Introduction
Type 2 diabetes mellitus (T2DM), formerly termed
non-insulin-dependent diabetes mellitus or adult-onset diabetes, is
a multifactor disease that involves complex interactions between
genes (1–3), abnormalities of the immune system
(4–6), environmental factors (7–10) and
health-impacting behavior (11,12), and
represents a serious public health problem in numerous developed
countries (13). Current
investigations have revealed a definite global increase in the
incidence and prevalence of diabetes. In 2013, 382 million people
worldwide were estimated to be diabetic by the International
Diabetes Federation, which is expected to rise to 592 million cases
in the year 2035 (14). As a result
of the increasing rate of diabetes and its widespread societal and
economic consequences, prevention of diabetes among people at high
risk is an important public health issue in clinic practice.
However, the extent to which multiple defects in the regulation of
lipids, insulin secretion and action, and the immune system
contribute to the pathogenesis of T2DM has yet to be
elucidated.
MicroRNAs (miRNAs) are a class of small,
single-stranded non-coding RNAs (~22 nucleotides) that are
transcribed from the DNA of a gene, and modulate the expression of
a network of mRNAs through binding to the 3′-untranslated region
(3′-UTR), 5′-UTR or to the open reading frame of target mRNAs
(15). Notably, each miRNA is able
to target multiple mRNAs. Growing evidence indicates that miRNAs
are involved in T2DM (16). However,
the specific role of miRNAs in T2DM has yet to be elucidated.
Previous studies have demonstrated that miR-34a directly targets
p53 and serves a crucial role in p53-mediated biological processes,
such as cell cycle arrest, apoptosis and senescence (17), functioning downstream of the p53
pathway and participating in the initiation and progression of
certain types of cancer (18).
Previous research has reported that p53 is involved in T cell
development (19), which is of note
as T cells mediate the immune response and inhibit autoimmune
disease (20,21). miR-125b is predicted to be able to
bind to B lymphocyte-induced maturation protein-1 (Blimp-1) and
interferon regulatory factor-4 (IRF-4) transcription factors, which
are essential for plasma cell differentiation (22). In CD4+ T cells, Blimp-1
attenuates T helper (Th)1 differentiation, and represses the
formation of follicular helper T cells (22). Furthermore, Villeneuve et al
(23) found that miR-125b is
upregulated in vascular smooth muscle cells cultured from
db/db mice with diabetes and increases expression of
inflammatory genes, monocyte chemoattractant protein-1 and
interleukin (IL)-6 via targeting Suv39h1 in db/db vascular smooth
muscle cells. Thus, we speculate that miR-34a and miR-125b may
contribute to the development of diabetes. In the present study,
the expression levels of miR-34a, miR-125b and their relevant genes
in peripheral blood mononuclear cells (PBMCs) were detected in
samples obtained from T2DM patients. The results suggest that
miRNAs may be used as biomarkers for the diagnosis or prognosis of
T2DM.
Materials and methods
Patients and healthy blood donors
In the present study, 73 patients with T2DM (age,
56.81±11.85 years), including 35 women (47.9%) and 38 men (52.1%),
were recruited between the March and December 2012 from the
Department of Laboratory Medicine at The Second People's Hospital
of Taicang. Patients were diagnosed according to the criteria of
the American Diabetes Association (24) Fifty-two healthy control subjects were
recruited from age- and gender-matched healthy blood donors.
Clinical parameters are summarized in Table I. All subjects provided written
informed consent prior to enrollment in the study, and approval for
this study was obtained from the Medical Ethics Committee of the
Jiangsu University (Jiangsu, China).
 | Table I.Clinical feature profile in T2DM and
control subjects. |
Table I.
Clinical feature profile in T2DM and
control subjects.
| Parameter | T2DM (n=73) | Control (n=52) |
|---|
| Disease duration
(years) | 4.54±5.41 | – |
| FBG (mM) | 8.62±2.91 | 5.42±0.45 |
| HbA1C (%) | 8.50±2.09 | 5.82±1.07 |
| TG (mmol/l) | 3.29±1.53 | 2.66±0.59 |
| TC (mmol/l) | 3.67±2.28 | 3.23±1.12 |
| HDL (mmol/l) | 1.15±0.45 | 1.30±0.36 |
| LDL (mmol/l) | 2.77±1.01 | 2.53±0.84 |
| TG/HDL | 3.36±1.89 | 2.05±0.76 |
| LDL/HDL | 0.13±0.06 | 1.95±0.68 |
| HOMA-IR | 3.05±2.28 | – |
| HOMA-β | 30.82±40.78 | – |
Lipid measurement
Low-density lipoprotein (LDL) and high-density
lipoprotein (HDL) levels were measured in the serum of patients
with T2DM and healthy donors using the LDL-C reagent and HDL-C
reagent, respectively (Weifang 3V Bioengineering Group Co., Ltd.,
Weifang, China). All lipids were measured using the Beckman Coulter
AU5800 chemistry analyzer (Beckman Coulter, Inc., Brea, CA,
USA).
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
PBMCs from 73 patients with T2DM and 52 healthy
donors were freshly isolated by density-gradient centrifugation at
600 × g at room temperature for 20 min over Ficoll-Hypaque (Tianjin
Haoyang Biological Manufacture Co., Ltd., Tianjin, China) media
(1.077±0.001 g/ml). Total RNA was extracted from PBMCs using a
TriPure Isolation Reagent kit (Roche Diagnostics, Shanghai, China)
and quantified using a spectrophotometer at 260 and 280 nm. The
integrity of total RNA was verified by analyzing ~1 µg RNA on 1%
(w/v) formaldehyde denaturing agarose gel. The cDNA templates for
each specific gene mRNA and miRNA were generated by reverse
transcription (RT) of 1 µg and 250 ng of total RNA, respectively
using a RevertAid™ First Strand cDNA Synthesis kits (Thermo Fisher
Scientific, Inc., Waltham, MA, USA). Briefly, the RT reaction
mixture contained 4.0 µl 5X RT buffer, 2 µl 10 mM of each dNTP, 1
µl 2 µM oligo (dT)18 or stem-loop RT primer, 1 µl 40 U/µl RNase
inhibitor, 1 µl 200 U/µl ReverTra Ace reverse transcriptase and 1
µg or 250 ng total RNA, and diethylpyrocarbonate-treated water was
added to provide a total volume of 20 µl. An Eppendorf Mastercycler
(Eppendorf, Hamburg, Germany) was used to perform the RT reaction
under the following conditions: 42°C for 60 min, 70°C for 15 min,
and finally, held at 4°C. RT-qPCR was performed using a Bio-Rad
Real-Time thermal cycler CFX 96 [Bio-Rad Laboratories (Shanghai)
Co., Ltd., Shanghai, China] with miScript SYBR Green PCR kit
(Qiagen China Co., Ltd., Shanghai, China). In accordance with the
manufacturer's instructions, the PCR reaction mixture contained 1
µl cDNA, 5 µl 2X SYBR Green Mix, 0.6 µl 10 µM primer mix and 3.4 µl
nuclease-free water. The reaction protocol was as follows: 94°C for
10 min, 40 amplification cycles of degeneration at 95°C for 10 sec,
annealing for 30 sec, and extension at 72°C for 30 sec. All
reactions were run in triplicate. The gene names, primer sequences
and the annealing temperature are listed in Table II. To account for possible
differences in the amount of starting RNA, RT-qPCR data were
represented by the quantification cycle (Cq) value. The relative
expression level (i.e., fold change) for each gene or miRNA was
calculated using the 2−ΔΔCq method (25). Relative mRNA expression levels of
retinoid-related orphan receptor γt (RORγt), Blimp-1, IRF-4, P53
and Foxp3 was normalized against the housekeeping gene β-actin. The
relative expression of miR-34a and miR-125b were normalized to U6
snRNA.
 | Table II.Sequences of primers used for reverse
transcription-quantitative polymerase chain reaction. |
Table II.
Sequences of primers used for reverse
transcription-quantitative polymerase chain reaction.
| Gene | Primer sequence
(5′-3′) | Cycles | Annealing (°C) |
|---|
| β-actin (genebank
#NM-001101) | Sense:
CACGAAACTACCTTCAACTCC | 40 | 57 |
|
| Antisense:
CATACTCCTGCTTGCTGATC |
|
|
| blimp-1 (genebank
#NM-182907.2) | Sense:
CCGGGACTCCTACGCTTACTT | 40 | 60 |
|
| Antisense:
CGTTGTACGAGGGGATGAAAG |
|
|
| RORγt (genebank
#NM-001001523.1) | Sense:
CAAGAGAGGTTCTGGGCAAG | 40 | 57 |
|
| Antisense:
CTGCTCTTGTTGTGGAGAAGG |
|
|
| IRF-4 (genebank
#NM-001195286.1) | Sense:
CATCGACAAGCCGGACCC | 40 | 59 |
|
| Antisense:
CTCCGCTCAACCAGTTCCT |
|
|
| P53 (genebank
#NM-001126818.1) | Sense:
GGCCCACTTCACCGTACTAA | 40 | 56 |
|
| Antisense:
GTGGTTTCAAGGCCAGATGT |
|
|
| Foxp3 (genebank
#NM-001114377.1) | Sense:
CAAGAGAGGTTCTGGGCAAG | 40 | 64 |
|
| Antisense:
CTGCTCTTGTTGTGGAGAAGG |
|
|
| U6 (genebank
#NM-001207056) | Sense:
CTCGCTTCGGCAGCACA | 40 | 60 |
|
| Antisense:
AACGCTTCACGAATTTGCGT |
|
|
| miR-34a (genebank
#MIMAT0000260) | RT:
GTCGTATCCAGTGCAGGGTCO | 40 | 60 |
|
|
GAGGTATTCGCACTGGATACGACAACAAC |
|
|
|
| Sense:
CGGTATCATTTGGCAGTGTCT |
|
|
|
| Antisense:
GTGCAGGGTCCGAGGT |
|
|
| miR-125b (genebank
#MIMAT0000680) | RT
GTCGTATCCAGTGCAGGGTC | 40 | 60 |
|
|
CGAGGTATTCGCACTGGATACGACTCACAA |
|
|
|
| Sense:
GCCGTAAAGTGCTGACAGT |
|
|
|
| Antisense:
GTGCAGGGTCCGAGGTAT |
|
|
Statistical analysis
Statistical analysis was performed with Prism 5
software (GraphPad Software, Inc., La Jolla, CA, USA). Correlations
between variables were determined by Pearson's correlation
coefficient. Target genes expression levels of T2DM patients were
compared with healthy donors using the Mann-Whitney U test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
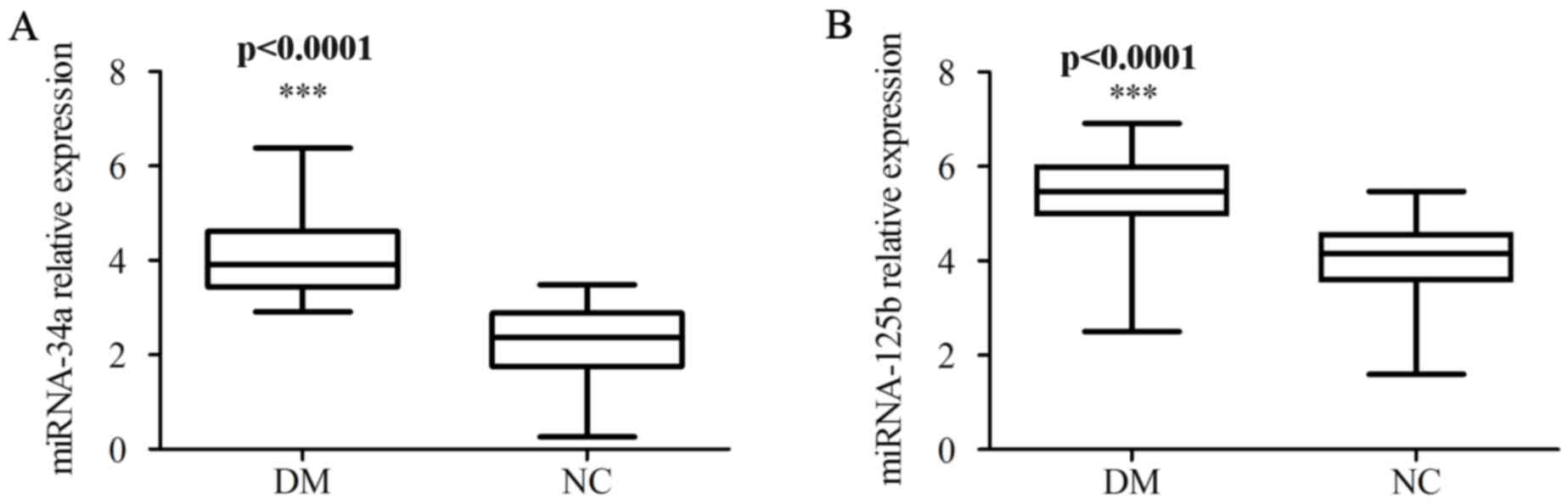
Higher expression of miR-34a and
miR-125b in patients with T2DM
Initially, RT-qPCR was employed to examine miR-34a
and miR-125b expression levels in PBMCs obtained from 73 patients
with T2DM and 52 healthy donors. The expression levels of miR-34a
and miR-125b were significantly higher in the PBMCs of patients
with T2DM compared with those of the PBMCs of healthy donors
(P<0.0001; Fig. 1).
Expression levels of IRF-4 and P53
were upregulated, whilst those of Blimp-1, RORγt and Foxp3 were
downregulated in the PBMCs of patients with T2DM
To further investigate the target or relevant genes
of miR-34a and miR-125b in the pathological process underlying
T2DM, the present study detected the expression levels of Blimp-1,
IRF-4, P53, RORγt and Foxp3, and found that the expression levels
of IRF-4 and P53 in the PBMCs from patients with T2DM were higher
compared with the PBMCs of healthy donors (P<0.05; Fig. 2), while Blimp-1, RORγt and Foxp3 were
lower compared with those of healthy donors (P<0.05).
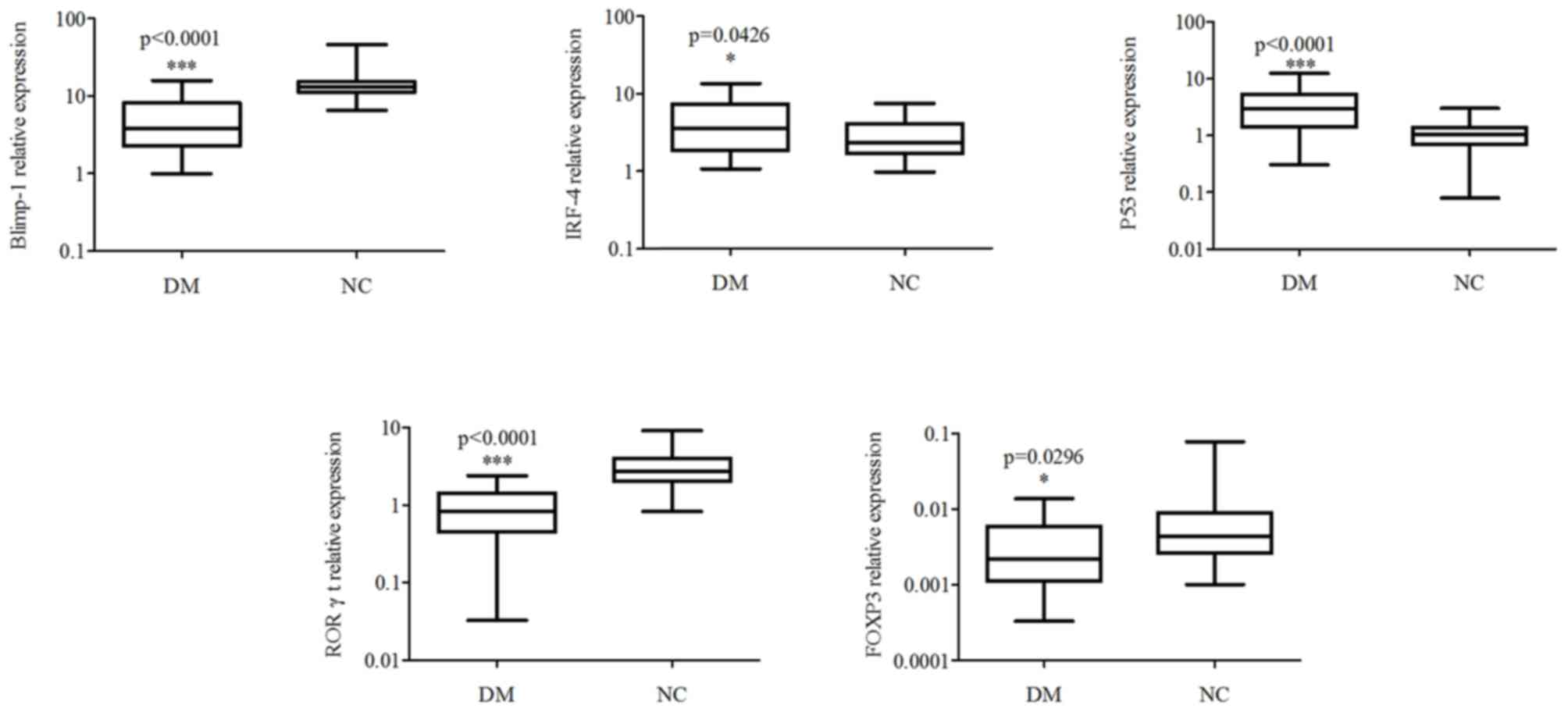 | Figure 2.Relative expression levels of
Blimp-1, IRF-4, P53, RORγt and Foxp3 in PBMCs of T2DM patients.
PBMCs were isolated from patients and the expression levels of
Blimp-1, IRF-4, P53, RORγt and Foxp3 were assessed by RT-qPCR.
Expression was normalized to the housekeeping gene β-actin.
*P<0.05 and ***P<0.001. IRF-4, interferon regulatory
protein-4; RORγt, retinoid-related orphan receptor γt, PBMCs,
peripheral blood mononuclear cells; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; T2DM, type 2
diabetes mellitus; DM, diabetes mellitus; NC, negative control. |
Association between the expression of
miR-34a, miR-125b and the levels of Blimp-1, IRF-4, P53, RORγt and
Foxp3
Subsequently an analysis was performed to determine
whether there was any correlation between miR-34a (Fig. 3A) and miR-125b (Fig. 3B) expression levels and Blimp-1,
IRF-4, P53, RORγt and Foxp3 genes in the PBMCs of patients with
T2DM. The present study found that there was a significant positive
correlation between the expression levels of miR-34a, miR-125b and
the level of Foxp3 (Fig. 3;
P<0.05). However, there was no significant correlation between
miR-34a and miR-125b levels and Blimp-1, IRF-4, P53 or RORγt
(Fig. 3; P>0.05).
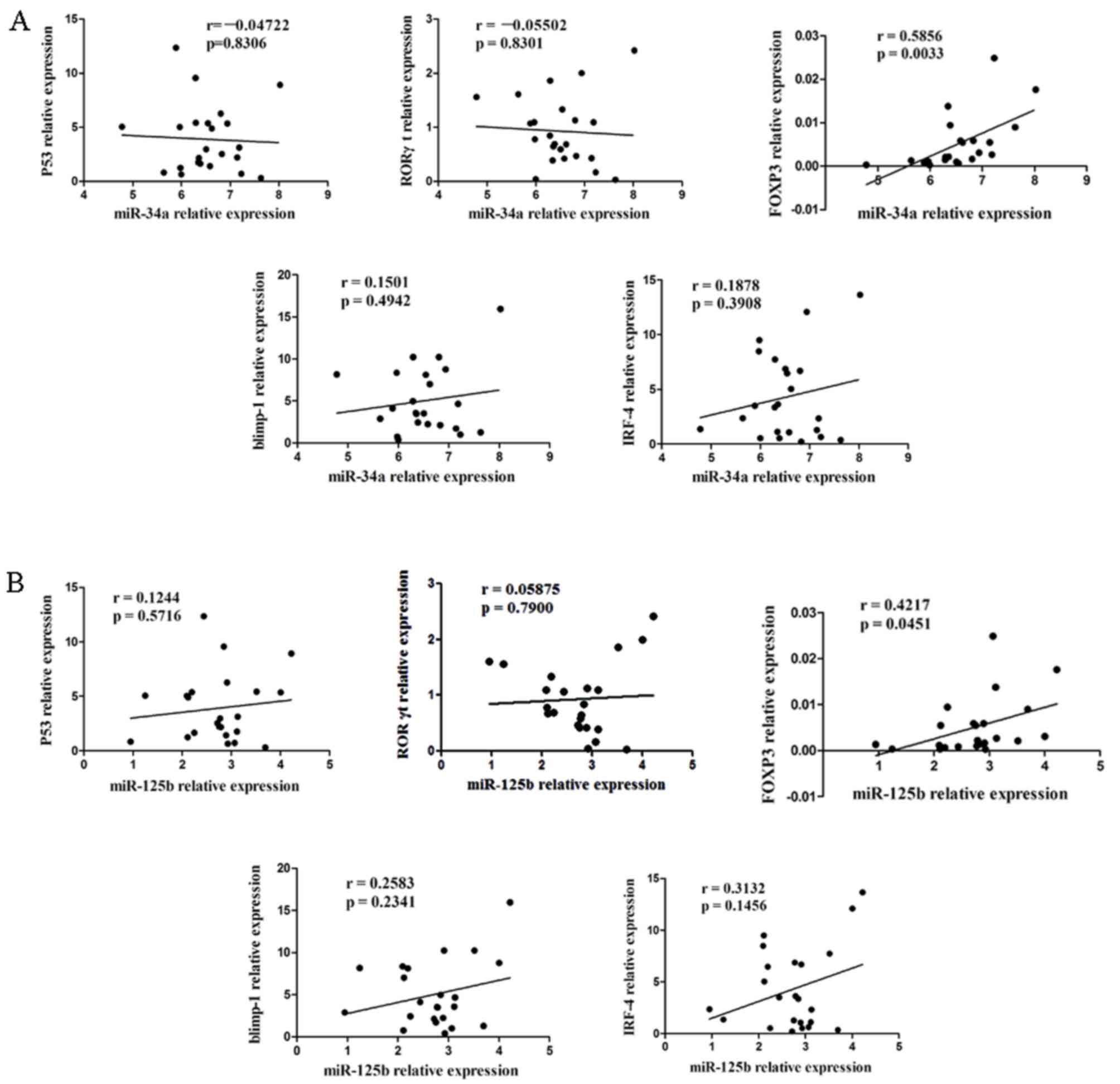 | Figure 3.Association between the expression
levels of (A) miR-34a or (B) miR-125b and the levels of Blimp-1,
IRF-4, p53, RORγt and Foxp3 in patients with T2DM. There was a
positive correlation between the upregulation of miR-34a and
miR-125b levels and genes of Foxp3 (P<0.05); There was no
observed correlation between miR-34a and miR-125b with Blimp-1,
IRF-4, P53 and RORγt (P>0.05). IRF-4, interferon regulatory
protein-4; RORγt, retinoid-related orphan receptor γt, PBMCs,
peripheral blood mononuclear cells; T2DM, type 2 diabetes
mellitus. |
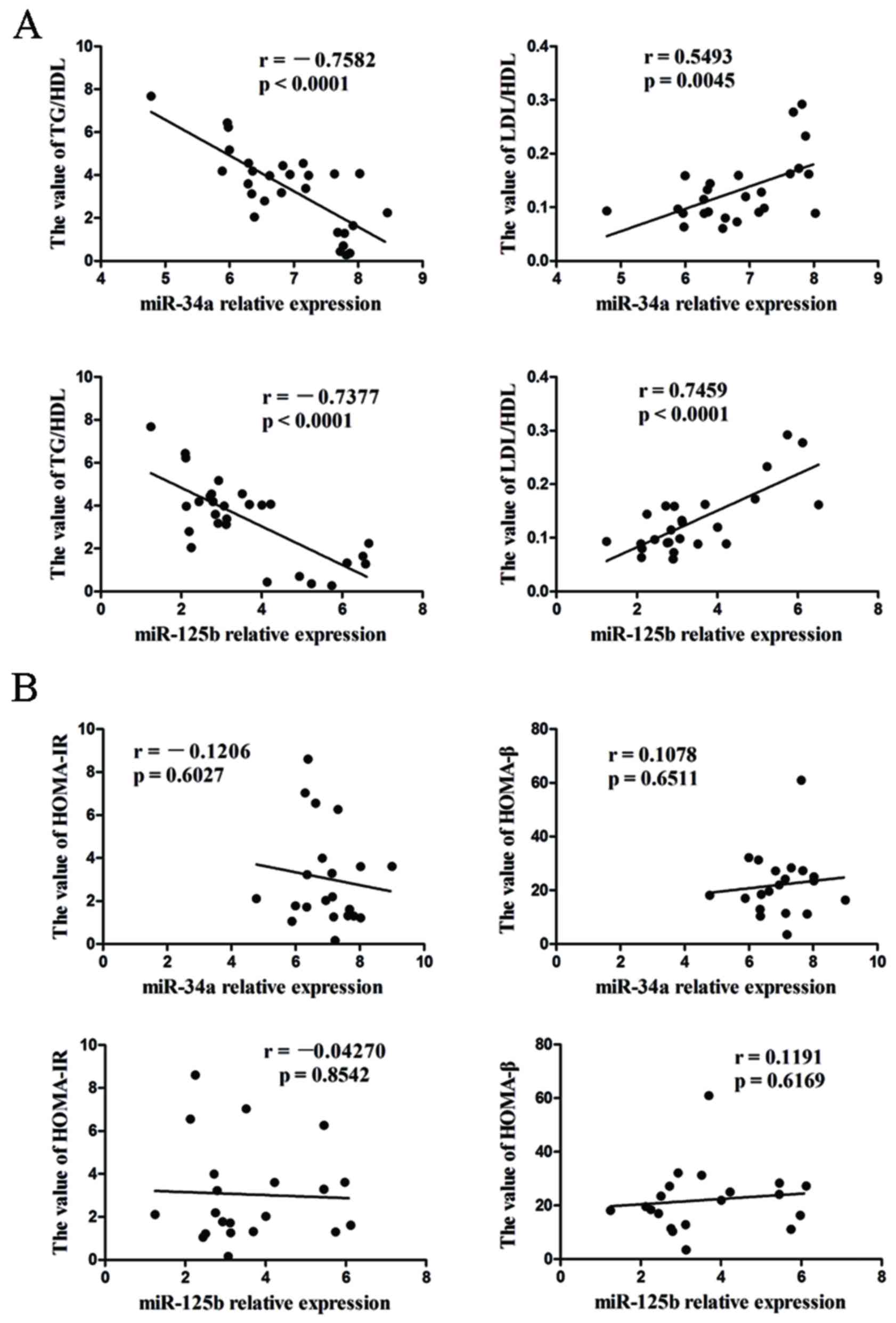
Relationship between miR-34a and
miR-125b levels and clinical features
In addition, the correlation between miR-34a and
miR-125b expression levels and clinical features was investigated.
Positive correlations were identified between the expression levels
of miR-34a, miR-125b and plasma (LDL/HDL) ratio, while the levels
of miR-34a and miR-125b are negatively correlated with TG/HDL ratio
(Fig. 4A; P<0.05), However, no
correlation was observed between miR-34a, miR-125b and homeostasis
model assessment of insulin resistance and homeostasis model
assessment of β-cell function (Fig.
4B, P>0.05). The current results suggest that miR-34a and
miR-125b may be involved in the pathogenesis of T2DM.
Discussion
Recently, considerable progress has been made with
regard to T2DM, which is a major health problem associated with
excess morbidity and mortality. Growing evidence indicates that
miRNAs are involved in the pathogenesis of T2DM, resulting in
increased glucagon secretion and reduced incretin response
(26–28). miRNAs have been implicated in the
epigenetic regulation of key metabolic, inflammatory, and
antiangiogenic pathways in T2DM. Zhang et al (29) demonstrated that miR-34a could
regulate mesangial proliferation and glomerular hypertrophy by
directly inhibiting GAS1 in early diabetic nephropathy (DN). A
further study has ascertained that miR-34a was able to inhibit
osteosarcoma growth through the downregulation of Eag1 expression
(17). Recently, miR-34a has been
shown to be one of the key mediators and downstream factors of p53
(30). It had been demonstrated that
miR-34a is involved in pancreatic development, and can inhibit
insulin secretion (31–33). Lovis et al reported that the
expression level of miR-34a was higher in the islets of diabetic
db/db mice, and was increased in the β-cell line,
MIN6B1 and in pancreatic islets following prolonged exposure to
saturated fatty acids (34). Kong
et al found that miR-34a was significantly upregulated in
the serum of patients with T2DM (35). An increase in miR-34a is associated
with the activation of p53, and results in sensitization to
apoptosis and impaired nutrient-induced secretion (34,36). The
latter effect is associated with inhibition of the expression of
vesicle-associated membrane protein 2, an important factor in
β-cell exocytosis (34).
T2DM results from progressive β-cell dysfunction in
the presence of chronic insulin resistance, leading to a
progressive decline in plasma glucose control. Previous studies
showed that the activation of T lymphocytes and cytokine-induce
inflammatory responses serve a considerable role in the development
of diabetes. The proinflammatory cytokines involved in β-cell
damage are those secreted by CD4+ T-cells (Th1 and Th17)
and those by macrophages, including tumor necrosis factor-α
(TNF-α), IL-1β, IFN-γ and IL-17 (37–39). It
also had been demonstrated that the Th1/Th2/Th17/T regulatory
(Treg) paradigm skewed to Th1 and Th17 in patients with T2DN
(5). Treg cells expressing Foxp3
have an anti-inflammatory role and maintain tolerance to
self-components by contact-dependent suppression, or by releasing
anti-inflammatory cytokines such as IL-10 and transforming growth
factor-β1 (40), while Th17 cells
expressing RORγt serve important roles in the development of
autoimmunity, allergic reactions and in T2DM by producing IL-17,
TNF-α and IL-6 (39,41). An imbalance of Th17/Treg is present
in T2DM patients (4,5). Thus, it may contribute to the
pathogenesis of inflammation and T2DM (42). miR-125b binds to the 3′-untranslated
region of p53, Blimp-1 and IRF-4 messenger RNAs, and inhibits cell
apoptosis by attenuating the expression of p53, or blocking B cell
differentiation in germinal centers through decreasing the
expression levels of Blimp-1 and IRF-4 (22,43).
Blimp-1 is able to attenuate type 1 diabetes mellitus (T1DM) via
suppressing the function of Th1 and Th17 (44). It has been demonstrated that the
expression of Blimp-1 is required for the production of IL-10 by
Treg cells that localize in mucosal sites, and for their tissue
homeostasis. Furthermore, IRF-4 was essential for Blimp-1
expression and for the differentiation of these Treg cells
(45). Thus, we speculated that
miR-125b may be involved in the pathological process underlying
T2DM, and Blimp-1, IRF-4 and Foxp3 may also be downregulated in the
disease.
In the present study, the expression levels of
miR-125b and miR-34a were upregulated in PBMCs obtained from
patients with T2DM. It had been demonstrated that miR-34a is the
product of downstream of p53, and that p53 activates the expression
of miR-34a, which subsequently attenuates E2F3 expression and
induces cell apoptosis (46). In
T2DM, miR-34a may contribute to β cell apoptosis and promote
insulin resistance (33).
Furthermore, miR-34a may be secreted in exosomes and migrate into
PBMCs. miR-125b inhibits p53 (43),
and prevents cells from undergoing apoptosis, and may serve an
important role in vascular endothelial cell proliferation. miR-125b
is also able to transfer from vascular endothelial cells to T cells
via exosomes. The migrated miR-125b inhibits the expression of
Blimp-1 and IRF-4, which drives the conversion of Tregs and Th2
cells into Th17 cells (45).
However, the expression levels of IRF-4 were observed to increase
in the present study, which may be a result of the fact that IRF-4
is the critical transcript in IL-21-mediated induction,
amplification, and stabilization of Th17 (47). In addition, it was observed that the
expression of Foxp3 was not consistent with previous studies. It is
probable that the expression of Foxp3 ocuurs in effector T cells as
well as in Tregs. It has been reported that activated T cells can
upregulate the expression of Foxp3 (48,49).
There are several activated T cells, such as Th1, involved in
PBMCs, which may indicate why the expression levels of Foxp3 are
upregulated in PBMCs. However, it had been shown that the p53
negatively regulates autoimmune diseases via attenuating the
STAT3-Th17 axis (21). In the
present study, p53 was upregulated, and may be associated with β
cell apoptosis.
In conclusion, the current investigation reveals
that miR-34a and miR-125b are involved in T2DM. In addition, there
was a positive correlation between the expression levels of miR-34a
and miR-125b with LDL/HDL, and a negative correlation with TG/HDL.
It has also been previously demonstrated that miR-34a was increased
in a high-carbohydrate diet and is associated in the nonalcoholic
fatty liver disease (50,51). The current findings may provide new
insights into how miRNA is associated with the cholesterol
metabolism of T2DM.
Acknowledgements
The present study was supported by the following
grants: The National Natural Science Foundation of China (grant
nos. 81273202, 31200676 and 31400773), the Clinical Medicine
Science & Technology Project of Jiangsu province of China
(grant no. BL2013024), the Natural Science Foundation of the
Jiangsu Higher Education Institutions of China (grant no.
14KJB320001), Jiangsu Province Postdoctoral Research Foundation,
China (grant no. 1402170C), Program of Jiangsu Province Innovative
Team, Project Funded by the Priority Academic Program Development
of Jiangsu Higher Education Institutions and Project Funded by the
Key Academic Program Development of Jiangsu University. The study
was also supported by Senior Talents Scientific Research Foundation
of Jiangsu University (grant no. 14JDG042).
References
|
1
|
Kommoju UJ, Maruda J, Kadarkarai Samy S,
Irgam K, Kotla JP and Reddy BM: Association of IRS1, CAPN10, and
PPARG gene polymorphisms with type 2 diabetes mellitus in the
high-risk population of Hyderabad, India. J Diabetes. 6:564–573.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang X, Deng Y, Gu H, Ren X, Li N, Lim A,
Snellingen T, Liu X, Wang N and Liu N: Candidate gene association
study for diabetic retinopathy in Chinese patients with type 2
diabetes. Mol Vis. 20:200–214. 2014.PubMed/NCBI
|
|
3
|
Hivert MF, Vassy JL and Meigs JB:
Susceptibility to type 2 diabetes mellitus-from genes to
prevention. Nat Rev Endocrinol. 10:198–205. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zeng C, Shi X, Zhang B, Liu H, Zhang L,
Ding W and Zhao Y: The imbalance of Th17/Th1/Tregs in patients with
type 2 diabetes: Relationship with metabolic factors and
complications. J Mol Med (Berl). 90:175–186. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang C, Xiao C, Wang P, Xu W, Zhang A, Li
Q and Xu X: The alteration of Th1/Th2/Th17/Treg paradigm in
patients with type 2 diabetes mellitus: Relationship with diabetic
nephropathy. Hum Immunol. 75:289–296. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jagannathan-Bogdan M, McDonnell ME, Shin
H, Rehman Q, Hasturk H, Apovian CM and Nikolajczyk BS: Elevated
proinflammatory cytokine production by a skewed T cell compartment
requires monocytes and promotes inflammation in type 2 diabetes. J
Immunol. 186:1162–1172. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dellamea BS, Leitao CB, Friedman R and
Canani LH: Nitric oxide system and diabetic nephropathy.
Diabetology and Metabolic Syndrome. 6:172014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Okada S, Saito M, Kazuyama E, Hanada T,
Kawaba Y, Hayashi A, Satoh K and Kanzaki S: Effects of
N-hexacosanol on nitric oxide synthase system in diabetic rat
nephropathy. Mol Cell Biochem. 315:169–177. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tang LL, Liu Q, Bu SZ, Xu LT, Wang QW, Mai
YF and Duan SW: The effect of environmental factors and DNA
methylation on type 2 diabetes mellitus. Yi Chuan. 35:1143–1152.
2013.(In Chinese). View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Anderssohn M, McLachlan S, Lüneburg N,
Robertson C, Schwedhelm E, Williamson RM, Strachan MW, Ajjan R,
Grant PJ, Böger RH and Price JF: Genetic and environmental
determinants of dimethylarginines and association with
cardiovascular disease in patients with type 2 diabetes. Diabetes
Care. 37:846–854. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nothwehr F and Stump T: Health-promoting
behaviors among adults with type 2 diabetes: Findings from the
health and retirement study. Prev Med. 30:407–414. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Green AJ, Bazata DD, Fox KM and Grandy S:
SHIELD Study Group: Health-related behaviours of people with
diabetes and those with cardiometabolic risk factors: Results from
SHIELD. Int J Clin Pract. 61:1791–1797. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Alberti KG and Zimmet PZ: Definition,
diagnosis and classification of diabetes mellitus and its
complications. Part 1: Diagnosis and classification of diabetes
mellitus provisional report of a WHO consultation. Diabet Med.
15:539–553. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guariguata L, Whiting DR, Hambleton I,
Beagley J, Linnenkamp U and Shaw JE: Global estimates of diabetes
prevalence for 2013 and projections for 2035. Diabetes Res Clin
Pract. 103:137–149. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mandke P, Wyatt N, Fraser J, Bates B,
Berberich SJ and Markey MP: MicroRNA-34a modulates MDM4 expression
via a target site in the open reading frame. PLoS One.
7:e420342012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guay C, Roggli E, Nesca V, Jacovetti C and
Regazzi R: Diabetes mellitus, a microRNA-related disease? Transl
Res. 157:253–264. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu X, Zhong D, Gao Q, Zhai W, Ding Z and
Wu J: MicroRNA-34a inhibits human osteosarcoma proliferation by
downregulating ether à go-go 1 expression. Int J Med Sci.
10:676–682. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tan J, Fan L, Mao JJ, Chen B, Zheng L,
Zhang T, Li T, Duan J, Duan Y, Jin Z and Kuang W: Restoration of
miR-34a in p53 deficient cells unexpectedly promotes the cell
survival by increasing NFkB activity. J Cell Biochem.
113:2903–2908. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Okazuka K, Wakabayashi Y, Kashihara M,
Inoue J, Sato T, Yokoyama M, Aizawa S, Aizawa Y, Mishima Y and
Kominami R: p53 prevents maturation of T cell development to the
immature CD4−CD8+ stage in
Bcl11b−/− mice. Biochem Biophys Res Commun. 328:545–549.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zheng SJ, Lamhamedi-Cherradi SE, Wang P,
Xu L and Chen YH: Tumor suppressor p53 inhibits autoimmune
inflammation and macrophage function. Diabetes. 54:1423–1428. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang S, Zheng M, Kibe R, Huang Y, Marrero
L, Warren S, Zieske AW, Iwakuma T, Kolls JK and Cui Y: Trp53
negatively regulates autoimmunity via the STAT3-Th17 axis. FASEB J.
25:2387–2398. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gururajan M, Haga CL, Das S, Leu CM,
Hodson D, Josson S, Turner M and Cooper MD: MicroRNA 125b
inhibition of B cell differentiation in germinal centers. Int
Immunol. 22:583–592. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Villeneuve LM, Kato M, Reddy MA, Wang M,
Lanting L and Natarajan R: Enhanced levels of microRNA-125b in
vascular smooth muscle cells of diabetic db/db mice lead to
increased inflammatory gene expression by targeting the histone
methyltransferase Suv39h1. Diabetes. 59:2904–2915. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
American Diabetes Association, . Diagnosis
and classification of diabetes mellitus. Diabetes Care. 35 Suppl
1:S64–S71. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tao W, Dong X, Kong G, Fang P, Huang X and
Bo P: Elevated circulating hsa-miR-106b, hsa-miR-26a, and
hsa-miR-29b in type 2 diabetes mellitus with diarrhea-predominant
irritable bowel syndrome. Gastroenterol Res Pract.
2016:92562092016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Latouche C, Natoli A, Reddy-Luthmoodoo M,
Heywood SE, Armitage JA and Kingwell BA: MicroRNA-194 modulates
glucose metabolism and its skeletal muscle expression is reduced in
diabetes. PLoS One. 11:e01551082016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sardu C, Barbieri M, Rizzo MR, Paolisso P,
Paolisso G and Marfella R: Cardiac resynchronization therapy
outcomes in type 2 diabetic patients: Role of MicroRNA changes. J
Diabetes Res. 2016:72925642016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang L, He S, Guo S, Xie W, Xin R, Yu H,
Yang F, Qiu J, Zhang D, Zhou S and Zhang K: Down-regulation of
miR-34a alleviates mesangial proliferation in vitro and glomerular
hypertrophy in early diabetic nephropathy mice by targeting GAS1. J
Diabetes Complications. 28:259–264. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chang TC, Wentzel EA, Kent OA,
Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M,
Ferlito M, Lowenstein CJ, et al: Transactivation of miR-34a by p53
broadly influences gene expression and promotes apoptosis. Mol
Cell. 26:745–752. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wei R, Yang J, Liu GQ, Gao MJ, Hou WF,
Zhang L, Gao HW, Liu Y, Chen GA and Hong TP: Dynamic expression of
microRNAs during the differentiation of human embryonic stem cells
into insulin-producing cells. Gene. 518:246–255. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chakraborty C, Doss CG, Bandyopadhyay S
and Agoramoorthy G: Influence of miRNA in insulin signaling pathway
and insulin resistance: Micro-molecules with a major role in type-2
diabetes. Wiley Interdiscip Rev RNA. 5:697–712. 2014.PubMed/NCBI
|
|
33
|
Chakraborty C, George Priya, Doss C and
Bandyopadhyay S: miRNAs in insulin resistance and
diabetes-associated pancreatic cancer: The ‘minute and miracle’
molecule moving as a monitor in the ‘genomic galaxy’. Curr Drug
Targets. 14:1110–1117. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lovis P, Roggli E, Laybutt DR, Gattesco S,
Yang JY, Widmann C, Abderrahmani A and Regazzi R: Alterations in
microRNA expression contribute to fatty acid-induced pancreatic
beta-cell dysfunction. Diabetes. 57:2728–2736. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kong L, Zhu J, Han W, Jiang X, Xu M, Zhao
Y, Dong Q, Pang Z, Guan Q, Gao L, et al: Significance of serum
microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: A
clinical study. Acta Diabetol. 48:61–69. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nesca V, Guay C, Jacovetti C, Menoud V,
Peyot ML, Laybutt DR, Prentki M and Regazzi R: Identification of
particular groups of microRNAs that positively or negatively impact
on beta cell function in obese models of type 2 diabetes.
Diabetologia. 56:2203–2212. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hotamisligil GS: Inflammation and
metabolic disorders. Nature. 444:860–867. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lasselin J and Capuron L: Chronic
low-grade inflammation in metabolic disorders: Relevance for
behavioral symptoms. Neuroimmunomodulation. 21:95–101. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Santos VR, Ribeiro FV, Lima JA, Napimoga
MH, Bastos MF and Duarte PM: Cytokine levels in sites of chronic
periodontitis of poorly controlled and well-controlled type 2
diabetic subjects. J Clin Periodontol. 37:1049–1058. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sakaguchi S, Ono M, Setoguchi R, Yagi H,
Hori S, Fehervari Z, Shimizu J, Takahashi T and Nomura T:
Foxp3+ CD25+ CD4+ natural
regulatory T cells in dominant self-tolerance and autoimmune
disease. Immunol Rev. 212:8–27. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bettelli E, Oukka M and Kuchroo VK:
T(H)-17 cells in the circle of immunity and autoimmunity. Nat
Immunol. 8:345–350. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Homey B: After TH1/TH2 now comes
Treg/TH17: Significance of T helper cells in immune response
organization. Hautarzt. 57:730–732. 2006.(In German). View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B,
Korzh V, Lodish HF and Lim B: MicroRNA-125b is a novel negative
regulator of p53. Genes Dev. 23:862–876. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lin MH, Chou FC, Yeh LT, Fu SH, Chiou HY,
Lin KI, Chang DM and Sytwu HK: B lymphocyte-induced maturation
protein 1 (BLIMP-1) attenuates autoimmune diabetes in NOD mice by
suppressing Th1 and Th17 cells. Diabetologia. 56:136–146. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cretney E, Xin A, Shi W, Minnich M, Masson
F, Miasari M, Belz GT, Smyth GK, Busslinger M, Nutt SL and Kallies
A: The transcription factors Blimp-1 and IRF4 jointly control the
differentiation and function of effector regulatory T cells. Nat
Immunol. 12:304–311. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Polager S and Ginsberg D: p53 and E2f:
Partners in life and death. Nat Rev Cancer. 9:738–748. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Huber M, Brüstle A, Reinhard K, Guralnik
A, Walter G, Mahiny A, von Löw E and Lohoff M: IRF4 is essential
for IL-21-mediated induction, amplification, and stabilization of
the Th17 phenotype. Proc Natl Acad Sci USA. 105:pp. 20846–20851.
2008, View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Allan SE, Crome SQ, Crellin NK, Passerini
L, Steiner TS, Bacchetta R, Roncarolo MG and Levings MK:
Activation-induced FOXP3 in human T effector cells does not
suppress proliferation or cytokine production. Int Immunol.
19:345–354. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kmieciak M, Gowda M, Graham L, Godder K,
Bear HD, Marincola FM and Manjili MH: Human T cells express CD25
and Foxp3 upon activation and exhibit effector/memory phenotypes
without any regulatory/suppressor function. J Transl Med. 7:892009.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Min HK, Kapoor A, Fuchs M, Mirshahi F,
Zhou H, Maher J, Kellum J, Warnick R, Contos MJ and Sanyal AJ:
Increased hepatic synthesis and dysregulation of cholesterol
metabolism is associated with the severity of nonalcoholic fatty
liver disease. Cell Metab. 15:665–674. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Li X, Lian F, Liu C, Hu KQ and Wang XD:
Isocaloric pair-fed high-carbohydrate diet induced more hepatic
steatosis and inflammation than high-fat diet mediated by
miR-34a/SIRT1 axis in mice. Sci Rep. 5:167742015. View Article : Google Scholar : PubMed/NCBI
|