Introduction
Hepatitis B virus (HBV) is an infectious disease
that poses a serious threat to human health. It is demonstrated
that sexual promiscuity, transfusion of unscreened blood, reusing
or sharing of syringes between injection in drug users are the
predominant associated risk factors (1,2). The
World Health Organization estimates that there are ~350 million
people worldwide infected with HBV, which may develop into chronic
hepatitis B, liver fibrosis, liver sclerosis or liver cancer
(3,4). The estimated worldwide mortality is 0.5
to 1.2 million fatalities a year (1). However, t here are currently no
effective treatments for HBV-related liver cancer.
Interferon-α (IFN-α) is an antiviral cytokine that
has a broad spectrum of action, exhibits high activity and indirect
and species specificity (5,6). IFN-α exerts its antiviral activity via
activation of the Janus kinase/signal transducer and activator of
transcription (JAK-STAT) signaling pathway (7,8). In
addition, IFN-α inhibits tumor development by decreasing cell
viability, promoting cell apoptosis and attenuating tumor
angiogenesis (9–11). IFN-α serves a role in immune
surveillance and regulation by enhancing the immune function of T-
and B-lymphocytes, natural killer cells and macrophages to enhance
the body's ability to kill cancer cells and tumor cells infected by
the virus (12,13). The effect of IFN-α on anti-viral,
anti-tumor and immune regulation indicates that IFN-α may be used
as to treat patients with HBV-related liver cancer. However, the
role served by IFN-α regulation in the development of HBV-related
liver cancer remains unknown.
Hepatitis B X protein (HBx), encoded by HBV DNA,
serves an important role during the development of chronic
hepatitis B, liver cirrhosis and liver cancer (14). Therefore, the current study
established a novel HBV-related liver cancer model by transfecting
the hepatoma cell line Huh-7 with HBx-expressing lentivirus, which
has been previously studied (15–18) and
subsequently investigated the effect of IFN-α on the growth of
cancer cells to identify its potential as a drug for treating
HBV-related liver cancer.
Materials and methods
Cell culture
The human hepatoma cell line Huh-7 (The Cell Bank of
Type Culture Collection of Chinese Academy of Sciences; Wuhan,
China) was cultured in Dulbecco's Modified Eagle medium (DMEM;
HyClone; GE Healthcare Life Sciences, Logan, UT, USA) supplemented
with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), 80 U/ml penicillin and 80 µg/ml streptomycin
(HyClone; GE Healthcare Life Sciences). The cells were incubated in
5% CO2 at 37°C. 1,000 IU/ml of IFN-α (Sigma-Aldrich;
Merck KGaA; Darmstadt, Germany) was used to treat the cells in the
following experiments.
Transfection of HBx-expressing
lentivirus into Huh-7 cells
HBx-expressing lentivirus was produced from
pLenti6.2/V5-DEST plasmid (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) with the second generational system.
Briefly, the packaging plasmids were transformed into 293T cells
with Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. Lentivirus was
harvested 72 h after transfection, and then the titer was
determined as described previously (19). Huh-7 cells were transfected with
3×107 infectious units per milliliter of HBx-expressing
lentivirus (Novobio Scientific, Inc., Shanghai, China) on a 96-well
plate. The control cells were transfected with the same
concentration of empty lentivirus. Medium was replaced 24 h
following transfection, and subsequent experiments began 24 h post
transfection.
Treatment groups
The following four groups were used in the present
study: Control (no treatment); IFN-α only, Huh-7 cells were treated
with 1,000 IU/ml of IFN-α for 24 h at 37°C; HBx infected cells,
Huh-7 cells were transfected with 3×107 infectious units
per milliliter of HBx-expressing lentivirus for 24 h at 37°C;
HBx+IFN-α group, after the transfection of HBx-expressing
lentivirus for 24 h, Huh-7 cells were treated with 1,000 IU/ml of
IFN-α for an additional 24 h.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Cells were washed with cold PBS three times and
harvested following centrifugation at 3,000 × g at 4°C for 5 min.
Total RNA was extracted using RNAiso Plus (Takara Bio., Dalian,
China) according to the manufacturer's instructions and subjected
to electrophoresis on 1.5% native agarose gel for integrity and
quality analysis. A total of 1 µg RNA was used for cDNA synthesis
with a PrimeScript RT reagent kit (Takara Bio, Inc., Otsu, Japan)
following the manufacturer's protocol. A total of 1 µl
20-fold-diluted cDNA was used as a template for qPCR, which was
performed using the Bio-Rad Detection system and SYBR Green qPCR
SuperMix (Invitrogen; Thermo Fisher Scientific, Inc.) following the
manufacturer's protocol with the corresponding primers (Table I). PCR conditions consisted of: 95°C
for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 30
sec. Relative expression was determined following normalization to
the reference gene GAPDH. RT-qPCR data were collected from three
independent biological replicates and analyzed with the
2−ΔΔCT method (20).
 | Table I.Primers for reverse
transcription-quantitative polymerase chain reaction. |
Table I.
Primers for reverse
transcription-quantitative polymerase chain reaction.
| Genes | Forward
(5′-3′) | Reverse
(5′-3′) |
|---|
| HBx |
GTCAACGCATAGTGGGTCT |
CCTTGTAGGTTGGCGAGA |
| IFNAR1 |
GTAAAGGTGGACATCAGTG |
AGAATCTGGTAAGGGAAA |
| IFNAR2 |
AAATGCACCCTCCTTCCA |
AGCCCTTAGCGAGACCTT |
| ISGF3 |
TGGCATTTCTGACTTTCTCC |
GGGCTATGGTAATGTGGGTA |
| PKR |
CGTGCCTGGATTGAGAAA |
CATCACTGCCGAACATTA |
| RNaseL |
GGAAGCGAGGAGCACAAG |
TGGGCACATTCAGGAACT |
| GAPDH |
AGGGCTGCTTTTAACTCTGG |
CCCCACTTGATTTTGGAGGG |
Western blot analysis
Antibodies against β-actin (catalogue no., ab8227),
IFN α and β receptor subunit 1 (IFNAR1) (catalogue no., ab45172),
IFNAR2, interferon-stimulated gene factor 3 (ISGF3) (catalogue no.,
ab56070), double-stranded RNA-activated protein kinase R (PKR)
(catalogue no., ab32506) and ribonuclease L (RNase L) (catalogue
no., ab191392) were purchased from Abcam (Cambridge, UK). Cells
were lysed with Cell lysis buffer (Beyotime Institute of
Biotechnology, Shanghai, China) and centrifuged at 10,000 × g at
4°C for 15 min. Supernatant lysates were harvested and the
concentration was determined by Bradford method with Bradford
Protein Assay Kit (Beyotime Institute of Biotechnology, China).
Subsequently, 30 µg of proteins were loaded per lane and subjected
to 10% SDS-PAGE. Proteins in the gel were transferred to a
nitrocellulose membrane and the membrane was blocked with 5% milk
powder at 4°C overnight. Primary antibodies were diluted 1:1,000 in
TBST buffer and incubated for 2 h at room temperature. Following
incubation with primary antibodies, the membrane was washed three
times with TBST. Horseradish peroxidase conjugated secondary
antibody (anti-rabbit IgG) (catalogue no., ab6721; Abcam) was
diluted 1:5,000 in 5% milk powder and incubated for 2 h at room
temperature. The blotting membrane was then washed three times with
TBST. Target bands were detected using ECL regents (Beyotime
Institute of Biotechnology; catalogue no., P0018) and
quantitatively analyzed using Quantity one software version 4.1
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). β-actin was used
as the reference gene.
Detection of cell viability using an
MTT assay
Following transfection, a total of 1×105
Huh-7 cells/well were seeded in a 96-well plate and allowed to
adhere and spread for 24 h. Huh-7 cells in the IFN-α alone and
HBVx-expressing lentivirus + treatment with IFN-α groups were
treated with IFN-α. A total of 10 µl MTT solution (5 mg/ml in PBS)
was subsequently added to the cell medium of cells in all groups
and incubated for 4 h at 37°C. The culture supernatant was
subsequently removed, 100 µl dimethylsulphoxide was added and the
solution was shaken for 10 min. Absorbance was measured at a
wavelength of 490 nm.
Cell scratch test
A total of 1.6×105 Huh-7 cells were
seeded in a 6-well plate and allowed to grow until 80% confluence
was reached. A micropipette tip was used to gently scratch a line
of cells off the plate. The cell plate was washed three times with
PBS to remove any remaining scratched cells. Following treatment,
cells were cultured for an additional 24 h in serum-free DMEM. Cell
migration into the scratch site was measured using an inverted IX81
Olympus microscope (Olympus Corporation of the Americas; Center
Valley, PA, USA) and ImageJ software version 1.41o (National
Institute of Health; Bethesda, MD, USA).
Cell invasion test
A suspension of 0.5×106 cells was placed
in each well of the lower chamber of a 24-well plate. Cells were
cultured in DMEM containing 10% fetal bovine serum. The upper
chamber Matrigel culture insert was then placed on top of the lower
chamber. A total of 1×105 of cells were added on top of
the Transwell membrane in the upper chamber, cultured in serum free
DMEM. Invasion chambers were incubated for 48 h in 5%
CO2 at 37°C. Following incubation, noninvasive cells
were removed by scraping the upper surface of the Matrigel membrane
using a cotton swab. Invading cells on the lower surface of the
membrane were fixed with 4% paraformaldehyde for 15 min at room
temperature, washed with PBS and stained with Giemsa solution for
10 min at room temperature. The stained membrane was photographed
using an inverted IX81 Olympus microscope (Olympus Corporation of
the Americas) and the number of cells was counted.
Statistical analysis
Data are expressed as the mean ± standard deviation
based on three independent biological replicates. Statistical
analysis was performed using SPSS 19.0 (IBM Corp., Armonk, NY,
USA). Student's t-test was used to conduct a pairwise comparison.
Multiple comparisons were tested using one-way analysis of
variance, followed by a Tukey honest significant difference test.
P<0.05 was determined to indicate statistically significant
difference.
Results
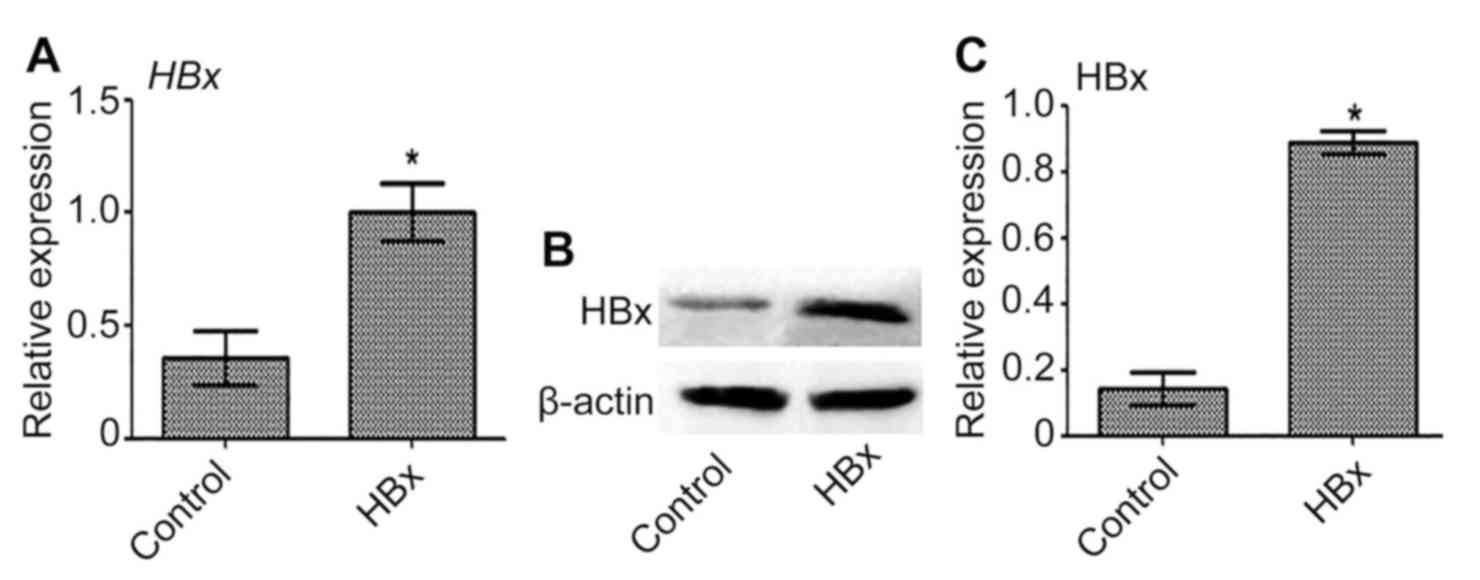
Transfection of Huh-7 cells with
HBx-expressing lentivirus
Transfection of Huh-7 cells with HBx-expressing
lentivirus was performed to establish a novel HBV-related liver
cancer model (Huh-7-HBx). The expression of HBx mRNA and protein in
Huh-7-HBx cells was significantly upregulated compared with the
control (P<0.05; Fig. 1).
Therefore, HBx was successfully overexpressed in Huh-7 cells.
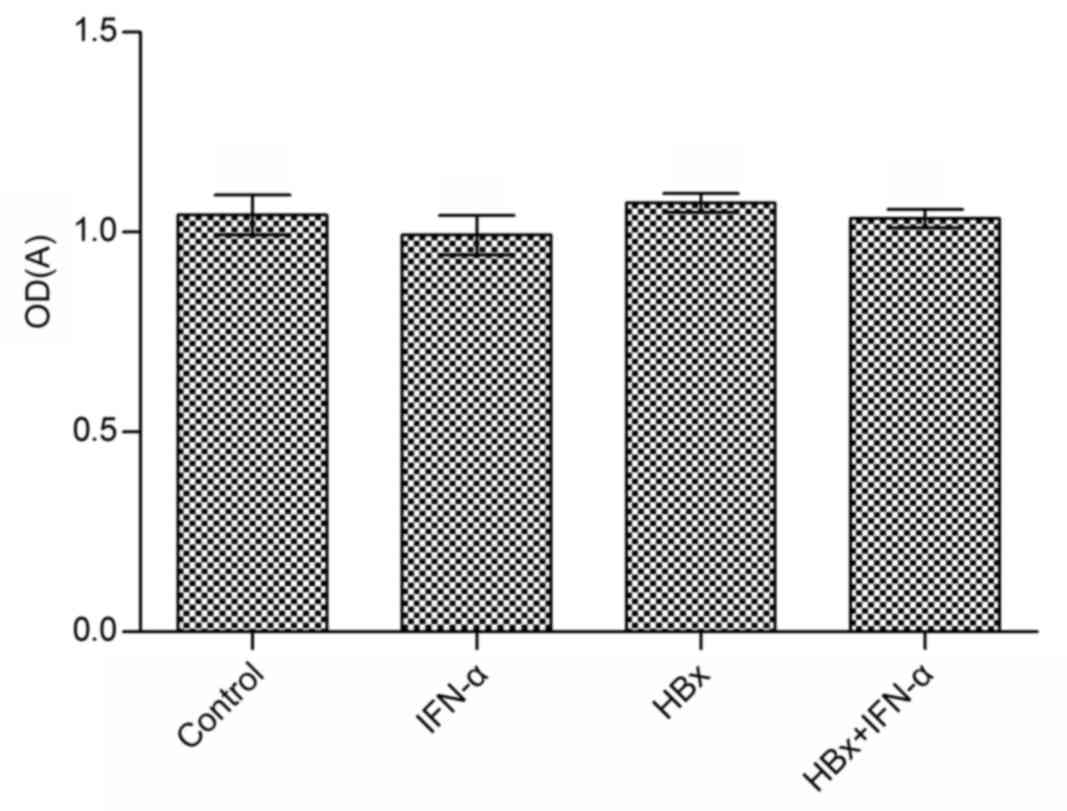
Cell viability is not affected by the
transfection of Huh-7 cells with HBx-expressing lentivirus or by
treatment with IFN-α
Huh-7 cell viability was measured using an MTT assay
following the transfection of Huh-7 cells with HBx-expressing
lentivirus and IFN-α treatment. The effect of IFN-α treatment alone
on Huh-7 cell viability compared with the control group was not
significant (Fig. 2). Similarly, the
difference in cell viability in the Huh-7-HBx and Huh-7-HBx+IFN-α
treatment groups compared with the control was not significant.
IFN-α treatment only slightly inhibited the viability of Huh-7-HBx
cells. These results suggest that IFN-α does not affect cell
viability in HBV-related liver cancer.
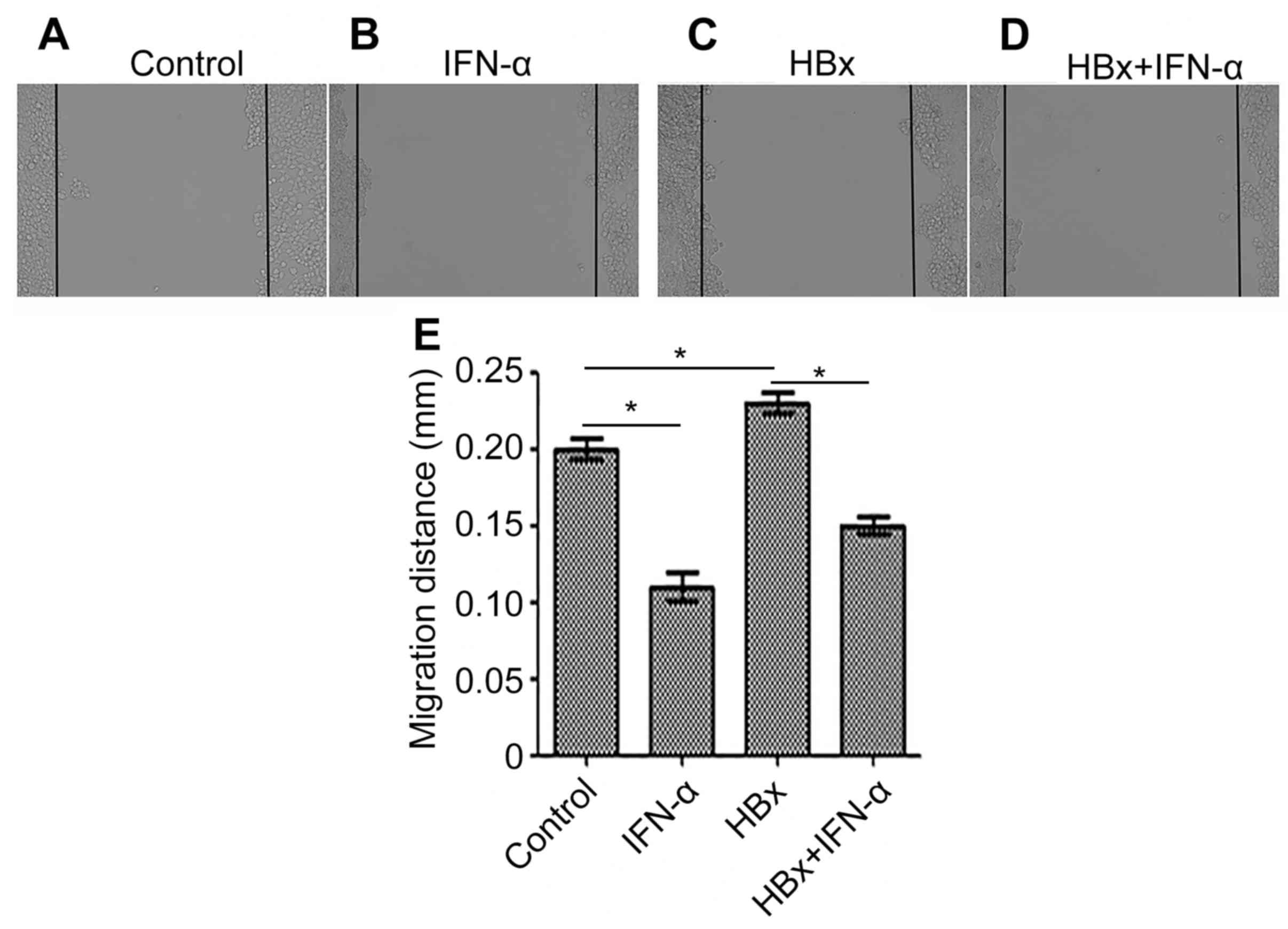
IFN-α inhibits HBx-induced cell
migration in Huh-7 cells
Migration is an important characteristic of cancer
cells; therefore cell migration was examined in the Huh-7-HBx and
Huh-7-HBx+IFN-α treatment groups to determine the curative function
of IFN-α in HBV-related liver cancer (Fig. 3). Cell migration was decreased
(P<0.05) in the IFN-α treatment alone group (Fig. 3B) and increased (P<0.05) in the
Huh-7-HBx group (Fig. 3C) compared
with the control (Fig. 3A). There
was a decrease (P<0.05) in cell migration in the Huh-7-HBx+IFN-α
treatment group (Fig. 3D) compared
with the control, but not to the same extent as the decrease
observed in the IFN-α treatment alone group. Quantitative analysis
of cell migration in each of the groups supported these
observations (Fig. 3E). This
suggests that IFN-α may inhibit cell migration in HBV-related liver
cancer.
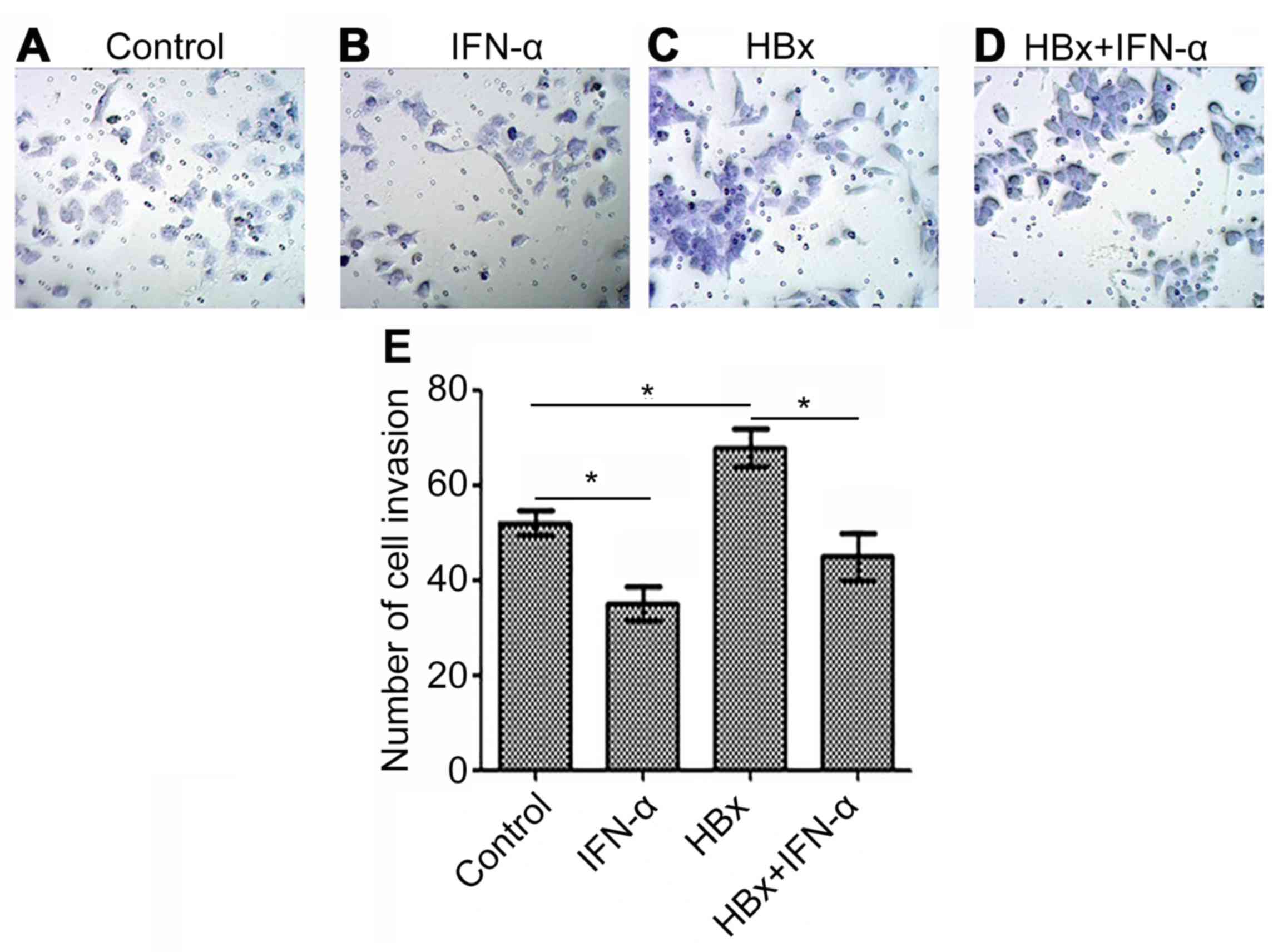
IFN-α inhibits HBx-induced Huh-7 cell
invasion
Cell invasion indicates tumor carcinogenesis, thus
the HBx-regulated liver cancer model was used to determine the
curative function of IFN-α in HBV-related liver cancer (Fig. 4). Huh-7 cell invasion was decreased
(P<0.05) in the IFN-α treatment alone group (Fig. 4B) and increased (P<0.05) in the
Huh-7-HBx group (Fig. 4C) compared
with the control (Fig. 4A). There
was a decrease (P<0.05) in cell invasion in the Huh-7-HBx+IFN-α
treatment group (Fig. 4D) compared
with the control, but not to the same extent as the decrease
observed in the IFN-α treatment alone group. Quantitative analysis
of cell invasion was consistent with these results (Fig. 4E). This indicates that IFN-α may
reduce cell invasion in HBV-related liver cancer.
IFN-α promotes the expression of
antiviral genes during transcription and translation in Huh-7-HBx
cells
IFNAR1, IFNAR2, ISGF3, PKR and RNase L are important
antiviral genes that exhibit anti-HBV effects (21–25).
Therefore, the expression of the antiviral genes was examined in a
HBx-regulated liver cancer model to determine the curative function
of IFN-α in HBV-related liver cancer. RT-qPCR demonstrated that the
expression of IFNAR1 (Fig. 5A),
IFNAR2 (Fig. 5B), PKR (Fig. 5C), RNaseL (Fig. 5D) and ISGF3 (Fig. 5E) mRNA was significantly increased
(P<0.05) in the IFN-α treatment only group compared to the
control. mRNA levels of these antiviral genes were upregulated
(P<0.05) in the Huh-7-HBx group compared with the control group
however, not to the extent of the increase demonstrated in the
IFN-α treatment only group. The most significant increase in
expression of antiviral gene mRNA, compared with the control, was
in the Huh-7-HBx+IFN-α treatment group. The protein expression of
these antiviral genes was consistent with this (Fig. 5F-K). These results suggest that IFN-α
increases the mRNA and protein levels of antiviral genes in
HBV-related liver cancer.
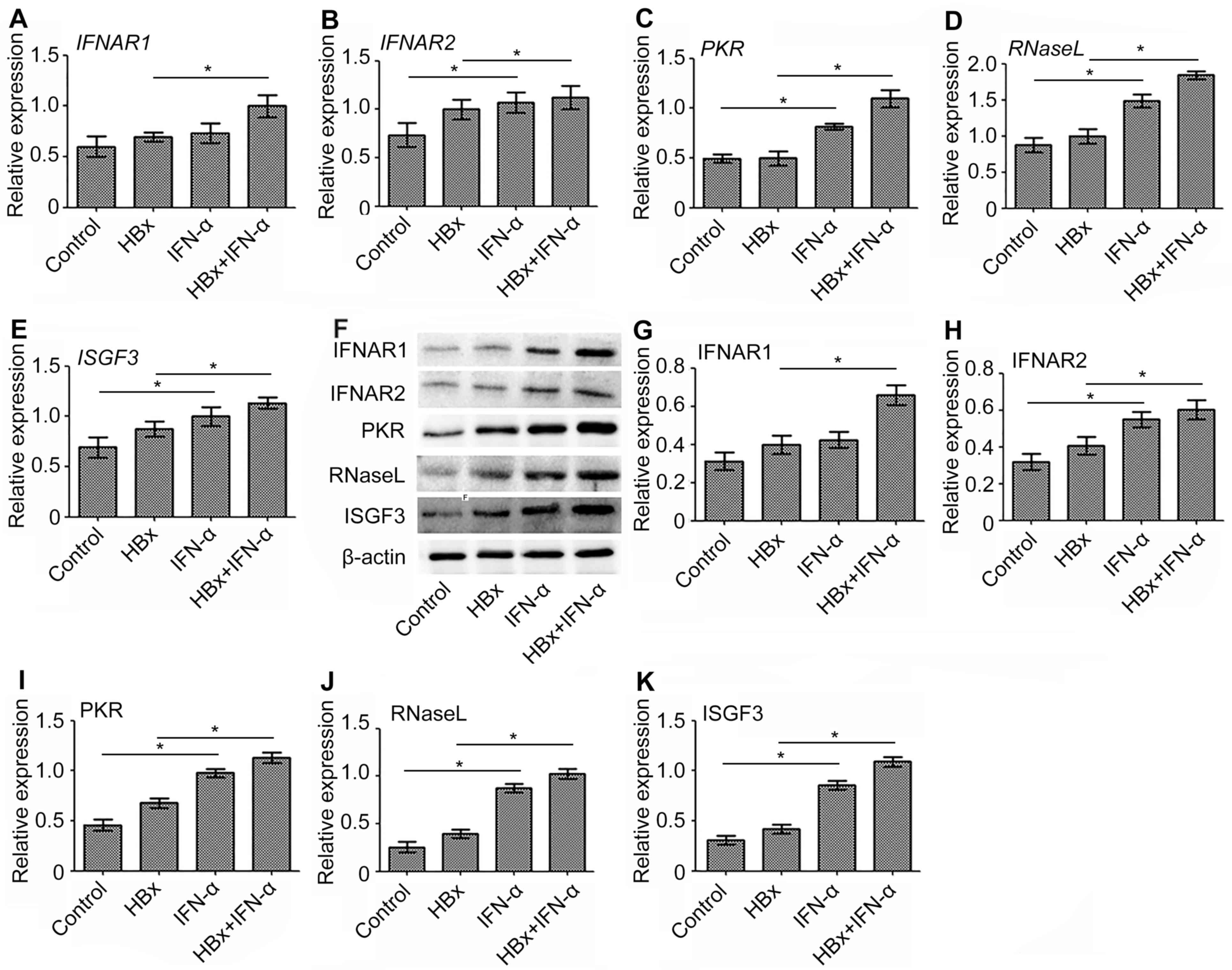 | Figure 5.Expression of antiviral gene mRNA and
protein following the transfection of Huh-7 cells with
HBx-expressing lentivirus and subsequent IFN-α treatment. The
expression of (A) IFNAR1, (B) IFNAR2, (C) PKR, (D) RNaseL, (E)
ISGF3 mRNA was determined using reverse transcription-quantitative
polymerase chain reaction. The protein expression of antiviral
genes was detected using (F) western blot analysis and quantified
for (G) IFNAR1, (H) IFNAR2, (I) PKR, (J) RNaseL and (K) ISGF3 using
Quantity one software. Data are expressed as the mean ± standard
deviation based on three independent biological replicates.
Statistically significant differences were determined using
analysis of variance, followed by the Tukey honest significant
difference test. *P<0.05 as indicated. HBx, hepatitis B X
protein; IFN-α, interferon-α; IFNAR1, interferon α and β receptor
subunit 1; IFNAR2, interferon α and β receptor subunit 2; PKR,
double-stranded RNA-activated protein kinase R; RNaseL,
ribonuclease L; ISGF3, interferon-stimulated gene factor 3. |
Discussion
HBV infection is a major cause of primary liver
cancer (12,26). The present study demonstrated that
the transfection of Huh-7 cells with HBx, a protein encoded by HBV
DNA, had no effect on cell viability but promoted cell migration
and invasion, which is consistent with the results of a previous
study (27).
IFN-α is currently used as a first-line antiviral
drug to treat chronic hepatitis B (CHB) (28,29). It
is an effective treatment of HBV due to its antiviral function and
immunomodulatory effects (30–34).
IFN-α does not directly kill or inhibit HBV; however, its antiviral
effect is facilitated by its binding to the cell membrane receptor
IFNAR1, which leads to the production of antiviral proteins that
inhibit HBV replication (35,36).
Therefore, IFNAR1 serves a role in the progression of CHB. The
binding of IFN-α to its receptor IFNAR1 activates Janus kinase-JAK1
and non-receptor tyrosine-protein kinase TYK2, which leads to the
phosphorylation of signal transducer and activator of transcription
(STAT) 1 and STAT2 (37,38). STAT1 and STAT2 then form a
heterodimer and bind to interferon regulatory factor 9 (IRF-9) to
form ISGF3. ISGF3 translocates from the cytoplasm to the nucleus
and binds to the IFN stimulated regulatory element to promote the
transcription of antiviral genes, such as PKR (39–41).
Previous studies have demonstrated that following treatment with
IFN-α, the expression of STAT2, IRF-9, and PKR is significantly
increased in HepG2 and HepG2.2.15 cells (42,43).
There was also a decrease in HBV DNA titer in HepG2.2.15 cells,
which suggests that the JAK-STAT pathway serves a major role in
IFN-α-inhibited HBV replication. Furthermore, the expression of
ISGF3 protein and PKR mRNA was significantly decreased following
inhibition of the IFN pathway and the titer of HBV DNA in the
supernatant of HepG2.2.15 cells was not significantly decreased.
These results indicate that ISGF3 is an important regulatory factor
of the pathway (25,44).
IFN-α activates the JAK-STAT signaling pathway by
binding to IFNARs on the cell surface, thereby facilitating the
transcription and expression of antiviral genes, including PKR
(45) and RNaseL (46). The present study demonstrated that
IFN-α significantly increases the expression of the antiviral genes
IFNAR1, IFNAR2, PKR, RNaseL and ISGF3 in Huh-7 cells transfected
with HBx-expressing lentivirus, suggesting that IFN-α may be
developed as a novel therapeutic strategy to treat patients with
HBV-related liver cancer.
In conclusion, the current study suggests that IFN-α
attenuates the development of HBV-related liver cancer by reducing
cell migration and invasion, as well as upregulating the expression
of antiviral proteins.
Acknowledgements
The present study was supported by the Specific Fund
of Clinical Medical Research of the Chinese Medical Association
(grant no. 14040350572; Beijing, China).
References
|
1
|
Lavanchy D: Hepatitis B virus
epidemiology, disease burden, treatment, and current and emerging
prevention and control measures. J Viral Hepat. 11:97–107. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liaw YF and Chu CM: Hepatitis B virus
infection. Lancet. 373:582–592. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hooshyar A, Habibzadeh S, Ghasemi N,
Yazdanbod A, Sohrabi S, Maleki N and Amani F: Females have a lower
liver histopathological score in HBeAg-negative chronic hepatitis B
than males. Arch Clin Infect Dis. 8:e179722013. View Article : Google Scholar
|
|
4
|
Yang N and Bertoletti A: Advances in
therapeutics for chronic hepatitis B. Hepatol Int. 10:277–285.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen J, Zhao J, Chen L, Dong N, Ying Z,
Cai Z, Ji D, Zhang Y, Dong L, Li Y, et al: STAT1 modification
improves therapeutic effects of interferons on lung cancer cells. J
Transl Med. 13:2932015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gerber SA, Yatsula B, Maier CL, Sadler TJ,
Whittaker LW and Pober JS: Interferon-gamma induces prolyl
hydroxylase (PHD)3 through a STAT1-dependent mechanism in human
endothelial cells. Arterioscler Thromb Vasc Biol. 29:1363–1369.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rehermann B and Bertoletti A:
Immunological aspects of antiviral therapy of chronic hepatitis B
virus and hepatitis C virus infections. Hepatology. 61:712–721.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bowick GC, Airo AM and Bente DA:
Expression of interferon-induced antiviral genes is delayed in a
STAT1 knockout mouse model of Crimean-Congo hemorrhagic fever.
Virol J. 9:1222012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
von Marschall Z, Scholz A, Cramer T,
Schäfer G, Schirner M, Oberg K, Wiedenmann B, Höcker M and Rosewicz
S: Effects of interferon alpha on vascular endothelial growth
factor gene transcription and tumor angiogenesis. J Natl Cancer
Inst. 95:437–448. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fazio N and Oberg K: Prospective,
randomized, multicenter trial on the antiproliferative effect of
lanreotide, interferon alfa and their combination for therapy of
metastatic neuroendocrine gastroenteropancreatic tumors. J Clin
Oncol. 22:573–575. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
De Palma M, Mazzieri R, Politi LS, Pucci
F, Zonari E, Sitia G, Mazzoleni S, Moi D, Venneri MA, Indraccolo S,
et al: Tumor-targeted interferon-alpha delivery by Tie2-expressing
monocytes inhibits tumor growth and metastasis. Cancer Cell.
14:299–311. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kim JY, Song EH, Lee HJ, Oh YK, Choi KH,
Yu DY, Park SI, Seong JK and Kim WH: HBx-induced hepatic steatosis
and apoptosis are regulated by TNFR1- and NF-kappaB-dependent
pathways. J Mol Biol. 397:917–931. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang HY, Yang SL, Liang HF and Li CH: HBx
protein promotes oval cell proliferation by up-regulation of cyclin
D1 via activation of the MEK/ERK and PI3K/Akt pathways. Int J Mol
Sci. 15:3507–3518. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Murakami S: Hepatitis B virus X protein: A
multifunctional viral regulator. J Gastroenterol. 36:651–660. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim CM, Koike K, Saito I, Miyamura T and
Jay G: HBx gene of hepatitis B virus induces liver cancer in
transgenic mice. Nature. 351:317–320. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lara-Pezzi E, Gomez-Gaviro MV, Galvez BG,
Mira E, Iñiguez MA, Fresno M, Martínez AC, Arroyo AG and
López-Cabrera M: The hepatitis B virus X protein promotes tumor
cell invasion by inducing membrane-type matrix metalloproteinase-1
and cyclooxygenase-2 expression. J Clin Invest. 110:1831–1838.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tanaka Y, Kanai F, Kawakami T, Tateishi K,
Ijichi H, Kawabe T, Arakawa Y, Kawakami T, Nishimura T, Shirakata
Y, et al: Interaction of the hepatitis B virus X protein (HBx) with
heat shock protein 60 enhances HBx-mediated apoptosis. Biochem
Biophys Res Commun. 318:461–469. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang X, Liu S, Hu T, Liu S, He Y and Sun
S: Up-regulated microRNA-143 transcribed by nuclear factor kappa B
enhances hepatocarcinoma metastasis by repressing fibronectin
expression. Hepatology. 50:490–499. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lizee G, Aerts JL, Gonzales MI, Chinnasamy
N, Morgan RA and Topalian SL: Real-time quantitative reverse
transcriptase-polymerase chain reaction as a method for determining
lentiviral vector titers and measuring transgene expression. Hum
Gene Ther. 14:497–507. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Frodsham AJ, Zhang L, Dumpis U, Taib NA,
Best S, Durham A, Hennig BJ, Hellier S, Knapp S, Wright M, et al:
Class II cytokine receptor gene cluster is a major locus for
hepatitis B persistence. Proc Natl Acad Sci USA. 103:pp. 9148–9153.
2006, View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou J, Smith DK, Lu L, Poon VK, Ng F,
Chen DQ, Huang JD, Yuen KY, Cao KY and Zheng BJ: A non-synonymous
single nucleotide polymorphism in IFNAR1 affects susceptibility to
chronic hepatitis B virus infection. J Viral Hepat. 16:45–52. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Han Q, Zhang C, Zhang J and Tian Z:
Involvement of activation of PKR in HBx-siRNA-mediated innate
immune effects on HBV inhibition. PLoS One. 6:e279312011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Park IH, Kwon YC, Ryu WS and Ahn BY:
Inhibition of hepatitis B virus replication by ligand-mediated
activation of RNase L. Antiviral Res. 104:118–127. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Q, Wang Y, Wei L, Jiang D, Wang JH,
Rao HY, Zhu L, Chen HS, Fei R and Cong X: Role of ISGF3 in
modulating the anti-hepatitis B virus activity of interferon-alpha
in vitro. J Gastroenterol Hepatol. 23:1747–1761. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Perz JF, Armstrong GL, Farrington LA,
Hutin YJ and Bell BP: The contributions of hepatitis B virus and
hepatitis C virus infections to cirrhosis and primary liver cancer
worldwide. J Hepatol. 45:529–538. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kong J, Kong F, Gao J, Zhang Q, Dong S, Gu
F, Ke S, Pan B, Shen Q, Sun H, et al: YC-1 enhances the anti-tumor
activity of sorafenib through inhibition of signal transducer and
activator of transcription 3 (STAT3) in hepatocellular carcinoma.
Mol Cancer. 13:72014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Terrault NA, Bzowej NH, Chang KM, Hwang
JP, Jonas MM and Murad MH: American Association for the Study of
Liver Diseases: AASLD guidelines for treatment of chronic hepatitis
B. Hepatology. 63:261–283. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yuen MF, Ahn SH, Chen DS, Chen PJ,
Dusheiko GM, Hou JL, Maddrey WC, Mizokami M, Seto WK, Zoulim F, et
al: Chronic hepatitis b virus infection: Disease revisit and
management recommendations. J Clin Gastroenterol. 50:286–294. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Belloni L, Allweiss L, Guerrieri F,
Pediconi N, Volz T, Pollicino T, Petersen J, Raimondo G, Dandri M
and Levrero M: IFN-α inhibits HBV transcription and replication in
cell culture and in humanized mice by targeting the epigenetic
regulation of the nuclear cccDNA minichromosome. J Clin Invest.
122:529–537. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lai CL and Yuen MF: Prevention of
hepatitis B virus-related hepatocellular carcinoma with antiviral
therapy. Hepatology. 57:399–408. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Robek MD, Boyd BS and Chisari FV: Lambda
interferon inhibits hepatitis B and C virus replication. J Virol.
79:3851–3854. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Piratvisuth T: Reviews for APASL
guidelines: Immunomodulator therapy of chronic hepatitis B. Hepatol
Int. 2:140–146. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sprengers D and Janssen HL:
Immunomodulatory therapy for chronic hepatitis B virus infection.
Fundam Clin Pharmacol. 19:17–26. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Uzé G, Di Marco S, Mouchel-Vielh E,
Monneron D, Bandu MT, Horisberger MA, Dorques A, Lutfalla G and
Mogensen KE: Domains of interaction between alpha interferon and
its receptor components. J Mol Biol. 243:245–257. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Peltekian C, Gordien E, Garreau F,
Meas-Yedid V, Soussan P, Willams V, Chaix ML, Olivo-Marin JC,
Bréchot C and Kremsdorf D: Human MxA protein participates to the
interferon-related inhibition of hepatitis B virus replication in
female transgenic mice. J Hepatol. 43:965–972. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Su L and David M: Distinct mechanisms of
STAT phosphorylation via the interferon-alpha/beta receptor.
Selective inhibition of STAT3 and STAT5 by piceatannol. J Biol
Chem. 275:12661–12666. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rani MR, Leaman DW, Han Y, Leung S, Croze
E, Fish EN, Wolfman A and Ransohoff RM: Catalytically active TYK2
is essential for interferon-beta-mediated phosphorylation of STAT3
and interferon-alpha receptor-1 (IFNAR-1) but not for activation of
phosphoinositol 3-kinase. J Biol Chem. 274:32507–32511. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ward SV and Samuel CE: The PKR kinase
promoter binds both Sp1 and Sp3, but only Sp3 functions as part of
the interferon-inducible complex with ISGF-3 proteins. Virology.
313:553–566. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
George CX, Das S and Samuel CE:
Organization of the mouse RNA-specific adenosine deaminase Adar1
gene 5′-region and demonstration of STAT1-independent,
STAT2-dependent transcriptional activation by interferon. Virology.
380:338–343. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Goh KC, Haque SJ and Williams BR: p38 MAP
kinase is required for STAT1 serine phosphorylation and
transcriptional activation induced by interferons. EMBO J.
18:5601–5608. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang X and Chan C: Repression of PKR
mediates palmitate-induced apoptosis in HepG2 cells through
regulation of Bcl-2. Cell Res. 19:469–486. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chai Y, Huang HL, Hu DJ, Luo X, Tao QS,
Zhang XL and Zhang SQ: IL-29 and IFN-α regulate the expression of
MxA, 2′,5′-OAS and PKR genes in association with the activation of
Raf-MEK-ERK and PI3K-AKT signal pathways in HepG2.2.15 cells. Mol
Biol Rep. 38:139–143. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Guan SH, Lu M, Grünewald P, Roggendorf M,
Gerken G and Schlaak JF: Interferon-alpha response in chronic
hepatitis B-transfected HepG2.2.15 cells is partially restored by
lamivudine treatment. World J Gastroenterol. 13:228–235. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Mathews JD, McCaw CT, McVernon J, McBryde
ES and McCaw JM: A biological model for influenza transmission:
Pandemic planning implications of asymptomatic infection and
immunity. PLoS One. 2:e12202007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ren S, Yu H, Zhang H, Liu Y, Huang Y, Ma
L, Wei L, Wu H and Chen XY: Polymorphisms of interferon-inducible
genes OAS associated with interferon-α treatment response in
chronic HBV infection. Antiviral Res. 89:232–237. 2011. View Article : Google Scholar : PubMed/NCBI
|