, morphological
and molecular markers in oral squamous cell tumors. Neoplasma.
52:95–102. 2005.PubMed/NCBI
|
|
7
|
He H, Huang J, Ping F, Chen G, Zhang S and
Dong Y: Anatomical and clinical study of lingual arterial
chemoembolization for tongue carcinoma. Oral Surg Oral Med Oral
Pathol Oral Radiol Endod. 103:e1–e5. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mano H: Discovery and clinical application
of mutations in the cancer genome. Nihon Rinsho. 73:1251–1255.
2015.(In Japanese). PubMed/NCBI
|
|
9
|
Ohtsuka M, Ling H, Doki Y, Mori M and
Calin GA: MicroRNA processing and human cancer. J Clin Med.
4:1651–1667. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li JD, Liang RY, Zhao YP and Xu YL:
Correlation analysis of VEGF and PTEN expression in gingival
carcinoma. Shanghai Kou Qiang Yi Xue. 23:619–623. 2014.(In
Chinese). PubMed/NCBI
|
|
11
|
Boldrup L, Coates PJ, Wahlgren M, Laurell
G and Nylander K: Subsite-based alterations in miR-21, miR-125b,
and miR-203 in squamous cell carcinoma of the oral cavity and
correlation to important target proteins. J Carcinog. 11:182012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang KC, Xi HQ, Cui JX, Shen WS, Li JY,
Wei B and Chen L: Hemolysis-free plasma miR-214 as novel biomarker
of gastric cancer and is correlated with distant metastasis. Am J
Cancer Res. 5:821–829. 2015.PubMed/NCBI
|
|
13
|
Wang F, Lv P, Liu X, Zhu M and Qiu X:
microRNA-214 enhances the invasion ability of breast cancer cells
by targeting p53. Int J Mol Med. 35:1395–1402. 2015.PubMed/NCBI
|
|
14
|
Wang J, Zhang X, Wang L, Yang Y, Dong Z,
Wang H, Du L and Wang C: MicroRNA-214 suppresses oncogenesis and
exerts impact on prognosis by targeting PDRG1 in bladder cancer.
PLoS One. 10:e01180862015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wan DY, Zhang Z and Yang HH:
Cardioprotective effect of miR-214 in myocardial ischemic
postconditioning by down-regulation of hypoxia inducible factor 1,
alpha subunit inhibitor. Cell Mol Biol (Noisy-le-grand). 61:1–6.
2015.PubMed/NCBI
|
|
16
|
Izawa T, Horiuchi T, Atarashi M, Kuwamura
M and Yamate J: Anti-fibrotic role of miR-214 in
Thioacetamide-induced liver cirrhosis in rats. Toxicol Pathol.
43:844–851. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Squarize CH, Castilho RM and Santos Pinto
D Jr: Immunohistochemical evidence of PTEN in oral squamous cell
carcinoma and its correlation with the histological malignancy
grading system. J Oral Pathol Med. 31:379–384. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li DM and Sun H: PTEN/MMAC1/TEP1
suppresses the tumorigenicity and induces G1 cell cycle arrest in
human glioblastoma cells. Proc Natl Acad Sci USA. 95:15406–15411.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Di Cristofano A and Pandolfi PP: The
multiple roles of PTEN in tumor suppression. Cell. 100:387–390.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Stiles B, Gilman V, Khanzenzon N, Lesche
R, Li A, Qiao R, Liu X and Wu H: Essential role of AKT-1/protein
kinase B in PTEN-controlled tumorigenesis. Mol Cell Biol.
22:3842–3851. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Davies MA, Koul D, Dhesi H, Berman R,
McDonnell TJ, McConkey D, Yung WK and Steck PA: Regulation of
Akt/PKB activity, cellular growth, and apoptosis in prostate
carcinoma cells by MMAC/PTEN1. Cancer Res. 59:2551–2556.
1999.PubMed/NCBI
|
|
22
|
Tamura M, Gu J, Takino T and Yamada KM:
Tumor suppressor PTEN inhibition of cell invasion, migration, and
growth: Differential involvement of focal adhesion kinase and
p130Cas. Cancer Res. 59:442–449. 1999.PubMed/NCBI
|
|
23
|
McCubrey JA, Steelman LS, Abrams SL, Lee
JT, Chang F, Bertrand FE, Navolanic PM, Terrian DM, Franklin RA,
D'Assoro AB, et al: Roles of the RAF/MEK/ERK and PI3K/PTEN/AKT
pathways in malignant transformation and drug resistance. Adv
Enzyme Regul. 46:249–279. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Punnoose EA, Ferraldeschi R,
Szafer-Glusman E, Tucker EK, Mohan S, Flohr P, Riisnaes R, Miranda
S, Figueiredo I, Rodrigues DN, et al: PTEN loss in circulating
tumor cells correlates with PTEN loss in fresh tumor tissue from
castration-resistant prostate cancer patients. Br J Cancer.
113:1225–1233. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li J, Yen C, Liaw D, Podsypanina K, Bose
S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, et al:
PTEN, a putative protein tyrosine phosphatase gene mutated in human
brain, breast, and prostate cancer. Science. 275:1943–1947. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mutter GL, Lin MC, Fitzgerald JT, Kum JB,
Baak JP, Lees JA, Weng LP and Eng C: Altered PTEN expression as a
diagnostic marker for the earliest endometrial precancers. J Natl
Cancer Inst. 92:924–930. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Duerr EM, Rollbrocker B, Hayashi Y, Peters
N, Meyer-Puttlitz B, Louis DN, Schramm J, Wiestler OD, Parsons R,
Eng C and von Deimling A: PTEN mutations in gliomas and
glioneuronal tumors. Oncogene. 16:2259–2264. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tsao H, Zhang X, Benoit E and Haluska FG:
Identification of PTEN/MMAC1 alterations in uncultured melanomas
and melanoma cell lines. Oncogene. 16:3397–3402. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rizvi MM, Alam MS, Ali A, Mehdi SJ, Batra
S and Mandal AK: Aberrant promoter methylation and inactivation of
PTEN gene in cervical carcinoma from Indian population. J Cancer
Res Clin Oncol. 137:1255–1262. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang XF, Xin Y and Mao LL:
Clinicopathological significance of PTEN and caspase-3 expressions
in breast cancer. Chin Med Sci J. 23:95–102. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bowen KA, Doan HQ, Zhou BP, Wang Q, Zhou
Y, Rychahou PG and Evers BM: PTEN loss induces
epithelial-mesenchymal transition in human colon cancer cells.
Anticancer Res. 29:4439–4449. 2009.PubMed/NCBI
|
|
32
|
Ren W, Qiang C, Gao L, Li SM, Zhang LM,
Wang XL, Dong JW, Chen C, Liu CY and Zhi KQ: Circulating
microRNA-21 (MIR-21) and phosphatase and tensin homolog (PTEN) are
promising novel biomarkers for detection of oral squamous cell
carcinoma. Biomarkers. 19:590–596. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-tie quantitative PCR and
the 2(−Delta Delta C (T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
León LE and Calligaris SD: Visualization
and analysis of MiRNA-targets interactions networks. Methods Mol
Biol. 1509:209–220. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Falconieri G, Luna MA, Pizzolitto S,
DeMaglio G, Angione V and Rocco M: Eosinophil-rich squamous
carcinoma of the oral cavity: A study of 13 cases and delineation
of a possible new microscopic entity. Ann Diagn Pathol. 12:322–327.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kalnins IK, Leonard AG, Sako K, Razack MS
and Shedd DP: Correlation between prognosis and degree of lymph
node involvement in carcinoma of the oral cavity. Am J Surg.
134:450–454. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Estimating the world cancer burden: Globocan 2000. Int J Cancer.
94:153–156. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rautava J, Luukkaa M, Heikinheimo K, Alin
J, Grenman R and Happonen RP: Squamous cell carcinomas arising from
different types of oral epithelia differ in their tumor and patient
characteristics and survival. Oral Oncol. 43:911–919. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yoshida R, Nakayama H, Nagata M, Hirosue
A, Tanaka T, Kawahara K, Nakagawa Y, Matsuoka Y, Sakata J, Arita H,
et al: Overexpression of nucleostemin contributes to an advanced
malignant phenotype and a poor prognosis in oral squamous cell
carcinoma. Br J Cancer. 111:2308–2315. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kirita T, Yamanaka Y, Imai Y, Yamakawa N,
Aoki K, Nakagawa Y, Yagyuu T and Hasegawa M: Preoperative
concurrent chemoradiotherapy for stages II–IV oral squamous cell
carcinoma: A retrospective analysis and the future possibility of
this treatment strategy. Int J Oral Maxillofac Surg. 41:421–428.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cheng YS, Rees T and Wright J: A review of
research on salivary biomarkers for oral cancer detection. Clin
Transl Med. 3:32014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Steck PA, Pershouse MA, Jasser SA, Yung
WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T,
et al: Identification of a candidate tumour suppressor gene, MMAC1,
at chromosome 10q23.3 that is mutated in multiple advanced cancers.
Nat Genet. 15:356–362. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li DM and Sun H: PTEN, encoded by a
candidate tumor suppressor locus, is a novel protein tyrosine
phosphatase regulated by transforming growth factor beta. Cancer
Res. 57:2124–2129. 1997.PubMed/NCBI
|
|
44
|
Wang YF and Tang WP: Progress of PTEN gene
and oral and maxillofacial tumors. Shi Yong Lin Chuang Yi Xue.
11:125–127. 2010.
|
|
45
|
Liu K, Liu S, Zhang W, Jia B, Tan L, Jin Z
and Liu Y: miR-494 promotes cell proliferation, migration and
invasion, and increased sorafenib resistance in hepatocellular
carcinoma by targeting PTEN. Oncol Rep. 34:1003–1010.
2015.PubMed/NCBI
|
|
46
|
Howe K: Extraction of miRNAs from
formalin-fixed paraffin-embedded (FFPE) tissues. Methods Mol Biol.
1509:17–24. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Deng H, Liu Q, Wang X, Huang R, Liu H, Lin
Q, Zhou X and Xing D: Quantum dots-labeled strip biosensor for
rapid and sensitive detection of microRNA based on target-recycled
nonenzymatic amplification strategy. Biosens Bioelectron.
87:931–940. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Rissland OS: The organization and
regulation of mRNA-protein complexes. Wiley Interdiscip Rev RNA.
8:2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chen K and Rajewsky N: The evolution of
gene regulation by transcription factors and microRNAs. Nat Rev
Genet. 8:93–103. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yang TS, Yang XH, Wang XD, Wang YL, Zhou B
and Song ZS: MiR-214 regulate gastric cancer cell proliferation,
migration and invasion by targeting PTEN. Cancer Cell Int.
13:682013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Schwarzenbach H, Milde-Langosch K,
Steinbach B, Müller V and Pantel K: Diagnostic potential of
PTEN-targeting miR-214 in the blood of breast cancer patients.
Breast Cancer Res Treat. 134:933–941. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yang H, Kong W, He L, Zhao JJ, O'Donnell
JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV and Cheng JQ:
MicroRNA expression profiling in human ovarian cancer: miR-214
induces cell survival and cisplatin resistance by targeting PTEN.
Cancer Res. 68:425–433. 2008. View Article : Google Scholar : PubMed/NCBI
|