Introduction
Cartilage defects seriously affect articulation, and
without their self-restoration capability even a minor cartilage
defect may be difficult to repair (1). In addition, articular cartilage defect
repair remains a big problem. Although some progress has been made,
current treatment options, including articular debridement, Pridie
drilling technique, microfracture surgery, tissue engineered
cartilage repair and periosteum, perichondrium, osteochondral and
cell transplantation are not of high quality due to poor efficacy,
high cost and limited resources, thus longer dated curative effects
remain to be found (2–6). The development of new biological
materials may assistant effective cartilage repair, however, in
order to improve our understanding of cartilage material bioactive
function, a selection of ideal seed cells and the promotion of cell
chondrogenic differentiation are particularly important (7,8).
Mesenchymal stem cells (MSCs) with self-renewal,
pluripotency and lower immunogenicity are considered to be ideal
biological material as seed cells (9). One type of MSC derived from mouse
embryos is the C3H10T1/2 cell line that has widely been used in
studies that concentrate on the differentiation and regulation of
stem cells (10–13).
MicroRNAs (miRNAs) are small non-coding
(~22-nucleotide) RNAs that exist in eukaryotes. The biological
functions of miRNA are diverse. For example, one miRNA can control
a number of different target genes and there are also several
miRNAs that together control one target gene (1). As a peculiar and endogenous small
molecule, miRNA has become a powerful tool in the field of genetic
engineering. Increasingly, studies indicate that miRNAs reveal stem
cell fate and regulation patterns of epigenetics in biological
behavior (2,3,14–17).
In the present study, miRNAs that significantly
changed during the differentiation of C3H10T1/2 cells toward
chondrocytes were screened. One of these was a novel miRNA,
miR-30b, whose expression was significantly decreased during
chondrogenic differentiation. The present study revealed that
miR-30b inhibited the differentiation of C3H10T1/2 cells towards
chondrocytes by modulating its expression, which indicates a
possible negative feedback loop. Furthermore, SOX9 was verified as
a direct target gene of miR-30b. Finally, the results of the
present study confirmed a previously uncharacterized function for
miR-30b as an inhibitor of chondrogenic differentiation.
Materials and methods
Cell culture
The mouse embryo-derived stem cells C3H10T1/2 were
purchased from American Type Culture Collection (Manassas, VA,
USA). C3H10T1/2 cells were cultured in low glucose Dulbecco's
modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS;
Thermo Fisher Scientific, Inc.) and incubated in a 5%
CO2 incubator at 37°C. Cells were then passaged until a
90% confluence level was reached. The seventh generation cells were
then used for establishing the pellet models of chondrogenic
differentiation in vitro. For chondrogenic differentiation,
C3H10T1/2 cells were cultured in pellets (2.5×105
cells/ml), and the culture medium was replaced every 2–3 days.
Pellet models were then divided into a non-induced and an induced
group. The pellets in the non-induced group were treated with
non-induced solution (500 ml high-glucose DMEM with 20% FBS) while
those in the induced group were treated with cartilage-induced
solution (500 ml high-glucose DMEM with 5 µg transforming growth
factor-β3 (TGF-β3) (Thermo Fisher Scientific, Inc.), 0.15 g vitamin
C, 50 µl dexamethasone, 0.5 ml Insulin/Transferrin/Selenium
solution and 20% FBS).
microRNA (miRNA) array analysis
Total RNA of the induced and non-induced groups of
chondrogenic differentiation models were cultured from C3H10T1/2
for 14 days in vitro, including miRNAs. These were extracted
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
following the manufacturer's protocol. NanoDrop® ND-1000
(Thermo Fisher Scientific, Inc.,) was used to validate the overall
quality of the total RNA and the RNAs that passed were used to
construct small RNA libraries using a TruSeq small RNA sample
preparation kit (Illumina Inc., San Diego, CA, USA). In addition,
the quality and fragment size of the small RNA libraries were
detected using Qubit 2.0 (Thermo Fisher Scientific, Inc.,) and
BioAnalyzer 2100 (Agilent Technologies, Inc., Santa Clara, CA,
USA), respectively. Subsequently, HiSeq 2500 (Illumina Inc.) was
performed to sequence the library and the sequencing results were
eventually confirmed by quantitative polymerase chain reaction
(qPCR). Following the quality control the original sequencing data
was pretreated by trim_galore (v0.3.3; Babraham Bioinformatics,
Babraham Institute, Cambridge, UK). The expression pattern of small
RNAs was statistically analyzed, classified and annotated further.
In addition, clustering analysis was performed to classify the
differentially expressed miRNAs between induced and non-induced
groups using a TruSeq SR Cluster Generation kit (cat no.
GD-300-2001, Illumina Inc.).
qPCR analysis
qPCR was performed to confirm the expression pattern
of miR-30b, which emerged in the microarray results and U6 was used
as an internal control. The total RNA was extracted from cell lines
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
following the manufacturer's protocol. Next, cDNA was generated
from total RNA using a qPCR detection kit (Takara Bio, Inc., Otsu,
Japan) according to manufacturer's protocol. Subsequently, the cDNA
was analyzed by qPCR using the following amplification parameters:
95°C for 10 min, followed by 40 cycles of 95°C for 10 sec and 60°C
for 60 sec. All the primers were designed by Shanghai Sangon
Biotech Co., Ltd. (Shanghai, China). The primers of qPCR were as
follows: U6, 5′CGC TTC ACG AAT TTG CGT GTC AT3′ and miR-30b, 5′GTC
GTA TCC AGT GCA GGG TCC GAG GTA TTC GCA CTG GAT ACG ACA GCT
GA3′.
The primers of qPCR are as follows: U6, forward,
5′GCT TCG GCA GCA CAT ATA CTA AAA T3′and U6, reverse, 5′CGC TTC ACG
AAT TTG CGT GTC AT3′ and miR-30b, forward, 5′GCG GCG GTG TAA ACA
TCC TAC AC3′ and miR-30b, reverse, 5′ATC CAG TGC AGG GTC CGA GG3′.
The 2−ΔΔCq method was used for the quantification of
gene expression (18).
Plasmid construction and
dual-luciferase reporter assays
Bioinformatics software (http://www.targetscan.org and http://www.microrna.org) was used to predict the
target gene of miR-30b and results suggested that SOX9 was a
potential target of miR-30b. To confirm the prediction,
miR-30b-SOX9-WT 3′untranslated region (UTR) was amplified using a
2X PCR Master mix (Fermentas; Thermo Fisher Scientific, Inc.) and
cloned into the psiCHECK-2 reporter plasmid (Shanghai Sangon
Biotech Co., Ltd.) using primers carrying restriction sites for
XhoI and NotI. Mutated miR-30b-SOX9 3′UTR was
generated by mutating the miR-30b seed sequence. The primers used
were as follows: SOX9-WT, forward,
5′-CCGCTCGAGACAAACTGGAAACCTGTCTCTC-3′ and SOX9-WT, reverse,
5′-ATTTGCGGCCGCTTAATGCAATGTATATTTATTGTAAACAATAATATACAA3′ and
SOX9-MUT, forward, 5′-CCGCTCGAGACAAACTGGAAACCTGTCTCTC-3′ and
SOX9-MUT, reverse,
5′-TTTGCGGCCGCTTAATGCAATGTATATTTATTCATATCTATAATATACAAAAAAGAAAG-3′.
In addition, the luciferase activity was assessed after
transfection of C3H10T1/2 cells using the dual-luciferase reporter
assay system (Promega Corporation, Madison, WI, USA) using the
pMIRREPORT luciferase reporter according to the manufacturer's
protocol. The luciferase activity was expressed as the mean ratio
of firefly luciferase to Renilla luciferase activity.
Cell transfection
To investigate the function of miR-30b, miR-30b
mimics, miR-30b inhibitor (600 pmol) or their negative controls
were transfected into C3H10T1/2 cells with 30 µl Lipofectamine 2000
transfection reagent (Invitrogen; Thermo Fisher Scientific Inc.)
following the manufacturer's protocol. Following incubation for 24
h, the transfected cells were used for chondrogenic differentiation
analysis, and cells were harvested for protein analysis at on days
1, 7 and 14, respectively.
Western blot analysis
Radioimmunoprecipitation assay lysis buffer (50 mM
Tris-Cl (pH 7.4), 150 mM NaCl, 1% Triton X-100, 0.1% SDS and 1%
sodium deoxycholate) was used to lyse the micromass cultured
C3H10T1/2 cells transfected with miR-30b mimics or miR-30b
inhibitor for SDS-PAGE analysis. The protein concentration was
measured using a bicinchoninic acid assay protein quantitative kit
(Thermo Fisher Scientific, Inc.) and protein samples were mixed
with 5X loading buffer (in a 1:4 ratio) and boiled for 6 min. The
protein samples (25 µg) were then resolved on a 12% SDS-PAGE gel
and transferred onto a polyvinylidene fluoride membrane (EMD
Millipore, Billerica, MA, USA). They were then incubated with a
primary antibody against SOX9 (anti-SOX9 antibody; cat no. Ab3697;
dilution: 1:1,000) or α-tubulin (anti-α-tubulin antibody; cat no.
Ab4074; dilution: 1:2,000) at 4°C overnight followed by incubation
with an horse radish peroxidase-conjugated secondary antibody
(anti-Rabbit immunoglobulin G VHH Single Domain, cat no. Ab191866;
dilution: 1:5,000; all Abcam, Cambridge, UK) at room temperature
for 1 h. α-tubulin was used as an internal control. Following
washing with Tris-buffered saline and Tween-20, enhanced
chemiluminescence was used for detection and they were developed
using X-ray film, The ImageTool version 3.0 gray-scale scanning
software (Microsoft Corporation, Redmond, WA, USA) was used to
quantify band density.
Alcian blue stain
To detect the deposition of cartilage matrix
proteoglycans, cells were collected on day 7 of induction and
Alcian blue staining was performed to measure sulfated cartilage
glycosaminoglycans. The pellets for Alcian blue staining were fixed
by 4% paraformaldehyde for 20 min, then dehydrated and paraffin
imbedded. Next, 4 µm sections were stained by 0.5% alcian blue 8GX
(Merck KGaA, Darmstadt, Germany) for 10 min.
Cell proliferation assay
The proliferation of C3H10T1/2 cells was detected by
directly counting cell numbers. miR-30b mimics, miR-30b inhibitor
and their negative controls were transfected into C3H10T1/2 cells
for 48 h. Subsequently, the transfected cells were trypsinized
using 0.25% trypsin and directly counted using an automated cell
counter. Each group repeated counting in triplicate.
Statistical analysis
SPSS 17.0 statistical software (SPSS, Chicago, IL,
United States) was used for all statistical analyses. Data are
expressed as the mean ± standard deviation. Statistical comparisons
were made between the two groups with the t-test and between
multiple groups with one-way analysis of variance. P<0.05 was
used to indicate a statistically significant difference.
Results
miR-30b is downregulated in C3H10T1/2
cells during TGF-β3-induced chondrogenic differentiation
In the present study, miRNA microarray technology
was applied to screen the upregulated and downregulated miRNAs
during in vitro chondrogenic differentiation of C3H10T1/2
cells. Clustering analysis was performed to classify the
differentially expressed miRNAs between the induced and non-induced
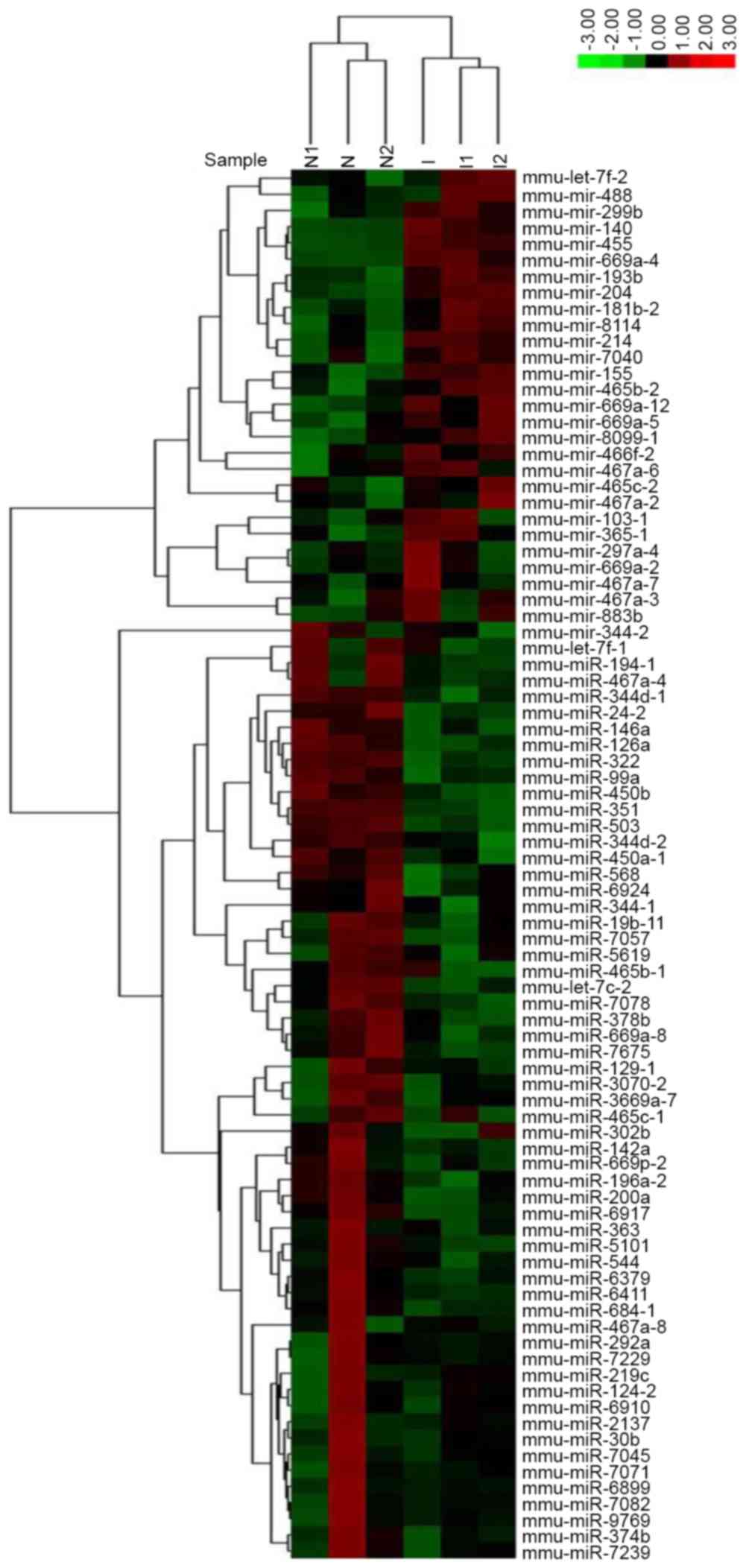
groups. A hierarchical clustering map demonstrated the
differentially expressed miRNAs (Fig.
1). In addition, 86 miRNAs were identified that showed higher
(28) or lower (58) expression in
induced C3H10T1/2 cells compared to the non-induced cells. Among
the differentially expressed miRNAs, miR-30b was significantly
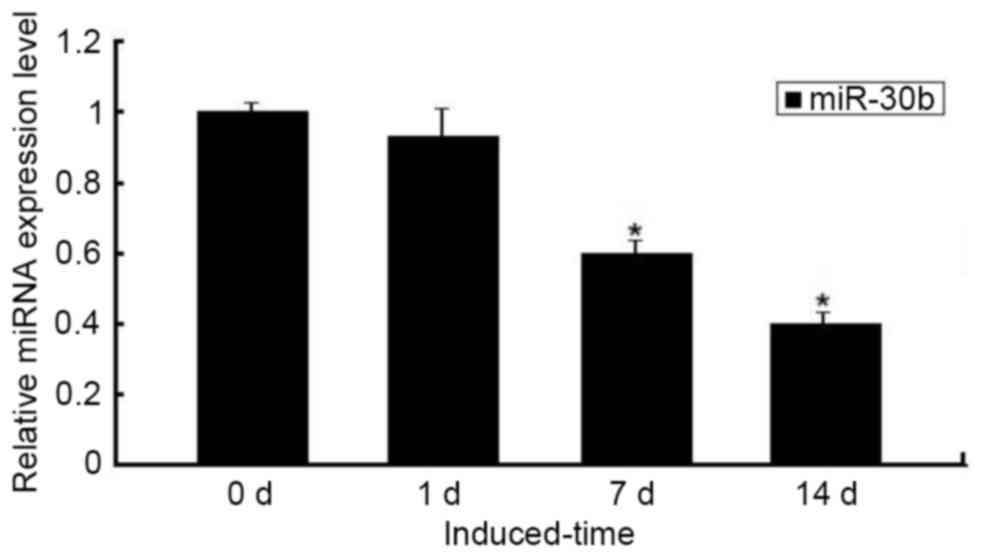
decreased during chondrogenic differentiation. Furthermore, the
downregulated expression of miR-30b was confirmed by qPCR, which
was significant on days 7–14 compared with day 0 (P<0.05;
Fig. 2).
miR-30b targets SOX9 by binding 3′-UTR
of SOX9 mRNA
miRNAs inhibit mRNA expression by binding the 3′-UTR
of target mRNA. Combined with bioinformatics software (http://www.targetscan.org and http://www.microrna.org) and literature analysis, it
was hypothesized that miR-30b may be combined with the SOX9 gene at
the 3′-UTR nucleotide site. In order to determine whether miR-30b
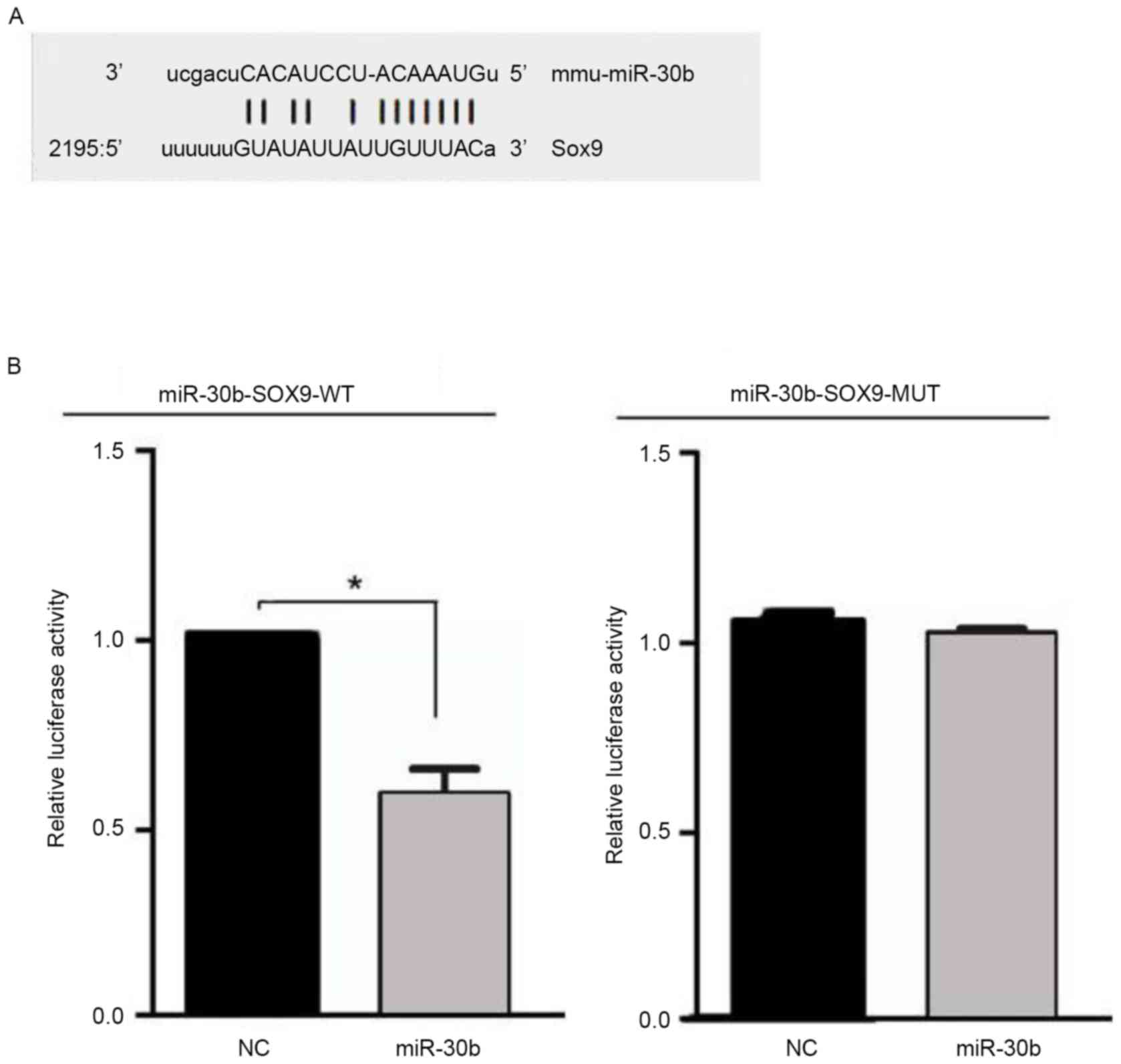
targets SOX9, a luciferase reporter gene assay was performed using
the pMIRREPORT luciferase reporter. Next, the miR-30b-SOX9-WT or
miR-30b-SOX9-MUT reporter plasmid was co-transfected into C3H10T1/2
cells with miR-30b or their negative control, and the results
demonstrated that luciferase activity was significantly decreased
in the C3H10T1/2 cells co-transfected with miR-30b with
miR-30b-SOX9-WT, whereas no effect was observed following
co-transfection of miR-30b with miR-30b-SOX9-MUT (Fig. 3). These data indicate that miR-30b
inhibits expression of transcripts containing an miR-30b binding
site.
miR-30b inhibits SOX9 expression
during chondrogenic differentiation
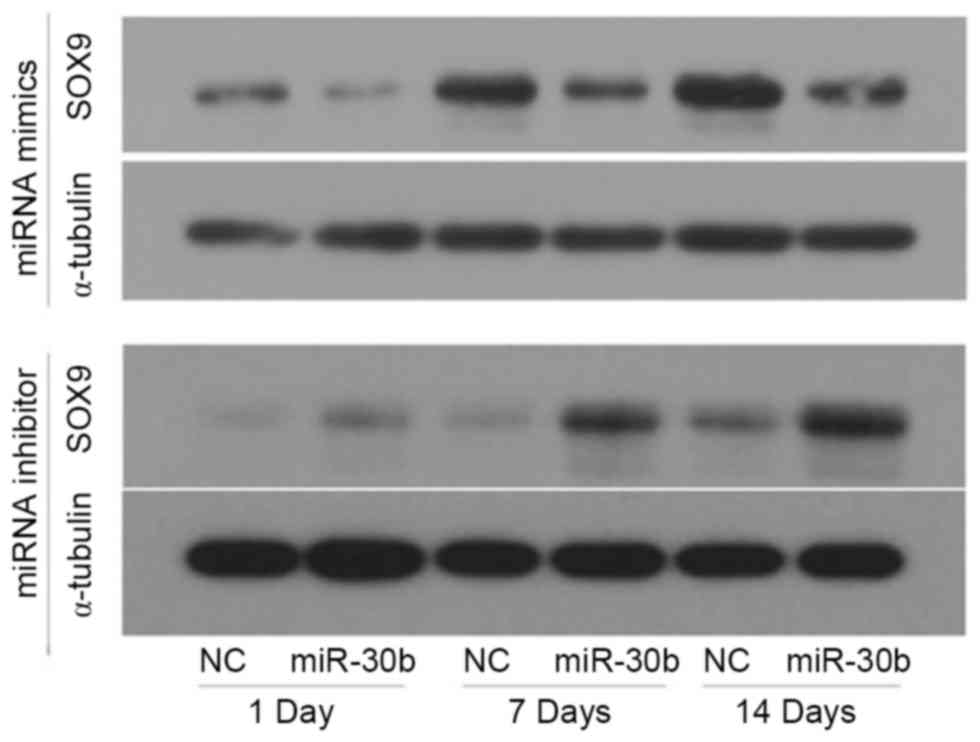
To determine whether miR-30b acts as an inhibitor of
SOX9 protein expression, C3H10T1/2 cells were transfected with
miR-30b mimics, miR-30b inhibitor or their negative controls for 24
h, respectively, then induced by TGF-β3. Furthermore, the
expression of SOX9 was detected using western blotting. Compared to
the negative control, the protein level of SOX9 was notably
decreased in the miR-30b mimics groups on day 1, 7 and 14,
respectively, whereas it was increased in the miR-30b inhibitor
groups (Fig. 4). Furthermore, the
protein expression level of SOX9 increased in a time-dependent
manner during chondrogenic differentiation. These results indicated
that miR-30b is important in suppressing SOX9 protein expression
during chondrogenic differentiation.
miR-30b inhibits early chondrogenic
differentiation
miR-30b mimics, miR-30b inhibitor or their negative
controls were transfected into C3H10T1/2 cells in order to
investigate whether miR-30b exhibited an effect on chondrogenic
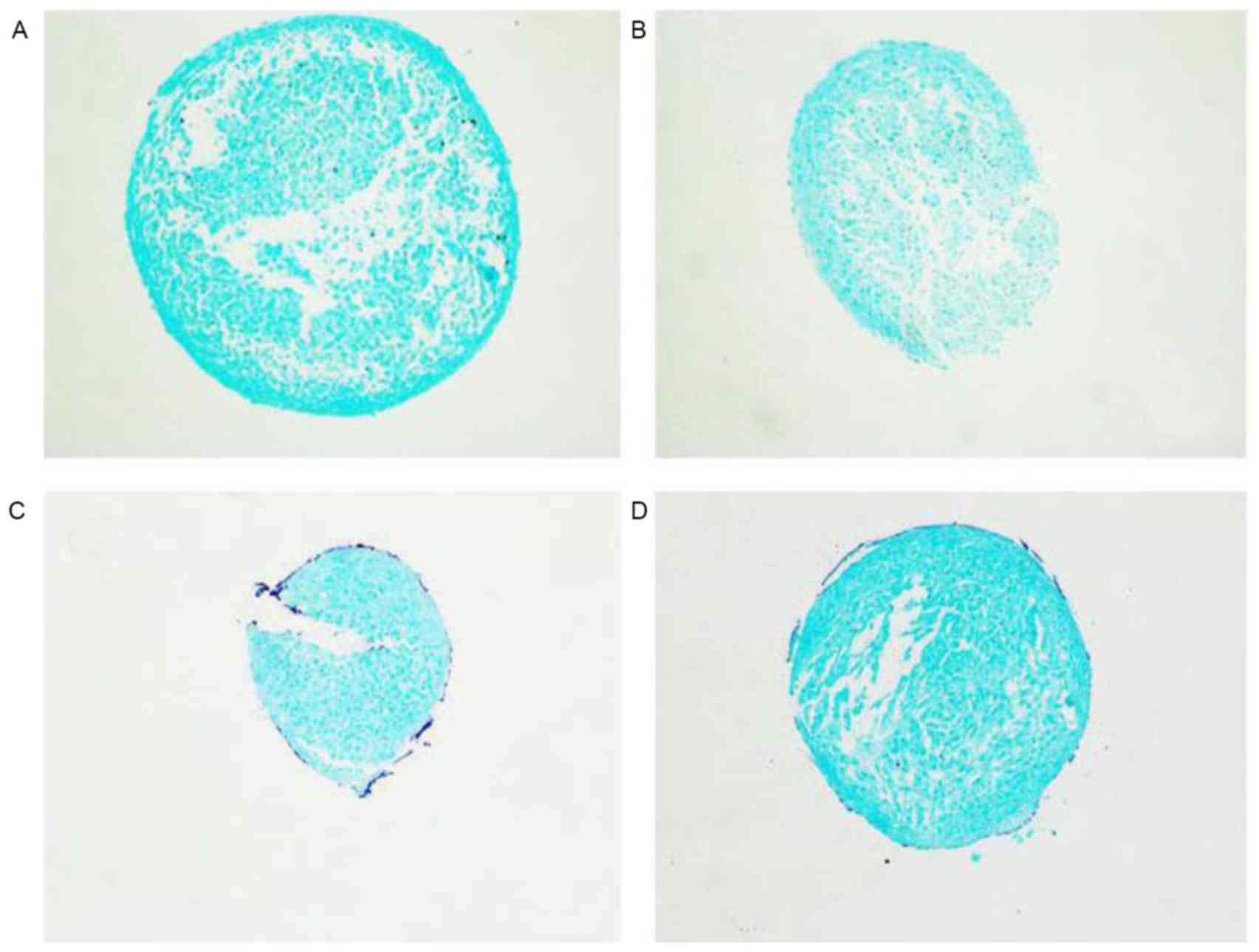
differentiation. Alcian blue staining intensity was detected
following the induction of chondrogenic differentiation by medium
containing TGF-β3 for 7 days. The alcian blue staining intensity
was decreased following miR-30b mimic treatment compared with the
negative group. By contrast, the intensity of the miR-30b inhibitor
treatment group was evidently enhanced (Fig. 5). Overall, the data of the present
study demonstrated that miR-30b suppresses chondrogenic
differentiation by affecting the secretion of characteristic
cartilage in the extracellular matrix.
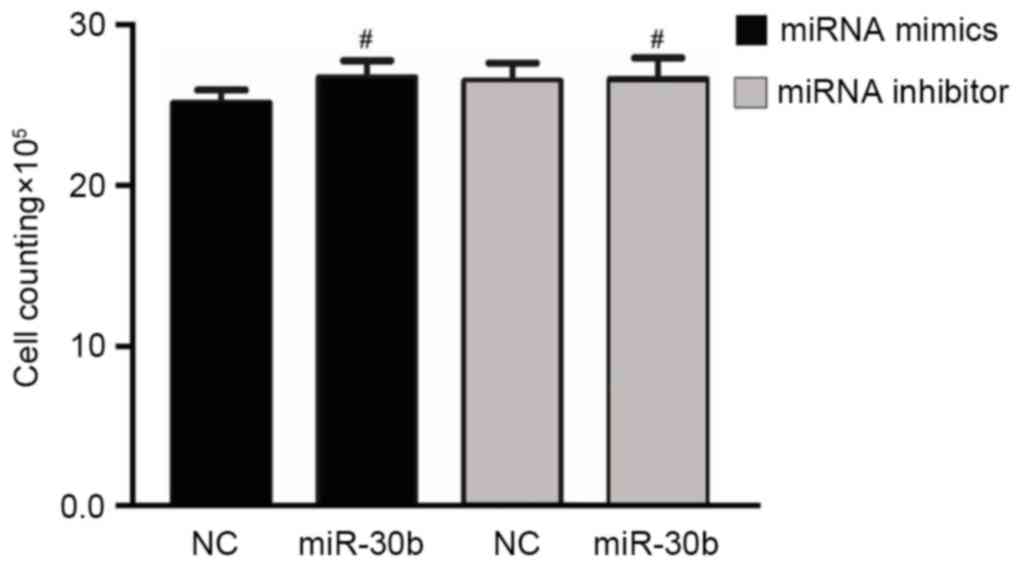
miR-30b has no influence on C3H10T1/2
cell line proliferation
In order to investigate the role of miR-30b in cell
proliferation, a direct cell count method was performed to detect
the proliferation of C3H10T1/2 cells. However, compared to the
negative control group, treatment of C3H10T1/2 cells with either
miR-30b mimics or miR-30b inhibitor did not significantly affect
cell proliferation (Fig. 6).
Discussion
Based on our previous results (19), high-throughput sequencing chip
technology was performed to investigate the expression of miR-30b
during TGF-β3-induced chondrogenic differentiation. Furthermore,
bioinformatics prediction and dual-luciferase reporter assays were
performed to verify whether SOX9 was a target gene of miR-30b.
However, due to false positive results that may appear in the
dual-luciferase reporter assays, protein expression results are
required in order to verify the results. In the present study,
C3H10T1/2 cells were transfected with miR-30b mimics or miR-30b
inhibitor and induced by TGF-β3. Furthermore, SOX9 protein
expression was detected on day 1, 7 and 14 during the chondrogenic
differentiation. Western blotting results demonstrated that
overexpression of miR-30b resulted in significant reduction of SOX9
protein; on the other hand, inhibition of the expression of miR-30b
demonstrated that SOX9 protein expression was increased. In
addition, the SOX9 protein expression level increased in a
time-dependent manner during the chondrogenic differentiation. The
protein test results once again verified that SOX9 was a target of
miR-30b.
miRNAs, a type of non-coding RNA, are 21–23
nucleotides in length and are a type of endogenous regulatory RNA.
Large quantities of miRNA have been identified in a variety of
organisms, and increasing numbers of studies of its function are
being published. Currently, studies in China and abroad demonstrate
that miRNAs are involved in numerous physiological and pathological
processes, including cell development, cell differentiation,
metabolism and cancer amongst others (20–24).
Specific to the relevant regulation of miRNA for chondrogenic
differentiation, research suggests that miRNA is involved in the
regulation of the cartilage formation, cartilage differentiation
phenotype maintenance and metabolism of chondrocytes (25–28).
Furthermore, miRNA is important in cartilage formation, a complex
regulatory process.
Initially, the target gene of miR-30b and the
interaction site were identified. A dual luciferase reporter assay
validated that miR-30b could recognize and interact with
SOX9-3′UTR. Being the target gene of miR-130b, SOX9 is an important
transcription factor of cartilage cell development and
differentiation that contains the high-mobility-group DNA-binding
domain (29). Previous studies have
demonstrated that SOX9 binds to and activate several genes,
including collagen type II alpha 1 chain, bone morphogenetic
protein 2, aggrecan and cartilage-derived retinoic acid-sensitive
protein amongst others (30–33). In addition, SOX9 is also important in
different stages of growth and development. On the one hand it
inhibits chondrocyte hypertrophy in advance while on the other hand
it promotes normal chondrocyte hypertrophy. Next, the present study
investigated whether SOX9 expression levels were affected by
miR-30b activation/inhibition in TGF-β3-induced C3H10T1/2 MSCs. The
results indicated that the protein levels of SOX9 evidently
overexpressed miR-30b but were markedly increased when inhibiting
miR-30b expression. Furthermore, the protein expression level of
SOX9 increased with time extension during chondrogenic
differentiation. These results indicated that miR-30b is important
role in suppressing SOX9 protein expression during chondrogenic
differentiation.
Furthermore, the proliferation of C3H10T1/2 cells
was detected using a direct cell count method. The results
demonstrated that there was no significant difference between the
miR-30b overexpression and miR-30b inhibition groups compared with
the negative control treatment group. At the same time,
glycosaminoglycan synthesis was measured by alcian blue staining
and the results revealed that, compared with the control group,
miR-30b overexpression can reduce dyeing. On the contrary, when
inhibited the expression of miR-30b dyeing can increase. The
results indicate that miR-30b affects the secretion of
characteristic cartilage in the extracellular matrix by directly
targeting SOX9 during chondrogenic differentiation, but had no
evident effect on cell proliferation. To the best of our knowledge
the present study demonstrated for the first time that during
chondrogenic differentiation induced by TGF-β3, miR-30b inhibited
C3H10T1/2 MSC chondrogenic differentiation by targeting SOX9.
However, no significant influence on C3H10T1/2 MSC proliferation
was observed.
In conclusion, the results of the present study
indicated that miR-30b was a key negative regulator of
TGF-β3-induced chondrogenic differentiation by affecting the
secretion of characteristic cartilage in the extracellular matrix
and by directly targeting SOX9 at the early stage of chondrogenic
differentiation. These results suggest that miR-30b may have
potentially important therapeutic implications for cartilage
regeneration.
Acknowledgements
The present study was supported by the National Key
Basic Research and Development Program (973 Program; grant no.
2012CB619105).
References
|
1
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kanellopoulou C, Muljo SA, Kung AL,
Ganesan S, Drapkin R, Jenuwein T, Livingston DM and Rajewsky K:
Dicer-deficient mouse embryonic stem cells are defective in
differentiation and centromeric silencing. Genes Dev. 19:489–501.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Luzi E, Marini F, Sala SC, Tognarini I,
Galli G and Brandi ML: Osteogenic differentiation of human adipose
tissue-derived stem cells is modulated by the miR-26a targeting of
the SMAD1 transcription factor. J Bone Miner Res. 23:287–295. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ansorge WJ: Next-generation DNA sequencing
techniques. N Biotechnol. 25:195–203. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang Z, Gerstein M and Snyder M: RNA-Seq:
A revolutionary tool for transcriptomics. Nat Rev Genet. 10:57–63.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Martin JA and Wang Z: Next-generation
transcriptome assembly. Nat Rev Genet. 12:671–682. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Studer D, Millan C, Öztürk E,
Maniura-Weber K and Zenobi-Wong M: Molecular and biophysical
mechanisms regulating hypertrophic differentiation in chondrocytes
and mesenchymal stem cells. Eur Cell Mater. 24:118–135. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hsiao SH, Lee KD, Hsu CC, Tseng MJ, Jin
VX, Sun WS, Hung YC, Yeh KT, Yan PS, Lai YY, et al: DNA methylation
of the Trip10 promoter accelerates mesenchymal stem cell lineage
determination. Biochem Biophys Res Commun. 400:305–12. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xian CJ and Foster BK: Repair of injured
articular and growth plate cartilage using mesenchymal stem cells
and chondrogenic gene therapy. Curr Stem Cell Res Ther. 1:213–29.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ji YH, Ji JL, Sun FY, Zeng YY, He XH, Zhao
JX, Yu Y, Yu SH and Wu W: Quantitative proteomics analysis of
chondrogenic differentiation of C3H10T1/2 mesenchymal stem cells by
iTRAQ labeling coupled with on-line two-dimensional LC/MS/MS. Mol
Cell Proteomics. 9:550–564. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ji C, Liu X, Xu L, Yu T, Dong C and Luo J:
RUNX1 plays an important role in mediating BMP9-Induced osteogenic
differentiation of mesenchymal stem cells line C3H10T1/2, murine
multi-lineage cells lines C2C12 and MEFs. Int J Mol Sci.
18:E13482017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Son HE, Kim TH and Jang WG: Curculactones
A and B induced the differentiation of C3H10T1/2 and MC3T3-E1 cells
to osteoblasts. Bioorg Med Chem Lett. 27:1301–1303. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hashimoto Y, Kobayashi M, Matsuzaki E,
Higashi K, Takahashi-Yanaga F, Takano A, Hirata M and Nishimura F:
Sphingosine-1-phosphate-enhanced Wnt5a promotes osteogenic
differentiation in C3H10T1/2cells. Cell Biol Int. 40:1129–1136.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Megosh HB, Cox DN, Campbell C and Lin H:
The role of PIWI and the miRNA machinery in drosophila germline
determination. Curr Biol. 16:1884–1894. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Park JK, Liu X, Strauss TJ, McKearin DM
and Liu Q: The miRNA pathway intrinsically controls self-renewal of
drosophila germline stem cells. Curr Biol. 17:533–538. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Förstemann K, Tomari Y, Du T, Vagin VV,
Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE and Zamore PD:
Normal microRNA maturation and germ-line stem cell maintenance
requires loquacious, a double-stranded RNA-binding domain protein.
PLoS Biol. 3:e2362005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Y, Medvid R, Melton C, Jaenisch R and
Blelloch R: DGCR8 is essential for microRNA biogenesis and
silencing of embryonic stem cell self-renewal. Nat Genet.
39:380–385. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wa Q, Gao M, Dai X, Yu T, Zhou Z, Xu D and
Zou X: Induction of chondrogenic differentiation of mouse embryonic
mesenchymal stem cells through an in vitro pellet model. Cell Biol
Int. 39:657–665. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Carmell MA, Girard A, van de Kant HJ,
Bourc'his D, Bestor TH, de Rooij DG and Hannon GJ: MIWI2 is
essential for spermatogenesis and repression of transposons in the
mouse male germline. Dev Cell. 12:503–514. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen CZ, Li L, Lodish HF and Bartel DP:
MicroRNAs modulate hematopoietic lineage differentiation. Science.
303:83–86. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hornstein E, Mansfield JH, Yekta S, Hu JK,
Harfe BD, McManus MT, Baskerville S, Bartel DP and Tabin CJ: The
microRNA miR-196 acts upstream of Hoxb8 and Shh in limb
development. Nature. 438:671–674. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pfeffer S and Voinnet O: Viruses,
microRNAs and cancer. Oncogene. 25:6211–6219. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Taft RJ, Pang KC, Mercer TR, Dinger M and
Mattick JS: Non-coding RNAs: Regulators of disease. J Pathol.
220:126–139. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fukumoto T, Sperling JW, Sanyal A,
Fitzsimmons JS, Reinholz GG, Conover CA and O'Driscoll SW: Combined
effects of insulin-like growth factor-1 and transforming growth
factor-beta1 on periosteal mesenchymal cells during chondrogenesis
in vitro. Osteoarthritis Cartilage. 11:55–64. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Schaefer JF, Millham ML, de Crombrugghe B
and Buckbinder L: FGF signaling antagonizes cytokine-mediated
repression of Sox9 in SW1353 chondrosarcoma cells. Osteoarthritis
Cartilage. 11:233–241. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Murakami S, Kan M, McKeehan WL and de
Crombrugghe B: Up-regulation of the chondrogenic Sox9 gene by
fibroblast growth factors is mediated by the mitogen-activated
protein kinase pathway. Proc Natl Acad Sci USA. 97:pp. 1113–1118.
2000, View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Woods A and Beier F: RhoA/ROCK signaling
regulates chondrogenesis in a context-dependent manner. J Biol
Chem. 281:13134–13140. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tamamura Y, Katsube K, Mera H, Itokazu M
and Wakitani S: Irx3 and Bmp2 regulate mouse mesenchymal cell
chondrogenic differentiation in both a Sox9-dependent and
-independent manner. J Cell Physiol. 232:3317–3336. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bridgewater LC, Lefebvre V and de
Crombrugghe B: Chondrocyte-specific enhancer elements in the
Col11a2 gene resemble the Col2a1 tissue-specific enhancer. J Biol
Chem. 273:14998–15006. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sekiya I, Tsuji K, Koopman P, Watanabe H,
Yamada Y, Shinomiya K, Nifuji A and Noda M: SOX9 enhances aggrecan
gene promoter/enhancer activity and is up-regulated by retinoic
acid in a cartilage-derived cell line, TC6. J Biol Chem.
275:10738–10744. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xie WF, Zhang X, Sakano S, Lefebvre V and
Sandell LJ: Trans-activation of the mouse cartilage-derived
retinoic acid-sensitive protein gene by Sox9. J Bone Miner Res.
14:757–763. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sakou T: Bone morphogenetic proteins: From
basic studies to clinical approaches. Bone. 22:591–603. 1998.
View Article : Google Scholar : PubMed/NCBI
|