Introduction
Esophageal cancer (EC) is a common malignant tumor
type originating from the esophageal mucosa (1). China has a high incidence of EC with a
rate of 23.38/100,000 for males and 14.03/100,000 for females
(2). EC is the fifth most common
type of cancer and esophageal squamous cell carcinoma (ESCC)
accounts for 90% of all EC cases (3,4).
According to data from the National Central Cancer Registry of
China, 477,900 new EC cases and 375,000 cases of mortality occurred
in 2015 (5). The majority of EC
patients are diagnosed at an advanced stage when the 5-year
survival rate is ~20%, though early detected EC can be treated with
endoscopic resection, and has a 5-year survival rate of up to 95%
(6). Therefore, early detection and
prompt intervention is an effective strategy to improve the
prognosis of patients with EC and reduce the disease burden.
Compared with endoscopic biopsy, detection of serum
tumor markers is a simple and minimally invasive method for early
diagnosis (7). Circulating microRNAs
(miRs) are stable in the blood and exhibit a high degree of tissue
and tumor specificity, which makes them suitable as potential serum
tumor markers (8). MiRs are also key
regulators of gene transcription, and are frequently involved in
the occurrence of cancers (9). In
particular, miR-148a has been reported to be a tumor suppressor in
multiple cancer types, including gastrointestinal (10), breast (11), ovarian (12), colorectal (13) and prostate cancer (14). Hummel et al (15) reported that miR-148a was
downregulated in patients with recurrent EC. They also demonstrated
that in EC cell lines, upregulation of miR-148a improved
sensitivity to chemotherapy (16).
However, whether miR-148a is downregulated in primary ESCC remains
unclear.
Previous studies have demonstrated that the human
leukocyte antigen-G (HLA-G) gene is one of the target genes
regulated by miR-148a. Tan et al (17) identified that miR-148a could target
HLA-G, and that differential expression of HLA-G interacted with
the mother's asthma status to determine a child's risk of asthma.
Manaster et al (18)
confirmed by luciferase assay that miR-148a could downregulate
HLA-G expression. A C/G polymorphism exists at position +3142 of
the HLA-G gene, which is in the seed sequence of miR-148a; Manaster
et al (18) identified that
this polymorphism had no influence on HLA-G expression.
HLA-G is a member of the non-classical major
histocompatibility complex class I antigens and serves a key
function in maternal-fetal tolerance during pregnancy (19). Aberrant expression of HLA-G has been
observed in multiple malignant cell types, which may be associated
with the strategy of evading host immunosurveillance (20). Epithelial cells of the normal
esophagus lack expression of HLA-G, while HLA-G is expressed to a
high level in primary ESCC tissues and is associated with prognosis
(21). Furthermore, the soluble
isoforms of HLA-G in the plasma may be useful molecules in the
differential diagnosis of patients with ESCC against healthy
controls (22).
Based on previous reports, it was hypothesized that
miR-148a may be differentially expressed in ESCC tissues, and this
was investigated in the present study. The regulatory role of
miR-148a on HLA-G expression and cell proliferation in ESCC cells
was also investigated.
Materials and methods
Tissue samples
A total of 20 pairs of ESCC tumor tissues and normal
adjacent tissues were used in the present study. Tissues were
isolated from newly diagnosed ESCC patients who had not received
chemotherapy or radiotherapy. A total of 20 patients (8 male, 12
female; mean age, 57.6 years) were recruited from the Department of
Cardiothoracic Surgery in the Third Affiliated Hospital of Soochow
University (Changzhou, China) from March 2012 to June 2013. The
diagnosis was confirmed based on histological examination according
to the American Joint Committee on Cancer 2010 TNM staging system
for EC (23,24). The samples were collected with
written informed consent from the patients and stored in liquid
nitrogen until assays were performed. The study protocol was
approved by the Ethics Board of The Third Affiliated Hospital of
Soochow University.
Cell line and culture
The human ESCC cell line EC9706 was purchased from
American Type Culture Collection (Manassas, VA, USA). The cells
were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplemented with 10% (V/V) fetal bovine
serum (Gibco; Thermo Fisher Scientific, Inc.) in a humidified hood
with 5% CO2 at 37°C.
Transfection of miR-148a mimic
The cells were seeded into 96-well plates to 60%
confluency in RMPI-1640 media supplemented with 10% FBS
(antibiotic-free medium) for 24 h at 37°C prior to transfection.
Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) was used to transfect 1,000–10,000 cells/well with 50 nM
miR-148a mimic. The experiment was performed in triplicate for each
group. Non-homologous RNA duplex was used as a negative control
(NC) and empty vector was used as a blank control (BC). The
nucleotides were chemically synthesized by Shanghai GenePharma Co.,
Ltd. (Shanghai, China). The nucleotide sequences used were as
follows: For miR-148a mimic, forward, 5′-UCAGUGCAUGACAGAACUUGG-3′
and reverse, 5′-AAGTTCUGUCAUGCACUGAUU-3′; and for NC, forward,
5′-UUCUCCGAACGUGUCACGUTT−3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′. The cells were harvested for reverse
transcription-quantitative polymerase chain reaction (RT-qPCR),
western blot analysis and apoptosis assays 72 h after transfection
under 37°C.
RNA extraction and RT-qPCR
Total RNA was extracted from tissues and cells using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. Synthesis of cDNA was
performed using a PrimeScript II First Strand cDNA Synthesis kit
(Takara Biotechnology Co., Ltd., Dalian, China). Target gene DNA
was amplified with specific primers and a Fast SYBR Green Master
mix (Applied Biosystems; Thermo Fisher Scientific, Inc.). Mature
miR-148a, U6 control and HLA-G were detected using the primers
reported previously (14). The
primers for the internal control GAPDH were: Forward,
5′-AGCGCGTGCCTTATACCAAG-3′ and reverse, 5′-GCCGCTCAGAGAGATTCGT-3′.
All amplifications were conducted with a 7500 Real-Time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling conditions were as follows: 94°C for 1 min, followed
by 45 cycles of 94°C for 20 sec, 58°C for 30 sec and 72°C for 30
sec. Relative mRNA expression was calculated using the
ΔΔCq method as described previously (25).
Western blot analysis
Cells were lysed with RIPA lysis buffer (R0020,
Beijing Solarbio Science and Technology Co., Ltd). Protein was
isolated by centrifuging the sample at 20,000 × g at 4°C for 15
min. Protein concentration was determined using a BCA Protein assay
kit (Beyotime Institute of Biotechnology, Haimen, China). Each lane
was filled with 1 ng protein and 10 µl 6× SDS loading dye. Proteins
were separated on 12.5% SDS-PAGE gels and transferred to
polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA,
USA. The membrane was blocked with 5% skimmed milk at room
temperature for 1 h, then probed with monoclonal mouse anti-HLA-G
(sc-21799; Santa Cruz Biotechnology, Inc., Dallas, TX, USA;
1:1,000), or monoclonal mouse anti-GAPDH (sc-32233; Santa Cruz
Biotechnology, Inc.; 1:1,000) for 1 h at room temperature. The
secondary antibody used was horseradish peroxidase-conjugated
rabbit anti-mouse Immunoglobulin G antibody (6120-05;
SouthernBiotech, Birmingham, AL, USA; 1:2,000) at room temperature
for 45 min. Signals were detected using an enhanced
chemiluminescence kit (BH4004, Shanghai Ke Min Biotechnology Co.,
Ltd., Shanghai, China).
Apoptosis assay
Cells (~5×105) were harvested and washed
with phosphate-buffered saline. Cells were stained with Annexin
V-fluorescein isothiocyanate (SouthernBiotech) for 5 min at room
temperature. A final concentration of 10 µg/ml propidium iodide was
added for 30 min at 4°C, away from light, then flow cytometry was
performed. Flow cytometry results were analyzed using CytoSpec
version 7 (Purdue University Cytometry Laboratories, West
Lafayette, IN, USA).
Bioinformatics prediction
TargetScan version 7.1 (http://www.targetscan.org/vert_71) online software was
used to predict the miRNA that could target HLA-G.
Statistical analysis
Results were analyzed using SPSS 16.0 software
(SPSS, Inc., Chicago, IL, USA). Expression data regarding patients
were expressed as the median (with maximum-minimum and
interquartile range), and all other continuous variables were
expressed as the mean ± standard deviation. In vitro
experiments were repeated at least three times. Comparisons among
groups were analyzed using one-way analysis of variance and the
least significant difference post hoc test. All tests were
two-tailed and P<0.05 was considered to indicate a statistically
significant difference.
Results
Analysis of miR-148a expression in
ESCC
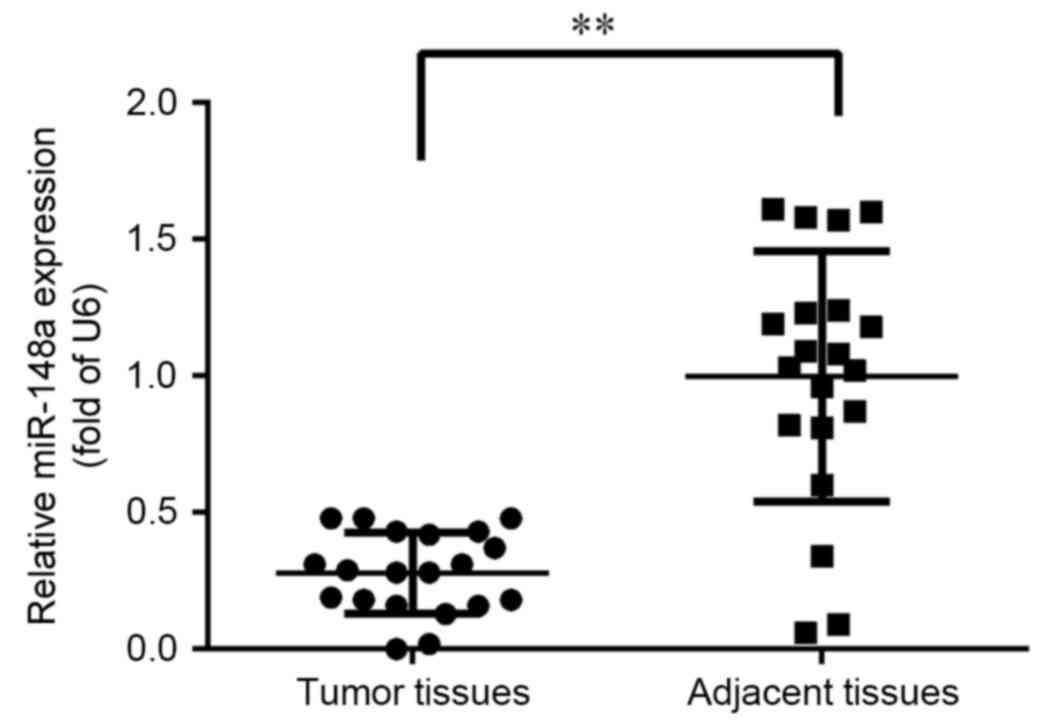
To investigate the tumor-specific expression pattern
of miR-148a, the expression of miR-148a was assessed in 20 pairs of
primary ESCC tissues and matched adjacent normal tissues. As
depicted in Fig. 1, expression of
miR-148a was significantly reduced to ~30% in the tumor tissue
samples when compared with normal adjacent tissues (P<0.01).
Therefore, a lower level of miR-148a expression was indicated to
occur in ESCC.
Effect of miR-148a on HLA-G
expression
Previous studies have indicated that HLA-G
expression level may be regulated by miR-148a, and that miR-148b
and HLA-G were upregulated in primary ESCC tissues (13,16).
Based on these reports, it was hypothesized that miR-148a may
contribute to the proliferation of ESCC cells by regulating HLA-G
expression. Therefore, the ESCC cell line EC9706 was transfected
with miR-148a mimic to identify the potential regulatory role of
miR-148a.
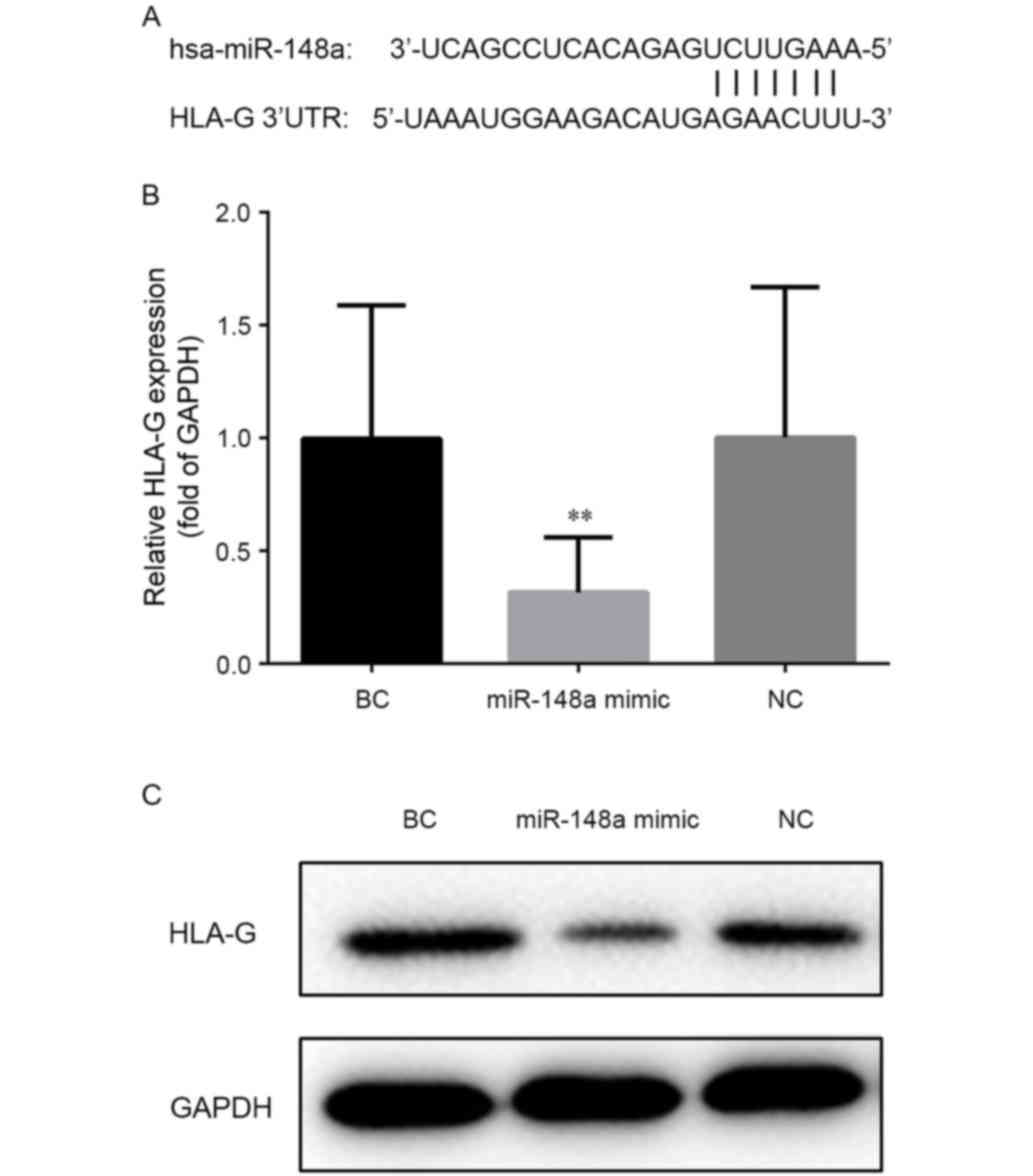
Position 376–382 of the HLA-G gene 3′-untranslated
region is predicted as the binding site of miR-148a according to
the TargetScan, as depicted in Fig.
2A. RT-qPCR confirmed that the level of HLA-G mRNA decreased
following transfection with miR-148a mimic (Fig. 2B). Compared with cells transfected
with nonsense (NC) or empty (BC) vector, the transcription level of
HLA-G was significantly decreased (to a relative expression of
~28%) in cells transfected with miR-148a mimic (P<0.01 vs. BC).
Accordingly, western blot analysis demonstrated that the level of
HLA-G protein was reduced in cells transfected with miR-148a mimic
when compared with NC or BC cells (Fig.
2C). These results confirmed that HLA-G expression was
regulated by miR-148a.
Effect of miR-148a on cell
apoptosis
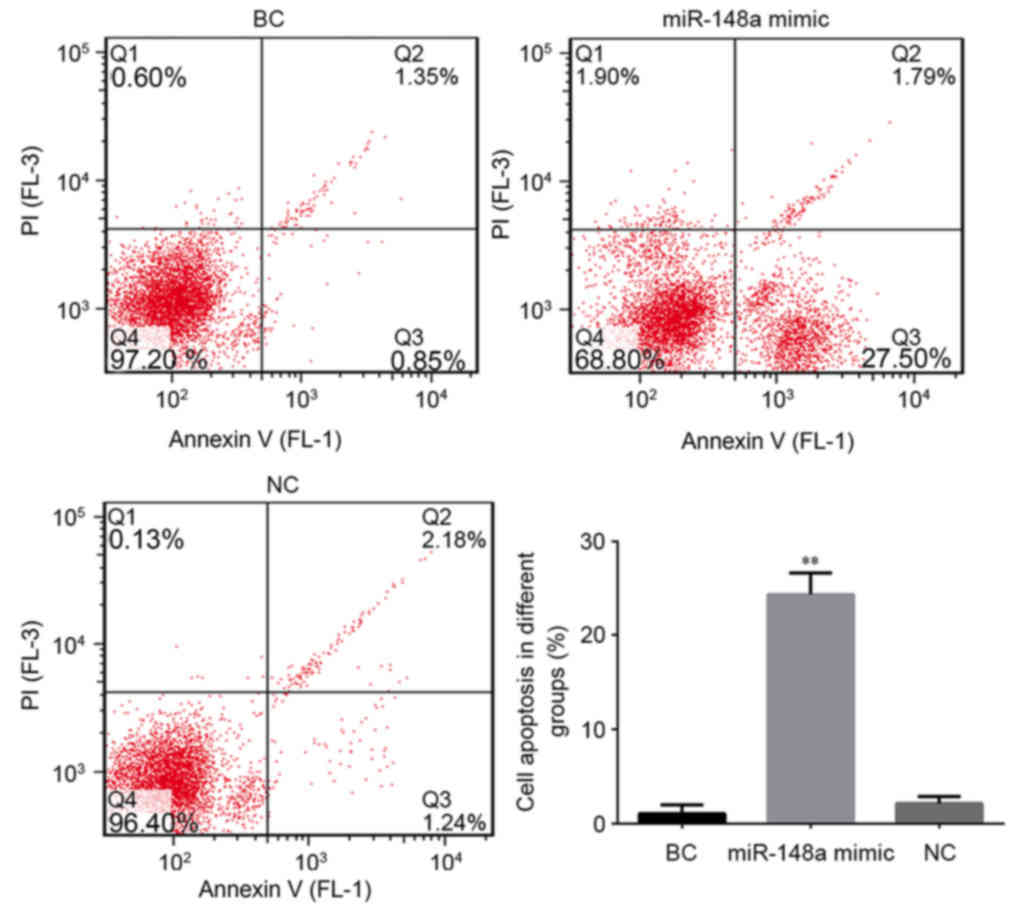
To further demonstrate whether miR-148a regulated
the growth of cells, the rates of cell apoptosis were compared
between the different cell groups. As depicted in Fig. 3, compared with the BC group,
transfection with miR-148a significantly increased the rate of
apoptosis in EC9706 cells (P<0.01); the apoptotic rate was
~13-fold higher in the cells transfected with miR-148a mimic. Rates
of apoptosis did not differ between the NC and BC groups,
indicating that the increase in apoptosis was due to transfection
with miR-148a mimic. Collectively these results suggest that
miR-148a may trigger apoptosis in ESCC cells. This pro-apoptotic
function may explain the low expression of miR-148a and
overexpression of its target HLA-G in ESCC tissues.
Discussion
Since the identification of miRs, large amounts of
data have demonstrated the critical role of miRs in numerous
biological processes including the occurrence, development and
differentiation of cancers (26).
Downregulation of miR-148a has been reported in multiple cancer
types. In gastrointestinal cancer tissues, the level of miR-148a
expression was inversely correlated with tumor size and
cholecystokinin B receptor was identified as a potential target
(10). It has also been reported
that miR-148a may promote the proliferation of gastric cancer cells
by directly targeting p27, as a key inhibitor of the cell cycle
(27). Another study demonstrated
that upregulation of miR-148a suppressed gastric cancer cell
invasion in vitro and lung metastasis formation in
vivo by downregulating the potential metastasis promoter
Rho-associated coiled-coil containing protein kinase 1 (28). Furthermore, upregulation of miR-148a
induced apoptosis in multiple colorectal cancer cell lines, and
Bcl-2 was confirmed as its direct target (13). MiR-148a has further been implicated
in hepatitis B-associated hepatocellular carcinoma through its
targeting of the tumor suppressor, phosphatase and tensin homolog
(29). Decreased expression of
miR-148a has also been correlated with recurrent EC (15). Similarly, the present study observed
that decreased expression of miR-148a was correlated with the
occurrence of primary ESCC. To the best of our knowledge, this is
the first report of a relationship between miR-148a and primary
ESCC tissues.
Among all the reported target genes of miR-148a,
HLA-G was chosen for further analysis in the current study, due to
previous indications that HLA-G is expressed to a high level in
primary ESCC tissues and may be regulated by miR-148a and miR-148b
(18,21). Further in vitro experiments
were performed that focused on miR-148a. By transfection with
miR-148a mimic, it was confirmed that the expression of HLA-G was
reduced by miR-148a in ESCC cells. These results indicated that
upregulation of HLA-G may be due to a low level of miR-148a in
ESCC.
HLA-G was first cloned and sequenced by Geraghty
et al (30). HLA-G may be
translated into seven isomers including transmembrane types (HLA-G1
to -G4) and soluble types (HLA-G5 to -G7) (31). It is generally accepted that HLA-G is
an immune tolerance molecule that prevents maternal-fetal assaults
through the inhibition of T- and natural killer cell-mediated
cytolysis (32). Other inhibitory
effects of HLA-G may include the induction of apoptosis of
activated CD8+ T-cells, downregulation of interferon
secretion and induction of the Th2 cytokine profile (33). As a consequence of miR-148a
downregulation, the increased expression of HLA-G by ESCC cells is
likely to be an evasion strategy against the anti-tumor
response.
It has been reported that an aberrant methylation
status of HLA-G may be involved in the pathogenesis of cancers. In
ovarian tumors, upregulation of HLA-G was associated with
spontaneous demethylation in the HLA-G promoter region, and
repression of the HLA-G gene could be reversed by demethylation
(34). Similar epigenetic
modification was observed in other tumor cell lines, including
JEG-3 and Tera-2 (35,36). DNA methyltransferase-1 (DNMT1) and
DNMT3b are the key enzymes in mammalian cells that maintain DNA
methylation, and both enzyme genes are direct targets of miR-148a
(37,38). Therefore, miR-148a may regulate HLA-G
expression via a dual function: By direct silencing of the HLA-G
gene and methylation regulation of the HLA-G promoter through DNMT1
and DNMT3b. However, this speculation requires further study.
Some limitations of this research should be
mentioned. Although an association between miR-148a expression and
primary ESCC tissues was reported, this result was observed in only
20 pairs of tumor tissues and adjacent tissues. The relationship
between miR-148a expression and the clinicopathological
characteristics and histologic grade of ESCC was not studied due to
the small sample size. Ideally, a luciferase activity assay would
have been performed to confirm the direct binding between miR-148a
and HLA-G. However, since Manaster et al (18) provided this data previously, this
assay was not performed in the present study.
In conclusion, the present study observed that
miR-148a was involved in the carcinogenesis of primary ESCC and
induced ESCC cell apoptosis by regulating HLA-G expression. The
results suggest that miR-148a is a potential biomarker of ESCC.
References
|
1
|
Napier KJ, Scheerer M and Misra S:
Esophageal cancer: A review of epidemiology, pathogenesis, staging
workup and treatment modalities. World J Gastrointest Oncol.
6:112–120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen WS, Zheng R, Zhang S, Zhao P, Zeng H,
Zou X and He J: Annual report on status of cancer in China, 2010.
Chin J Cancer Res. 26:48–58. 2014.PubMed/NCBI
|
|
3
|
Arnold M, Soerjomataram I, Ferlay J and
Forman D: Global incidence of oesophageal cancer by histological
subtype in 2012. Gut. 64:381–387. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen W, He Y, Zheng R, Zhang S, Zeng H,
Zou X and He J: Esophageal cancer incidence and mortality in China,
2009. J Thorac Dis. 5:19–26. 2013.PubMed/NCBI
|
|
6
|
Merkow RP, Bilimoria KY, Keswani RN, Chung
J, Sherman KL, Knab LM, Posner MC and Bentrem DJ: Treatment trends,
risk of lymph node metastasis, and outcomes for localized
esophageal cancer. J Natl Cancer Inst. 106:pii: dju133. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Andrew C, Fergus N and Timothy U:
Strategies to improve outcomes in esophageal adenocarcinoma. Expert
Rev Anticancer Ther. 14:677–687. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cheng GF: Circulating miRNAs: Roles in
cancer diagnosis, prognosis and therapy. Adv Drug Deliv Rev.
81:75–93. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M
and Croce CM: Human microRNA genes are frequently located at
fragile sites and genomic regions involved in cancers. P Natl Acad
Sci USA. 101:2999–3004. 2004. View Article : Google Scholar
|
|
10
|
Chen Y, Song Y, Wang Z, Yue Z, Xu H, Xing
C and Liu Z: Altered expression of miR-148a and miR-152 in
gastrointestinal cancers and its clinical significance. J
Gastrointest Surg. 14:1170–1179. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu Q, Jiang Y, Yin Y, Li Q, He J, Jing Y,
Qi YT, Xu Q, Li W, Lu B, et al: A regulatory circuit of
miR-148a/152 and DNMT1 in modulating cell transformation and tumor
angiogenesis through IGF-IR and IRS1. J Mol Cell Biol. 5:3–13.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou X, Zhao F, Wang ZN, Song YX, Chang H,
Chiang Y and Xu HM: Altered expression of miR-152 and miR-148a in
ovarian cancer is related to cell proliferation. Oncol Rep.
27:447–454. 2012.PubMed/NCBI
|
|
13
|
Zhang H, Li Y, Huang Q, Ren X, Hu H, Sheng
H and Lai M: MiR-148a promotes apoptosis by targeting Bcl-2 in
colorectal cancer. Cell Death Differ. 18:1702–1710. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Murata T, Takayama K, Katayama S, Urano T,
Horie-Inoue K, Ikeda K, Takahashi S, Kawazu C, Hasegawa A, Ouchi Y,
et al: miR-148a is an androgen-responsive microRNA that promotes
LNCaP prostate cell growth by repressing its target CAND1
expression. Prostate Cancer Prostatic Dis. 13:356–361. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hummel R, Hussey DJ, Michael MZ, Haier J,
Bruewer M, Senninger N and Watson DI: MiRNAs and their association
with locoregional staging and survival following surgery for
esophageal carcinoma. Ann Surg Oncol. 18:253–260. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hummel R, Watson DI, Smith C, Kist J,
Michael MZ, Haier J and Hussey DJ: Mir-148a improves response to
chemotherapy in sensitive and resistant oesophageal adenocarcinoma
and squamous cell carcinoma cells. J Gastrointest Surg. 15:429–438.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tan Z, Randall G, Fan J, Camoretti-Mercado
B, Brockman-Schneider R, Pan L, Solway J, Gern JE, Lemanske RF,
Nicolae D and Ober C: Allele-specific targeting of microRNAs to
HLA-G and risk of asthma. Am J Hum Genet. 81:829–834. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Manaster I, Goldman-Wohl D, Greenfield C,
Nachmani D, Tsukerman P, Hamani Y, Yagel S and Mandelboim O:
MiRNA-mediated control of HLA-G expression and function. PLoS One.
7:e333952012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Carosella ED, Moreau P, LeMaoult J and
Rouas-Freiss N: HLA-G: From biology to clinical benefits. Trends
Immunol. 29:125–132. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chang CC, Campoli M and Ferrone S: HLA
class I antigen expression in malignant cells: Why does it not
always correlate with CTL-mediated lysis? Curr Opin Immunol.
16:644–650. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yie SM, Yang H, Ye SR, Li K, Dong DD and
Lin XM: Expression of HLA-G is associated with prognosis in
esophageal squamous cell carcinoma. Am J Clin Pathol.
128:1002–1009. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cao M, Yie SM, Liu J, Ye S, Xia D and Gao
E: Plasma soluble HLA-G is a potential biomarker for diagnosis of
colorectal, gastric, esophageal and lung cancer. Tissue Antigens.
78:120–128. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rice TW: Esophageal cancer staging. Korean
J Thorac Cardiovasc Surg. 48:157–163. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Napier KJ, Scheerer M and Misra S:
Esophageal cancer: A Review of epidemiology, pathogenesis, staging
workup and treatment modalities. World J Gastrointest Oncol.
6:112–120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Berindan-Neagoe I, Monroig P, Pasculli B
and Calin GA: MicroRNAome genome: A treasure for cancer diagnosis
and therapy. CA Cancer J Clin. 64:311–336. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guo SL, Peng Z, Yang X, Fan KJ, Ye H, Li
ZH, Wang Y, Xu XL, Li J, Wang YL, et al: miR-148a promoted cell
proliferation by targeting p27 in gastric cancer cells. Int J Biol
Sci. 7:567–574. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zheng B, Liang L, Wang C, Huang S, Cao X,
Zha R, Liu L, Jia D, Tian Q, Wu J, et al: Microrna-148a suppresses
tumor cell invasion and metastasis by downregulating rock1 in
gastric cancer. Clin Cancer Res. 17:7574–7583. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yuan K, Lian Z, Sun B, Clayton MM, Ng Io
and Feitelson MA: Role of miR-148a in hepatitis B associated
hepatocellular carcinoma. PLoS One. 7:e353312012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Geraghty DE, Koller BH and Orr HT: A human
major histocompatibility complex class I gene that encodes a
protein with a shortened cytoplasmic segment. Proc Natl Acad Sci
USA. 84:9145–9149. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Paul P, Cabestre Adrian FA, Ibrahim EC,
Lefebvre S, Khalil-Daher I, Vazeux G, Quiles RM, Bermond F, Dausset
J and Carosella ED: Identification of HLA-G7 as a new splice
variant of the HLA-G mRNA and expression of soluble HLA-G5, -G6 and
-G7 transcripts in human transfected cells. Hum Immunol.
61:1138–1149. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rizzo R, Vercammen M, van de Velde H, Horn
PA and Rebmann V: The importance of HLA-G expression in embryos,
trophoblast cells and embryonic stem cells. Cell Mol Life Sci.
68:341–352. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rouas-Freiss N, Moreau P, Ferrone S and
Carosella D: HLA-G proteins in cancer: Do they provide tumor cells
with an escape mechanism? Cancer Res. 65:10139–10144. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Menendez L, Walker LD, Matyunina LV,
Totten KA, Benigno BB and McDonald JF: Epigenetic changes within
the promoter region of the HLA-G gene in ovarian tumors. Mol
Cancer. 7:1–11. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Moreau P, Mouillot G, Rousseau P, Marcou
C, Dausset J and Carosella ED: HLA-G gene repression is reversed by
demethylation. Proc Natl Acad Sci USA. 100:1191–1196. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mouillot G, Marcou C, Rousseau P,
Rouas-Freiss N, Carosella ED and Moreau P: HLA-G gene activation in
tumor cells involves cis-acting epigenetic changes. Int J Cancer.
113:928–936. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Braconi C, Huang N and Patel T:
MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor
suppressor gene expression by interleukin-6 in human malignant
cholangiocytes. Hepatology. 51:881–890. 2010.PubMed/NCBI
|
|
38
|
Duursma AM, Kedde M, Schrier M, le Dage C
and Agami R: miR-148 targets human DNMT3b protein coding region.
RNA. 14:872–877. 2008. View Article : Google Scholar : PubMed/NCBI
|