Introduction
Hepatocellular carcinoma (HCC), the fifth-most
common malignancy and second-most frequent cause of cancer-related
deaths worldwide (1,2), accounts for approximately 780,000 new
cases and 745,000 deaths attributed to HCC each year (3). Multiple risk factors, including
hepatitis B virus or hepatitis C virus infection, dietary aflatoxin
B1 contamination, chronic alcohol abuse and tobacco consumption,
lack of dietary antioxidants, arsenic exposure, obesity and
non-alcoholic fatty liver disease, involved in HCC formation and
progression have been identified (2,4). Chronic
infection with hepatitis B virus or hepatitis C virus causes
approximately 75% of all HCC cases (5). Although advanced treatments for
patients with HCC have been developed, the long-term prognosis for
these patients remains poor, and their current 5-year survival rate
is approximately 30% (6). In
addition, molecular mechanisms underlying HCC development have yet
to be fully elucidated (7).
Therefore, the molecular mechanisms of HCC initiation and
progression should be further investigated to establish novel
prognostic biomarkers and therapeutic methods for patients with
this disease.
MicroRNAs (miRNAs/miRs), an endogenous group of
short (18–25 nucleotides), non-coding and single-stranded RNA
molecules, have emerged as a major regulator of tumourigenesis and
tumour development (8,9). miRNAs negatively regulate gene
expression by typically binding to complementary sequences in
3′-untranslated regions (3′-UTRs) of their target genes and
therefore stimulate mRNA degradation or translational inhibition
(10). Altered miRNA expression has
been described in various types of human malignancies, such as HCC
(11), renal cell carcinoma
(12), gastric cancer (13) and colorectal cancer (14). miRNA dysregulation has been
demonstrated to be involved in various cellular biological
processes, including cell proliferation, cell cycle, apoptosis,
angiogenesis, invasion, migration, epithelial-to-mesenchymal
transition and metastasis (15–17). In
human cancer, miRNAs may serve as tumour suppressors or oncogenes
which mainly depend on the biological roles of their target genes
(13). Highly expressed miRNAs act
as oncogenes by inhibiting tumour suppressor gene function, whereas
downregulated miRNAs function as tumour suppressor genes through
oncogene downregulation (14,15).
Therefore, the expression and molecular mechanisms of miRNAs in
human cancers should be further investigated to propose therapeutic
targets for anticancer treatments.
miR-495, mapped in the 14q32.31 region, is
abnormally expressed in multiple types of human cancers, such as
upregulated in bladder cancer (18),
and downregulated in osteosarcoma (19), melanoma (20), endometrial cancer (21), acute myeloid leukaemia (22). However, the expression level and
roles of miR-495 in HCC have yet to be completely elucidated. In
our study, the expression levels of miR-495 in HCC tissues and cell
lines were examined, and the effects of this miRNA on cell
proliferation and invasion were evaluated in vitro. The
underlying mechanism of miR-495 in HCC cells was also determined.
Our study might provide novel insights into HCC occurrence and
development.
Materials and methods
Human tissue specimens
A total of 47 individual primary HCC tissues and
matched adjacent non-tumor tissues were collected from patients who
underwent surgical resection at the Seventh People's Hospital of
Shanghai University of Traditional Chinese Medicine (Shanghai,
China) between May 2014 and January 2016. None of these HCC
patients had been treated with chemotherapy or radiotherapy before
surgery. All tissues specimens were flash-frozen in liquid nitrogen
immediately after collection and stored at −80°C. This study was
approved by the Ethics Committee of the Seventh People's Hospital
of Shanghai University of Traditional Chinese Medicine, and written
informed consent was also obtained from all participants prior to
their participation in the study.
Cell lines and culture conditions
Three human HCC cell lines (SMMC-7721, Hep3B, and
Huh7) and immortalized normal liver epithelial cell line (THLE-3)
were purchased from the Cell Bank of Type Culture Collection of
Chinese Academy of Sciences (Shanghai, China).
All HCC cell lines were cultured in Dulbecco's
modified Eagle's medium (DMEM) supplemented with 10% fetal bovine
serum (FBS; bth from Gibco, Grand Island, NY, USA), 100 U/ml
penicillin and 100 µg/ml streptomycin (Sigma-Aldrich, Merck KGaA,
Darmstadt, Germany). THLE-3 cell line was maintained in Bronchial
Epithelial Cell Growth Medium (Clonetics Corporation, Walkersville,
MD, USA) containing 10% FBS, 5 ng/ml EGF and 70 ng/ml
Phosphoethanolamine (both from Sigma-Aldrich, Merck KGaA). All cell
lines were grown at 37°C in humidified atmosphere with 5%
CO2.
Oligonucleotide transfection
Mature miR-495 mimics and miRNA mimics negative
control (miR-NC) were chemically synthesized by GeneCopoeia
(Shanghai, China). Insulin-like growth factor receptor-1 (IGF1R)
overexpression plasmid (pcDNA3.1-IGF1R) and empty plasmid
(pcDNA3.1) were acquired from Shanghai GenePharma Co., Ltd.
(Shanghai, China). For cell transfection, cells were seeded into
6-well plates at a density of 5×105 cells per well.
After incuabtion overnight, cell transfection was performed using
Lipofectamine 2000 (Invitrogen Life Technologies, Carlsbad, CA,
USA), according to the manufacturer's instructions.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from tissue specimens or
cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) in accordance with the manufacturer's
instructions. For miR-495 quantification, complementary DNA (cDNA)
was synthesized from 1 µg of total RNA using the TaqMan microRNA
Reverse Transcription kit (Applied Biosystems, Foster City, CA,
USA). Quantification of miR-495 was performed using a TaqMan
microRNA Assay kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.). U6 small nuclear RNA was used as an endogenous control. To
quantify IGF1R mRNA, corresponding cDNA was obtained form total RNA
with a PrimeScript RT Reagent kit (Takara Biotechnology, Co., Ltd.,
Dalian, China), according to the manufacturer's protocols. The
quantitative PCR was carried out in an Applied
Biosystems® 7500 Real-Time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) using SYBR Premix Ex
Taq™ kit (Takara Biotechnology, Co., Ltd.), with GAPDH as an
internal control. All reactions were performed in triplicate and
fold changes were calculated based on relative quantification using
the 2−ΔΔCq method (23).
Cell Counting kit-8 (CCK-8) assay
CCK-8 assay was utilized to determine the HCC cell
proliferative ability. Transfected cells were collected at 24 h
post-transfection, and mechanically dissociated into single cell
suspension. Afterwards, transfected cells were seeded into 96-well
plates at a density of 3,000 cells/well, and incubated at 37°C with
5% CO2 for 0, 24, 48, and 72 h. At each time point,
CCK-8 assay was performed according to the manufacturer's
protocols. Briefly, each well was treated with 10 ul CCK-8 reagent
(Dojindo Molecular Technologies, Inc., Kumamoto, Japan). After
incubating at 37°C for additional 2 h, the optical density (OD) at
a wavelength of 450 nm was determined using a Spectramax M5
microplate reader (Molecular Devices LLC, Sunnyvale, CA, USA). Each
assay was performed with 5 replications.
Cell invasion assay
Cell invasion assay was performed using Transwell
insert chambers (8-µm pore size; Corning, Inc., Corning, NY, USA)
coated with Matrigel (BD Biosciences, San Jose, CA, USA). After
transfection 48 h, cells were harvested and suspended in FBS-free
DMEM. Transfected cells (5×104) in 300 µl FBS-free DMEM
were seeded into the upper chamber. The lower chambers were then
filled with DMEM containing 10% FBS. Following a 24 h incubation at
37°C with 5% CO2, non-invaded cells were removed using a
cotton swab. Invasive cells were fixed with 4% polyoxymethylene,
stained with 0.5% crystal violet, washed with PBS and dried in air.
Finally, invasive cells were counted in five randomly selected
visual fields under an inverted microscope (magnification, ×200;
Olympus Corporation, Tokyo, Japan).
Target prediction
The TargetScan (http://www.targetscan.org/) and PicTar (http://pictar.mdc-berlin.de/) were used to predict the
potential targets of miR-495.
Luciferase reporter assay
Bioinformatic analysis indicated a potential miR-495
binding site in the 3′-UTR region of IGF1R. Lciferase reporter
plasmids, pMIR-IGF1R-3′-UTR wild-type (Wt) and pMIR-IGF1R-3′-UTR
wild mutant (Mut), were chemically synthesized by Shanghai
GenePharma Co., Ltd. Cells were seeded into 24-well plates at a
density of 1.5×105 cells per well. After incubaiton
overnight, cells were co-transfected with the wild-type or mutant
3′-UTR of IGF1R plasmid, and miR-495 mimics or miR-NC using the
Lipofectamine 2000 reagent, according to the manufacturer's
protocol. After transfection 48 h, the relative luciferase activity
was detected using the Dual-Luciferase Reporter assay system
(Promega Corporation, Madison, WI, USA), according to the
manufacturer's instructions. Renilla luciferase activity
served as an internal control. Each assay was performed in 3
replicates and repeated at least three times.
Western blot analysis
Total protein was isolated from tissue specimens or
cells with Radioimmunoprecipitation assay lysis buffer (Beyotime
Institute of Biotechnology, Haimen, China) containing 0.1 mg/ml
phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate and 1
mg/ml aprotinin. The concentration of total protein was quantified
with BCA assay kit (Beyotime Institute of Biotechnology). Equal
amount of protein was separated by 10% SDS-PAGE and electronically
transferred onto a polyvinylidene difluoride membrane (EMD
Millipore, Billerica MA, USA). After blocking with 5% nonfat milk
in TBST for 2 h at room temperature, the membranes were incubated
with primary antibodies at 4°C overnight. The primary antibodies
used in this study include mouse anti-human monoclonal IGF1R
antibody (sc-81464; 1:1,000 dilution), mouse anti-human monoclonal
p-AKT (sc-271966; 1:1,000 dilution), mouse anti-human monoclonal
AKT (sc-81434; 1:1,000 dilution), mouse anti-human monoclonal p-ERK
(sc-81492; 1:1,000 dilution), mouse anti-human monoclonal ERK
(sc-514302; 1:1,000 dilution), and mouse anti-human monoclonal
GAPDH antibody (sc-47724; 1:1,000 dilution; all from Santa Cruz
Biotechnology, Santa Cruz, CA, USA). Subsequent washing three times
with TBST, the membranes were probed with goat anti-mouse
horseradish peroxidase (HRP)-conjugated secondary antibody
(sc-2005; 1:5,000 dilution; Santa Cruz Biotechnology) at room
temperature for 2 h. Protein bands were visualized using an
enhanced chemiluminescence kit (EMD Millipore), and analyzed using
ImageJ 1.49 (National Institutes of Health, Bethesda, MD, USA).
GAPDH served as a loading control.
Statistical analysis
Data are shown as the mean ± standard error of at
least three independent experiments, and analyzed with Student's
t-test or one-way analysis of variance. SPSS 17.0 software (SPSS,
Inc., Chicago, IL, USA) was used to perform statistical analysis.
Spearman's correlation analysis was adopted to investigate the
association between miR-495 and IGF1R mRNA expression level in HCC
tissues. P<0.05 was considered statistically significant.
Results
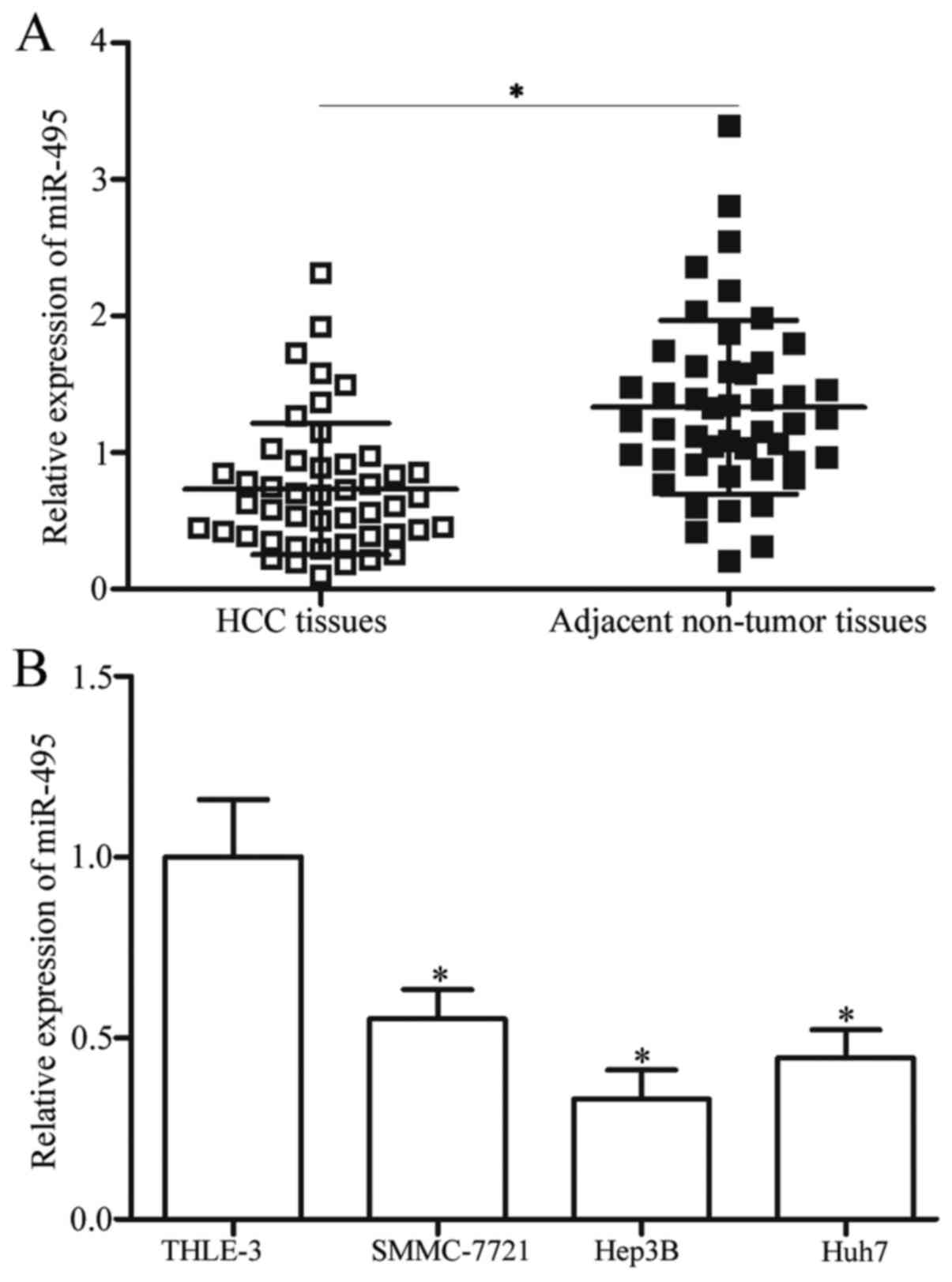
miR-495 is downregulated in HCC
tissues and cell lines
To determine the potential roles of miR-495 in HCC,
RT-qPCR was performed to detect miR-495 expression levels in 47
primary HCC tissues and matched adjacent non-tumour tissues. Our
results demonstrated that miR-495 was significantly downregulated
in HCC tissues compared with that in matched adjacent non-tumour
tissues (P<0.05; Fig. 1A). The
miR-495 expression was also examined in three HCC cell lines,
namely, SMMC-7721, Hep3B and Huh7, and in immortalized normal liver
epithelial cell line, namely, THLE-3. In Fig. 1B, the miR-495 expression levels in
the four HCC cell lines were lower than that of THLE-3 (P<0.05).
Among these HCC cell lines, Hep3B yielded the lowest miR-495
expression levels and thus were selected for further functional
studies. These results suggested that miR-495 may be involved in
the formation and progression of HCC.
Low miR-495 expression is correlated
with adverse clinical features of patients with HCC
The patients with HCC were subsequently divided into
either miR-495 low-expression group (n=24) or miR-495
high-expression group (n=23) to elucidate the clinical significance
of miR-495 in HCC. The median expression levels of miR-495 in HCC
tissues were regarded as cut-off. As shown in Table I, the low miR-495 expression level
was correlated with tumour size (P=0.028), tumor-node-metastasis
(TNM) stage (P=0.013) and lymph node metastasis (P=0.011).
Conversely, miR-495 expression was not correlated with other
clinical features, including age (P=0.642), sex (P=0.471), HBsAg
(P=0.413), and differentiated (P=0.471). These results implied that
miR-495 may be a prognostic indicator for patients with HCC.
 | Table I.Association between miR-495
expression and clinical features of hepatocellular carcinoma
patients. |
Table I.
Association between miR-495
expression and clinical features of hepatocellular carcinoma
patients.
|
|
| miR-495
expression |
|
|---|
|
|
|
|
|
|---|
| Clinical
features | Case number | Low | High | P-value |
|---|
| Age |
|
|
| 0.642 |
| <60
years | 20 | 11 | 9 |
|
| ≥60
years | 27 | 13 | 14 |
|
| Sex |
|
|
| 0.471 |
|
Male | 31 | 17 | 14 |
|
|
Female | 16 | 7 | 9 |
|
| Tumour size |
|
|
| 0.028a |
| <5
cm | 25 | 9 | 16 |
|
| ≥5
cm | 22 | 15 | 7 |
|
| HBsAg |
|
|
| 0.413 |
|
Negative | 6 | 4 | 2 |
|
|
Positive | 41 | 20 | 21 |
|
| TNM stage |
|
|
| 0.013a |
|
I–II | 22 | 7 | 15 |
|
|
III–IV | 25 | 17 | 8 |
|
| Lymph node
metastasis |
|
|
| 0.011a |
|
Negative | 26 | 8 | 18 |
|
|
Positive | 21 | 16 | 5 |
|
| Differentiated |
|
|
| 0.471 |
| Well
and moderate | 25 | 14 | 11 |
|
|
Poor | 22 | 10 | 12 |
|
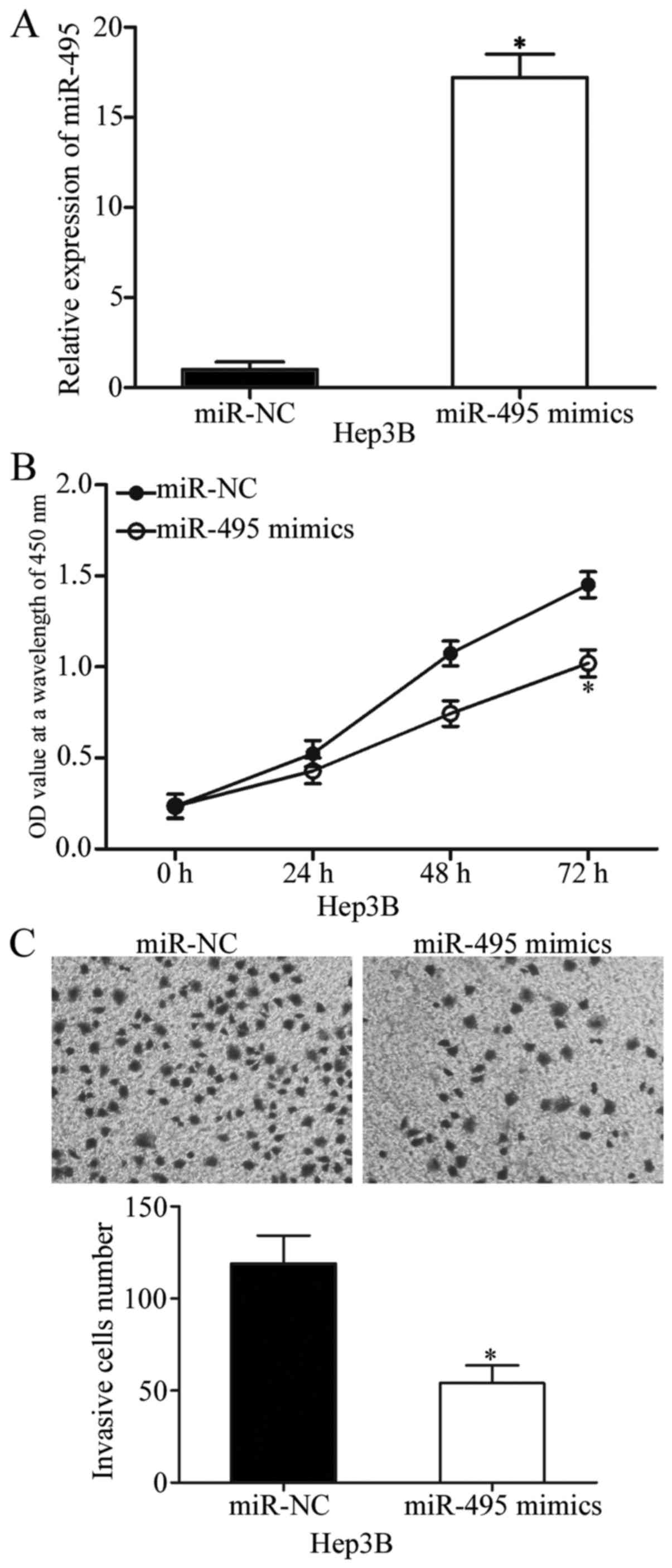
miR-495 inhibits cell proliferation
and invasion in HCC
Hep3B cells were transfected with miR-495 mimics or
miR-NC to examine the effects of miR-495 on the biological
characteristics of tumours. Transfection efficiency was determined
through RT-qPCR, and the results indicated that miR-495 was
markedly upregulated in Hep3B cells transfected with miR-495 mimics
(P<0.05; Fig. 2A). CCK-8 assay
was conducted to verify the effect of miR-495 overexpression on HCC
cell proliferation. In Fig. 2B,
upregulation of miR-495 inhibited Hep3B cell proliferation
(P<0.05). Cell invasion assay was also performed to show the
effect of miR-495 on HCC cell invasion abilities. The results
demonstrated that the restored miR-495 expression reduced the
invasive capabilities of Hep3B cells (P<0.05; Fig. 2C). These results illustrated the
tumour-suppressive effects of miR-495 on HCC cell proliferation and
invasion.
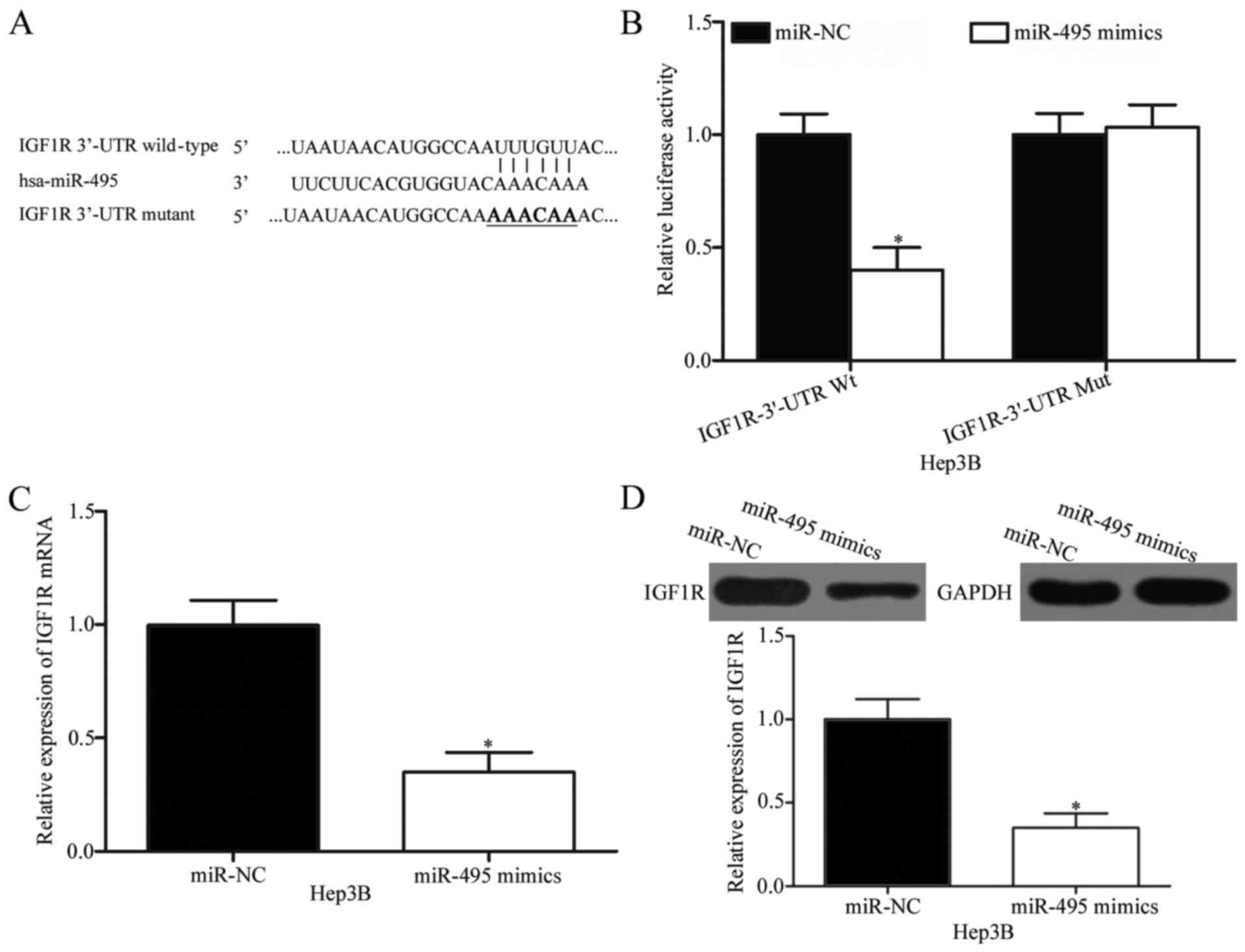
IGF1R is a direct downstream target of
miR-495 in HCC
To investigate the mechanisms by which miR-495 plays
its tumour-suppressing roles in HCC, bioinformatics analysis was
conducted to predict the candidate targets of miR-495. IGF1R, which
is highly expressed in HCC and possibly involved in HCC formation
and progression (24,25), was predicted as a potential target of
miR-495 (Fig. 3A) and consequently
selected for further experimental validation. To confirm this
hypothesis, we carried out a luciferase reporter assay in Hep3B
cells co-transfected with wild-type or mutant 3′-UTR of IGF1R
plasmid and miR-495 mimics or miR-NC. In Fig. 3B, ectopic miR-495 expression
significantly decreased the luciferase activity of
pMIR-IGF1R-3′-UTR Wt in Hep3B cells (P<0.05) but did not affect
the luciferase activity of pMIR-IGF1R-3′-UTR Mut. To confirm
whether IGF1R is a direct target of miR-495, we performed RT-qPCR
and Western blot analysis and then determined the regulatory
effects of miR-495 on endogenous IGF1R expression in HCC cells.
RT-qPCR and Western blot analysis revealed that the restored
miR-495 expression suppressed the IGF1R expression in Hep3B cells
at mRNA (P<0.05; Fig. 3C) and
protein (P<0.05; Fig. 3C) levels.
These results suggested that IGF1R is a direct target of miR-495 in
HCC.
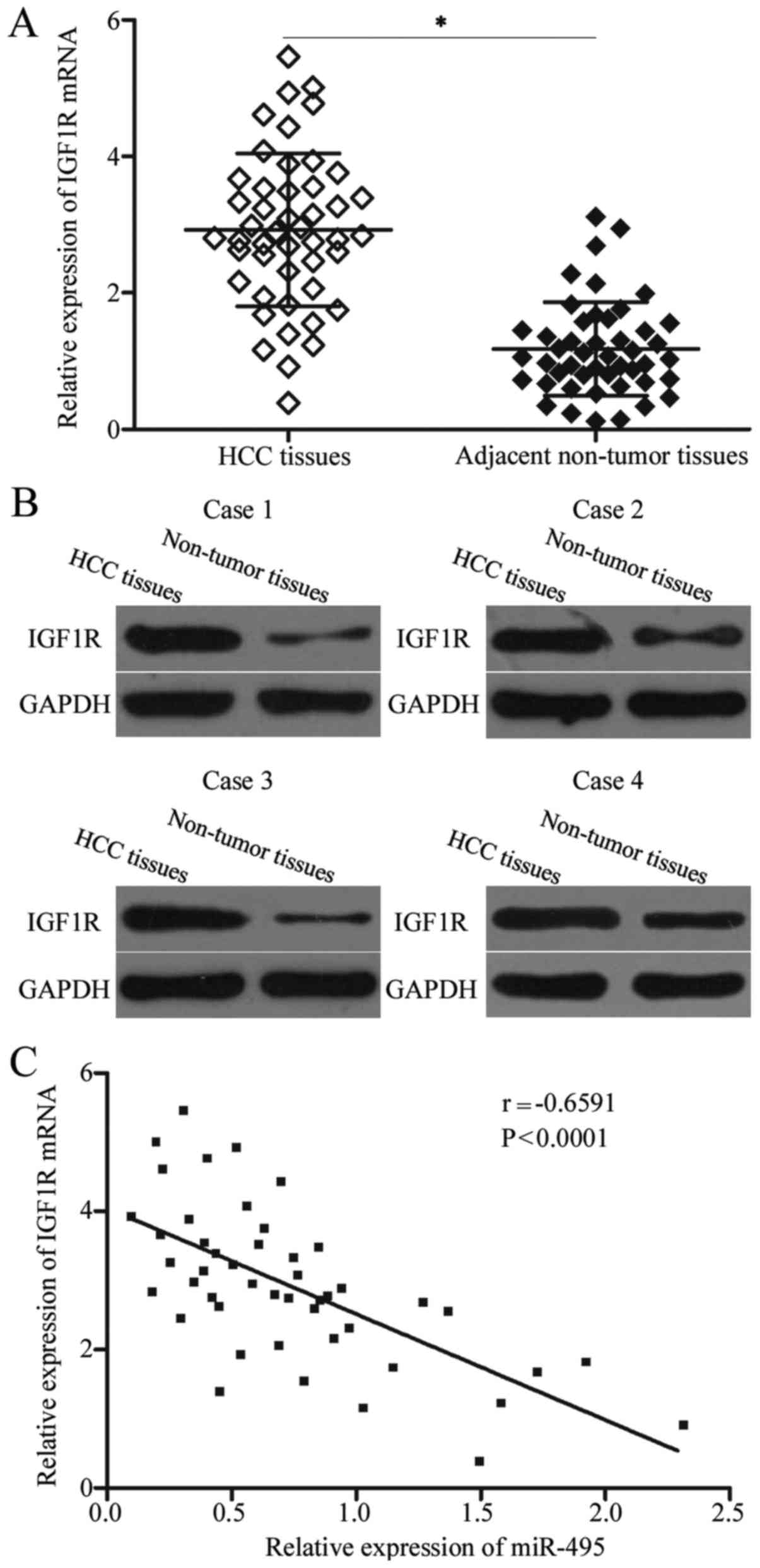
IGF1R is upregulated in HCC tissues
and negatively correlated with miR-495 expression levels
The IGF1R level was examined in HCC tissues and
matched adjacent non-tumour tissues to analyse the correlation
between miR-495 and IGF1R. In Fig. 4A
and B, the IGF1R expression levels were significantly higher in
HCC tissues than in matched adjacent non-tumour tissues at mRNA
(P<0.05) and protein (P<0.05) levels. Spearman's correlation
analysis revealed that miR-495 was negatively associated with the
mRNA expression level of IGF1R in HCC tissues (r=−0.6591,
P<0.001; Fig. 4C).
IGF1R upregulation rescues the
tumour-suppressive roles induced by miR-495 overexpression in
HCC
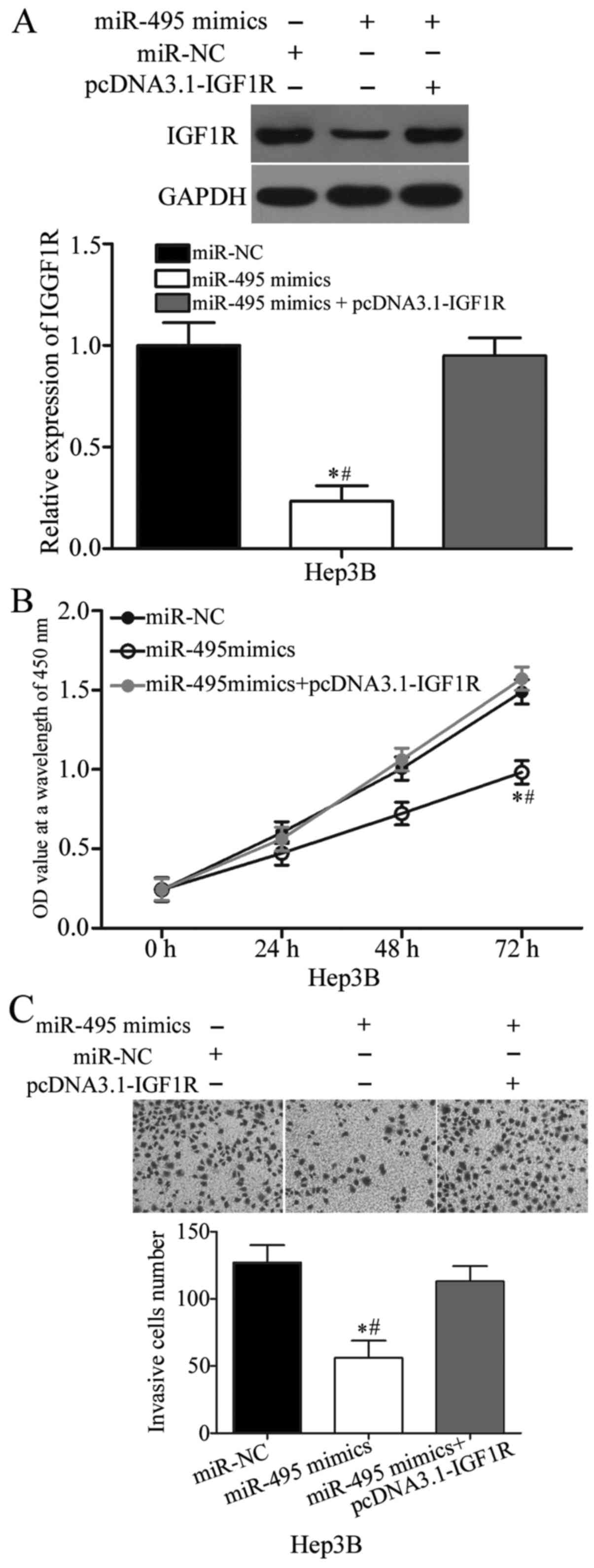
Rescue experiments were performed to confirm whether
the biological roles of miR-495 in HCC are mediated by IGF1R. Hep3B
cells were transfected with miR-495 mimics with or without IGF1R
overexpression plasmid (pcDNA3.1-IGF1R). Western blot analysis
indicated that miR-495 overexpression decreased the IGF1R protein
expression, whereas pcDNA3.1-IGF1R co-transfection could rescue the
IGF1R expression in Hep3B cells (P<0.05; Fig. 5A). Subsequent functional assays
demonstrated that the restored IGF1R expression rescued the
suppressive effects of miR-495 on the proliferation (P<0.05;
Fig. 5B) and invasion of Hep3B
cells. These results implied that miR-495 plays its
tumour-suppressing roles in HCC partly by downregulating IGF1R.
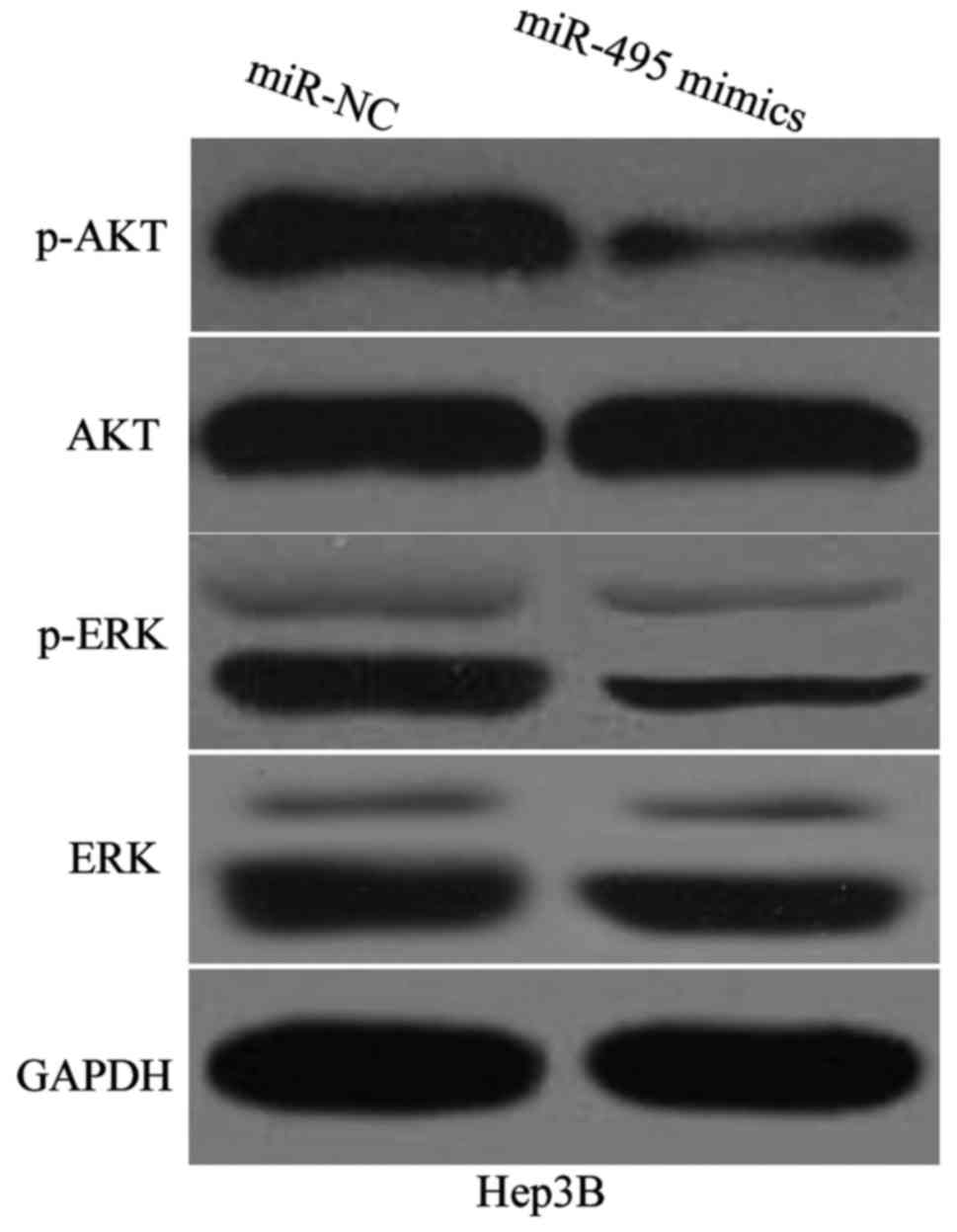
miR-495 attenuates AKT and ERK
signalling pathways in HCC
IGF1R likely performs its functions partly by
participating in the regulation of AKT and ERK signalling pathways
(26–28). To determine whether miR-495 is
involved in the regulation of AKT and ERK pathways in HCC, we
detected the expression levels of AKT, p-AKT, ERK and p-ERK in
Hep3B cells transfected with miR-495 mimics or miR-NC. Western blot
analysis demonstrated that miR-495 overexpression reduced the
expression of p-AKT and p-ERK in Hep3B cells (Fig. 6). Conversely, the expression of total
AKT and ERK in these two cell lines did not significantly change.
These results suggested that miR-495 inactivates AKT and ERK
signalling pathways in HCC.
Discussion
Recently, increasing studies reported that miRNAs
are abnormally expressed in various types of human cancers
(29–31) and can participate in the
tumourigenesis and tumour development of HCC (32,33).
miRNA-based targeted therapy is effective against different
molecular targets and can increase the sensitisation of cancer
cells to therapy by several folds (34). Therefore, further validation of
potentially important miRNAs involved in HCC initiation and
progression may provide valuable insights into the treatment of
patients with HCC. In our study, miR-495 was significantly
downregulated in HCC tissues and cell lines. Low miR-495 expression
levels were correlated with adverse clinical features of the
patients with HCC. The restored expression of miR-495 inhibited the
proliferation and invasion of HCC cells in vitro. IGF1R was
identified as a direct target of miR-495 in HCC, and miR-495
upregulation attenuated the activation of AKT and ERK signalling
pathways in HCC. Therefore, miR-495 might serve as a tumour
suppressor in HCC by directly targeting IGF1R and regulating AKT
and ERK signalling pathways and might be developed as a novel
therapeutic target for the treatment of patients with this fatal
malignancy.
miR-495 is aberrantly expressed in numerous types of
human cancer. For example, miR-495 is upregulated in bladder cancer
tissues and cell lines. High miR-495 expression levels are
correlated with tumour size, TNM stage and lymph node metastasis of
patients with bladder cancer (18).
However, miR-495 in medulloblastoma is downregulated in tumour
tissues compared with that in normal cerebellum tissues. Log-rank
analysis demonstrated that the average survival time of patients
with medulloblastoma and with low miR-495 levels is shorter than
that of patients with high miR-495 expression levels. Multivariate
analysis also demonstrates miR-495 as an independent predictor of
the overall survival of patients with medulloblastoma (35). miR-495 is weakly expressed in
prostate cancer and associated with prostate-specific antigen
levels, lymph node invasion and Gleason scores (36). miR-495 is also downregulated in
osteosarcoma (19), melanoma
(20), endometrial cancer (21), acute myeloid leukaemia (22), renal cell carcinoma (37), breast cancer (38,39) and
lung cancer (40,41). These findings suggested that miR-495
expression exhibits tissue specificity and may be a biomarker for
these human cancers.
miR-495 plays tumour-suppressive roles in multiple
types of human malignancy. For instance, Jiang et al
revealed that miR-495 overexpression suppresses osteosarcoma cell
proliferation, colony formation, invasion and increased apoptosis
in vitro (19). Studies
showed that miR-495 upregulation inhibits cell proliferation and
invasion and induces a metabolic shift in glioma (42–44).
Formosa et al (36) and Li et
al (45) indicated that the restored
miR-495 expression attenuates prostate cancer cell growth and
metastasis and promotes apoptosis in vitro. Liu et al
reported that ectopic miR-495 expression decreases cell
proliferation and metastasis and triggers cell apoptosis of
melanoma (20). Xu et al
demonstrated that miR-495 re-expression represses endometrial
cancer cell growth and migration, promotes apoptosis and reduces
growth in vivo (21). Lv
et al found that restored miR-495 expression inhibits cell
proliferation and motility and induces G0/G1 phase arrest in renal
cell carcinoma (37). In lung
cancer, miR-495 is involved in the regulation of cell
proliferation, migration, epithelial-mesenchymal transition,
chemosensitivity and chemoresistance (40,41,46).
However, Tan et al indicated that miR-495 serves as an
oncogene in bladder cancer through the regulation of cell
proliferation and invasion (18).
These conflicting findings suggested that the functional roles of
miR-495 in human malignancies may be multifaceted and mainly
dependent on involved tissues and their target genes.
The following target miR-495 genes have been
identified: HMGN5 (19) in
osteosarcoma; GFI1 (35) in
medulloblastoma; Glut1 (42), MYB
(43) and CDK6 (44) in glioma; FZD4 (36), Akt (45) and mTOR (45) in prostate cancer; SATB1 (37) in renal cell carcinoma; STAT-3
(38) and Bmi-1 (39) in breast cancer; epithelial and
endothelial tyrosine kinase (40),
MTA3 (41) and ATP7A (46) in lung cancer; and PTEN (18) in bladder cancer. In our study, IGF1R
was demonstrated to be a direct functional downstream target of
miR-495 in HCC. IGF1R, a transmembrane tyrosine kinase receptor of
the insulin receptor family, has been reported to be upregulated in
multiple types of cancer, such as osteosarcoma (47), colorectal cancer (48), prostate cancer (49), gastric cancer (50), endometrial cancer (51) and bladder cancer (52). A previous study revealed that IGF1R
is significantly upregulated in HCC and correlated with TNM stage.
Univariate analysis indicated that high IGF1R expression predicted
poor overall and disease-free survival for patients with HCC. In
multivariate analysis, IGF1R is significant in the overall survival
of patients with HCC (24).
Functional experiments demonstrated that IGF1R downregulation
inhibits HCC cell proliferation and invasion but increases
apoptosis (25). Therefore, IGF1R
could be developed as a novel therapeutic target in HCC because of
its cancer-related functions.
In conclusion, this study is the first to
demonstrate that miR-495 inhibited the proliferation and invasion
of HCC cells by directly targeting IGF1R and regulating AKT and ERK
signalling pathways. These results provide novel insights into the
molecular mechanism underlying HCC progression and suggest that
miR-495 may be investigated as a novel therapeutic target for
patients with HCC.
Acknowledgements
The study was supported by grants from Key
Disciplines Group Construction Project of Pudong Health Burea of
Shanghai (PWZxq2014-12), Natural Science Foundation of China (no.
81571718), Shanghai Science and Technology Committee Foundation
(no. 17ZR1421600), Science and Technology Development Fund of
Shanghai Pudong New Area (PKJ2016-Y50) and Talents Training Program
of Seventh People's Hospital of Shanghai University of Traditional
Chinese Medicine (grant no. QMX2017-01).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yu MC and Yuan JM: Environmental factors
and risk for hepatocellular carcinoma. Gastroenterology. 127 5
Suppl 1:S72–S78. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zeng Z: Human genes involved in hepatitis
B virus infection. World J Gastroenterol. 20:7696–7706. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dhanasekaran R, Limaye A and Cabrera R:
Hepatocellular carcinoma: Current trends in worldwide epidemiology,
risk factors, diagnosis, and therapeutics. Hepat Med. 4:19–37.
2012.PubMed/NCBI
|
|
7
|
Aravalli RN, Steer CJ and Cressman EN:
Molecular mechanisms of hepatocellular carcinoma. Hepatology.
48:2047–2063. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gu S, Jin L, Zhang F, Sarnow P and Kay MA:
Biological basis for restriction of microRNA targets to the 3′
untranslated region in mammalian mRNAs. Nat Struct Mol Biol.
16:144–150. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yan JJ, Chang Y, Zhang YN, Lin JS, He XX
and Huang HJ: miR-195 inhibits cell proliferation via targeting
AEG-1 in hepatocellular carcinoma. Oncol Lett. 13:3118–3126.
2017.PubMed/NCBI
|
|
12
|
Pengcheng S, Ziqi W, Luyao Y, Xiangwei Z,
Liang L, Yuwei L, Lechen L and Wanhai X: MicroRNA-497 suppresses
renal cell carcinoma by targeting VEGFR-2 in ACHN cells. Biosci
Rep. 37:pii: BSR201702702017. View Article : Google Scholar
|
|
13
|
Li F, Guo Y, Liu J and Zhang R: The
significance of elevated plasma expression of microRNA 106b~25
clusters in gastric cancer. PLoS One. 12:e01784272017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fateh A, Feizi MAH, Safaralizadeh R and
Azarbarzin S: Importance of miR-299-5p in colorectal cancer. Ann
Gastroenterol. 30:322–326. 2017.PubMed/NCBI
|
|
15
|
Lujambio A and Lowe SW: The microcosmos of
cancer. Nature. 482:347–355. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ebert MS and Sharp PA: Roles for microRNAs
in conferring robustness to biological processes. Cell.
149:515–524. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Iorio MV and Croce CM: microRNA
involvement in human cancer. Carcinogenesis. 33:1126–1133. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tan M, Mu X, Liu Z, Tao L, Wang J, Ge J
and Qiu J: microRNA-495 promotes bladder cancer cell growth and
invasion by targeting phosphatase and tensin homolog. Biochem
Biophys Res Commun. 483:867–873. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiang W, Zheng J, Yu T and Wang J:
Overexpression of microRNA-495 suppresses the proliferation and
invasion and induces the apoptosis of osteosarcoma cells by
targeting high-mobility group nucleosome-binding domain 5. Oncol
Rep. 38:1099–1107. 2017.PubMed/NCBI
|
|
20
|
Liu P, Hu Y, Ma L, Du M, Xia L and Hu Z:
miR-425 inhibits melanoma metastasis through repression of PI3K-Akt
pathway by targeting IGF-1. Biomed Pharmacother. 75:51–57. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu YY, Tian J, Hao Q and Yin LR:
MicroRNA-495 downregulates FOXC1 expression to suppress cell growth
and migration in endometrial cancer. Tumour Biol. 37:239–251. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jiang X, Huang H, Li Z, He C, Li Y, Chen
P, Gurbuxani S, Arnovitz S, Hong GM, Price C, et al: MiR-495 is a
tumor-suppressor microRNA down-regulated in MLL-rearranged
leukemia. Proc Natl Acad Sci USA. 109:pp. 19397–19402. 2012;
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lin SB, Zhou L, Liang ZY, Zhou WX and Jin
Y: Expression of GRK2 and IGF1R in hepatocellular carcinoma:
Clinicopathological and prognostic significance. J Clin Pathol.
70:754–759. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yao WF, Liu JW, Sheng GL and Huang DS:
Blockade of IGF-IR exerts anticancer effects in hepatocellular
carcinoma. Mol Med Rep. 4:719–722. 2011.PubMed/NCBI
|
|
26
|
Zhang W, Liu K, Liu S, Ji B, Wang Y and
Liu Y: MicroRNA-133a functions as a tumor suppressor by targeting
IGF-1R in hepatocellular carcinoma. Tumour Biol. 36:9779–9788.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guo T, Feng Y, Liu Q, Yang X, Jiang T,
Chen Y and Zhang Q: MicroRNA-320a suppresses in GBM patients and
modulates glioma cell functions by targeting IGF-1R. Tumour Biol.
35:11269–11275. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen G, Fang T, Huang Z, Qi Y, Du S, Di T,
Lei Z, Zhang X and Yan W: MicroRNA-133a inhibits osteosarcoma cells
proliferation and invasion via targeting IGF-1R. Cell Physiol
Biochem. 38:598–608. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yuan J, Ji H, Xiao F, Lin Z, Zhao X, Wang
Z, Zhao J and Lu J: MicroRNA-340 inhibits the proliferation and
invasion of hepatocellular carcinoma cells by targeting JAK1.
Biochem Biophys Res Commun. 483:578–584. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Duan X, Bai J, Wei J, Li Z, Liu X and Xu
G: MicroRNA-508-5p suppresses metastasis in human gastric cancer by
targeting S-phase kinase-associated protein 2. Mol Med Rep.
16:2163–2171. 2017.PubMed/NCBI
|
|
31
|
Ma F, Li W, Liu C, Li W, Yu H, Lei B, Ren
Y, Li Z, Pang D and Qian C: MiR-23a promotes TGF-beta1-induced EMT
and tumor metastasis in breast cancer cells by directly targeting
CDH1 and activating Wnt/beta-catenin signaling. Oncotarget.
8:69538–69550. 2017.PubMed/NCBI
|
|
32
|
Han K, Li J, Zhao H, Liang P, Huang X,
Zheng L, Li Y, Yang T and Wang L: Identification of the typical
miRNAs and target genes in hepatocellular carcinoma. Mol Med Rep.
10:229–235. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Afonso MB, Rodrigues PM, Simão AL and
Castro RE: Circulating microRNAs as potential biomarkers in
non-alcoholic fatty liver disease and hepatocellular carcinoma. J
Clin Med. 5:pii: E302016. View Article : Google Scholar
|
|
34
|
Gandhi NS, Tekade RK and Chougule MB:
Nanocarrier mediated delivery of siRNA/miRNA in combination with
chemotherapeutic agents for cancer therapy: Current progress and
advances. J Control Release. 194:238–256. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang C, Yun Z, Zhao T, Liu X and Ma X:
MiR-495 is a predictive biomarker that downregulates GFI1
expression in medulloblastoma. Cell Physiol Biochem. 36:1430–1439.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Formosa A, Markert EK, Lena AM, Italiano
D, Finazzi-Agro' E, Levine AJ, Bernardini S, Garabadgiu AV, Melino
G and Candi E: MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c,
miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p,
mapped to the 14q32.31 locus, regulate proliferation, apoptosis,
migration and invasion in metastatic prostate cancer cells.
Oncogene. 33:5173–5182. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lv C, Bai Z, Liu Z, Luo P and Zhang J:
MicroRNA-495 suppresses human renal cell carcinoma malignancy by
targeting SATB1. Am J Transl Res. 7:1992–1999. 2015.PubMed/NCBI
|
|
38
|
Chen Y, Luo D, Tian W, Li Z and Zhang X:
Demethylation of miR-495 inhibits cell proliferation, migration and
promotes apoptosis by targeting STAT-3 in breast cancer. Oncol Rep.
37:3581–3589. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang L, Liu JL, Yu L, Liu XX, Wu HM, Lei
FY, Wu S and Wang X: Downregulated miR-495 [Corrected] inhibits the
G1-S phase transition by targeting Bmi-1 in breast cancer.
Medicine. 94:e7182015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wei T, Zhu W, Fang S, Zeng X, Huang J,
Yang J, Zhang J and Guo L: miR-495 promotes the chemoresistance of
SCLC through the epithelial-mesenchymal transition via Etk/BMX. Am
J Cancer Res. 7:628–646. 2017.PubMed/NCBI
|
|
41
|
Chu H, Chen X, Wang H, Du Y, Wang Y, Zang
W, Li P, Li J, Chang J, Zhao G and Zhang G: MiR-495 regulates
proliferation and migration in NSCLC by targeting MTA3. Tumour
Biol. 35:3487–3494. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nie S, Li K, Huang Y, Hu Q, Gao X and Jie
S: miR-495 mediates metabolic shift in glioma cells via targeting
Glut1. J Craniofac Surg. 26:e155–e158. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang B, Yuan F, Liu J, Li Y, Zhou F, Liu
X, Hao Z, Li Q, Zheng Y and Wang W: Hsa-miR-495 acts as a tumor
suppressor gene in glioma via the negative regulation of MYB. Mol
Med Rep. 14:977–982. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chen SM, Chen HC, Chen SJ, Huang CY, Chen
PY, Wu TW, Feng LY, Tsai HC, Lui TN, Hsueh C and Wei KC:
MicroRNA-495 inhibits proliferation of glioblastoma multiforme
cells by downregulating cyclin-dependent kinase 6. World J Surg
Oncol. 11:872013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li JZ, Wang ZL, Xu WH, Li Q, Gao L and
Wang ZM: MicroRNA-495 regulates migration and invasion in prostate
cancer cells via targeting Akt and mTOR signaling. Cancer Invest.
34:181–188. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Song L, Li Y, Li W, Wu S and Li Z: miR-495
enhances the sensitivity of non-small cell lung cancer cells to
platinum by modulation of copper-transporting P-type adenosine
triphosphatase A (ATP7A). J Cell Biochem. 115:1234–1242. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang YH, Han XD, Qiu Y, Xiong J, Yu Y,
Wang B, Zhu ZZ, Qian BP, Chen YX, Wang SF, et al: Increased
expression of insulin-like growth factor-1 receptor is correlated
with tumor metastasis and prognosis in patients with osteosarcoma.
J Surg Oncol. 105:235–243. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Shiratsuchi I, Akagi Y, Kawahara A,
Kinugasa T, Romeo K, Yoshida T, Ryu Y, Gotanda Y, Kage M and
Shirouzu K: Expression of IGF-1 and IGF-1R and their relation to
clinicopathological factors in colorectal cancer. Anticancer Res.
31:2541–2545. 2011.PubMed/NCBI
|
|
49
|
Ma Y, Cheng Q, Ren Z, Xu L, Zhao Y, Sun J,
Hu S and Xiao W: Induction of IGF-1R expression by EGR-1
facilitates the growth of prostate cancer cells. Cancer Lett.
317:150–156. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Gryko M, Kiśluk J, Cepowicz D, Zińczuk J,
Kamocki Z, Guzińska-Ustymowicz K, Pryczynicz A, Czyżewska J, Kemona
A and Kędra B: Expression of insulin-like growth factor receptor
type 1 correlate with lymphatic metastases in human gastric cancer.
Pol J Pathol. 65:135–140. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Pengchong H and Tao H: Expression of
IGF-1R, VEGF-C and D2-40 and their correlation with lymph node
metastasis in endometrial adenocarcinoma. Eur J Gynaecol Oncol.
32:660–664. 2011.PubMed/NCBI
|
|
52
|
Xie QX, Lin XC, Zhang MF, Han CX and Guo
YH: Expression of IGF-I and IGF-IR in bladder cancer. Ai Zheng.
23:707–709. 2004.(In Chinese). PubMed/NCBI
|