Introduction
Antibiotic-resistant bacteria are reported as the
greatest threaten to human health in the world (1,2). Overuse
and misuse of antibiotics contributes to the occurrence of
antibiotic resistance genes and the evolution of
antibiotic-resistant bacteria according to the discipline of
genesis and evolution (3,4). In the past several decades, antibiotic
resistance has been accelerated by the indiscriminate application
of antibiotics leading to the vital problem of antibiotic
resistance and significant public health concerns (5). Antibiotic resistance is a significant
challenge for microbiology labs and clinicians (6). Previous studies have suggested that the
major driving force in the occurrence of antibiotic-resistant
pathogens is the evolution of metabolic function caused by the
rapid antibiotic consumption in the world (7,8).
Extended-spectrum β-lactamase (ESBL) produced by
antibiotic-resistant bacteria is another mechanism underlying the
phenomenon of the evolution of antibiotic-resistant bacteria
(9,10). Clinically, antibiotic resistance in
patients has made treatment approaches for bacterial infections
insufficient and resulted in a markedly increased morbidity and
mortality (11,12). Therefore, understanding the molecular
mechanisms of resistance to antimicrobial agents is imminent to
develop novel intervention strategies to improve the survival of
patients.
In recent years, antimicrobial resistance and
susceptibility of Klebsiella pneumoniae have been observed
in clinical practice (13).
Klebsiella pneumoniae has represented an intractable
pathogen in pulmonary disease departments, which partly attributed
to an increased economic burden, promoted intra-hospital
transmission and challenged infection control practices (14). Previous studies have suggested that
nosocomial infections of Klebsiella pneumoniae frequently
erupted due to increased production of ESBL produced by
Klebsiella pneumoniae, which attributed to multiple
mechanisms of drug resistance (15,16). In
addition, antibacterial drug susceptibility tests of isolated
strains of Klebsiella pneumoniae revealed that strains have
been disseminated with simultaneous resistance to various types of
antimicrobial agents, mediated by loss of porins on the
cytomembrane, the production of ESBL, carbapenemases and even
metallo-β-lactamases to enhance antibiotic resistance (17,18). In
these studies, ESBL was reported to have the most important role in
the progression of the antimicrobial resistance of Klebsiella
pneumoniae. Furthermore, clinical research has indicated that
ESBL production is frequently upregulated in Klebsiella
pneumoniae isolated from patients with long-term infection, who
are likely to be infected with other bacteria, making it difficult
for clinicians to select appropriate antibiotics for treatment
(19). However, despite the
different types of antibiotics that have been developed, the
molecular mechanisms of Klebsiella pneumoniae resistance to
antimicrobial drugs have remained to be fully elucidated (20).
At present, persistent microbial resistance of
Klebsiella pneumoniae represents a clinical problem
(21). Development of alternative
drugs to inhibit anti-microbial resistance for treating
hospital-acquired Klebsiella pneumoniae infections urgently
requires novel approaches. The present study investigated the role
of β-arrestin in the β-lactamase signaling pathway in Klebsiella
pneumoniae. A previous study reported that β-arrestin
translocates from the cytosol to the activated receptor and
regulates the G protein-coupled receptor signaling pathway
(22). In addition, the
β-arrestin-mediated pathway is mutually independent by selective
regulation of certain ligands in the transmembrane domain (23). Furthermore, studies have suggested
that the β-arrestin-mediated signaling pathway may be associated
with an unknown accommodation mode (24–26). In
agreement with this, the present study found that the
β-arrestin-mediated signaling pathway regulates ESBL production in
Klebsiella pneumoniae with antibiotic resistance.
The present study investigated the correlation
between β-arrestin and ESBL production to assess the mechanisms of
antimicrobial drug resistance of Klebsiella pneumoniae. The
results indicated that interference with β-arrestin recruitment or
synthesis decreased antimicrobial drug resistance, which may
provide an approach for controlling nosocomial infectious,
transmission and cross-infection. The association between
β-arrestin recruitment, ESBL and mechanisms of antibiotic
resistance of Klebsiella pneumoniae were studied in
vitro and in vivo. The results revealed that inhibition
of the recruitment of β-arrestin inhibited ESBL synthesis and
decreased the potential of antimicrobial drug resistance of
Klebsiella pneumoniae, which validated the role of the
β-arrestin-induced β-lactamase signaling pathway in the β-lactam
resistance of Klebsiella pneumoniae.
Materials and methods
Bacterial culture and reagents
A native Klebsiella pneumoniae strain
(NB-K.P.) was purchased from the American Type Culture
Collection (Manassas, VA, USA). The antibiotics-resistant strain
AR-K.P. was isolated from a 56-year male patient with
pneumonia who suffered from pneumonia for ~30 years. Klebsiella
pneumoniae cells were grown in Lysogeny broth (Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C
humidified atmosphere containing 5% CO2 for 24 h.
Growth potential assay
The growth potential was assessed according to the
protocol of a previous study (27).
The Klebsiella pneumoniae cells were cultured in medium
containing 10 mg/ml penicillin for 24 h. The number of
Klebsiella pneumoniae colonies was calculated on agar
plates.
Antimicrobial susceptibility
testing
Antimicrobial susceptibility tests of Klebsiella
pneumoniae were performed using the disk diffusion method,
according to the Clinical and Laboratory Standards Institute
recommendations (28). The final
results were obtained according to the respective standards for
antimicrobial susceptibility testing using Mueller-Hinton Broth
medium in agar plates (Merck KGaA; Darmstadt, Germany).
Small interfering (si)RNA-mediated
knockdown of β-arrestin in Klebsiella pneumoniae
A siRNA targeting β-arrestin gene sequences was
designed and synthesized by Invitrogen (Thermo Fisher Scientific,
Inc.). The siRNA oligonucleotides had the following sequences: p-1,
5′-AGCTTCTGTCCGGATCTAA-3′ with the sequence
5′-ACGTAGATCCTTCAGCACC-3′ was designed as a negative control.
siRNA-β-arrestin or siRNA-control were transfected into
Klebsiella pneumoniae cells for further analysis according
to the protocol of a previous study (29).
β-arrestin activity
Klebsiella pneumoniae clones were seeded in
Mueller–Hinton Broth medium in agar plates for 48 h at 37°C.
Following the addition of the Flash detection reagent (10 mg/ml;
Discoverx; Birmingham, UK), β-arrestin-luciferase signal was read
using a fluorescence imaging plate reader (FLIPR Tetra; Molecular
Devices, LLC; Sunnyvale, CA, USA).
β-arrestin endogenous or ESBL
expression
Klebsiella pneumonia cells were cultured in
Mueller-Hinton Broth medium in agar plates (Darmstadt, Germany) for
24 h at 37°C. Klebsiella pneumonia cells were prepared and
used to make competent cells according to previous report (30). β-arrestin or ESBL gene was cloned and
inserted into pET-28a plasmids using a transformation method
described previously (31). Cells
were checked by colony PCR and confirmed by sequence analysis of
the PCR products as described previously (32).
Reverse-transcription quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was obtained from Klebsiella
pneumoniae cells by using the RNAeasy Mini kit (Qiagen, Hilden,
Germany). Expression of β-arrestin and ESBL in cells was measured
by applying an RT-qPCR kit (Invitrogen; Thermo Fisher Scientific,
Inc.) (33). All of the forward and
reverse primers were synthesized by Thermo Fisher Scientific, Inc.
(Table I). Relative mRNA expression
changes were calculated by the 2−ΔΔCq method (34). The results are expressed as the fold
of the β-actin control.
 | Table I.Primer sequences used in the present
study. |
Table I.
Primer sequences used in the present
study.
|
| Sequence |
|---|
|
|
|
|---|
| Gene name | Reverse | Forward |
|---|
| β-arrestin |
5′-GGGAGGACGATGCGGA-3′ |
5′-CGCTGAGGATCCGAGA-3′ |
| ESBL |
5′-ATACGGGAGGGCTTACCATC-3′ |
5′-CGCCGCATACACTATTCTC-3′ |
| β-actin |
5′-CGGAGTCAACGGATTTGGTC-3′ |
5′-AGCCTTCTCCATGGTCGTGA-3′ |
Animal study
The study was approved by the Ethics Committee of
Tianjin First Center Hospital (Tianjin, China). A total of 60
female BALB/c nude mice (age, 6 weeks; body weight, 30–35 g) were
purchased from Charles River Laboratories GmbH (Sulzfeld, Germany).
All animals were fed under pathogen-free conditions. Mice were
maintained under a 12-h light/dark cycle with free access to food
and water. The mice were randomly divided into four groups, into
which different types of Klebsiella pneumoniae cells
(AR-K.P., AR-K.P.-β-arrestin-knockdown,
NB-K.P. or NB-K.P.-arrestin-knockdown) were injected
into the vena caudalis (108 infective particles per
mouse). The onset of illness was observed for each mouse in every
group. All mice received penicillin (0.3 mg/kg; Sigma-Aldrich;
Merck KGaA) treatment on day 24. The penicillin treatment was
continued for 15 days with administration once daily. On day 40,
lungs were isolated and used for protein analysis by
immunohistological staining. The efficacy of β-arrestin knockdown
in the progression of pneumonia was determined by PSI score of
pneumonia mice (35).
Immunofluorescence
Klebsiella pneumoniae cells or lung tissues
were fixed with formaldehyde solution (10%) and processed according
to standard procedures. Rehydrated slides or cells were incubated
with primary antibodies: β-arrestin (1:200 dilution; 30036; Cell
Signaling Technology, Inc., Danvers, MA, USA), ESBL (1:200
dilution; 90122; Cell Signaling Technology, Inc.) and β-actin
(1:1,000 dilution; ab8227; Abcam; Cambridge, MA, USA) for 60 min at
37°C in a humidified chamber. Detection of primary antibodies was
performed with horseradish peroxidase-conjugated anti-rabbit IgG
(1:200 dilution; 71623; Bio-Rad Laboratories, Inc., Hercules, CA,
USA) for 60 min at 37°C. Samples were washed and mounted with
VectaShield mounting media with DAPI (Vector Laboratories, Inc.,
Burlingame, CA, USA) and kept in the dark at 4°C until microscopic
analysis.
Immunohistochemistry
Lung tumors from experimental mice and were fixed in
10% formaldehyde for 20 min at 37°C, embedded in paraffin wax and
cut into serial sections of 4 µm in thickness. Tumor sections were
subjected to antigen retrieval using an Antigen Retrieval
Reagent-Universal kit (CTS015; R&D Systems, Inc., Minneapolis,
MN, USA) for 1 h at 25°C. Lung tumor sections were incubated with
rabbit anti-mouse antibody: BlaR1 (1:1,000 dilution; 91763; Cell
Signaling Technology, Inc.), Blal (1:1,000 dilution; 97364; Cell
Signaling Technology, Inc.), β-arrestin (1:200 dilution; 30036;
Cell Signaling Technology, Inc.) and ESBL (1:200 dilution; 90122;
Cell Signaling Technology, Inc.) for 12 h at 4°C. Lung sections
were then incubated with immunoperoxidase by means of a
3,3′-diaminobenzidine kit (Thermo Fischer Scientific, Inc.)
according to the manufacturer's instructions. The sections were
examined under a light microscope at a magnification of ×400.
Statistical analysis
Values are expressed as the mean ± standard error of
the mean. Unpaired data were analyzed by Student's t-test.
Comparisons of data between multiple groups were performed by
analysis of variance. All data were analyzed using SPSS Statistics
19.0 (IBM Corp., Armonk, NY, USA). P<0.05 and was considered to
indicate a statistically significant difference.
Results
β-arrestin and ESBL gene expression
and antimicrobial drug resistance of Klebsiella pneumoniae
In order to assess the drug resistance of
Klebsiella pneumoniae to β-lactams, β-arrestin and ESBL gene
expression were evaluated in AR-K.P. As presented in
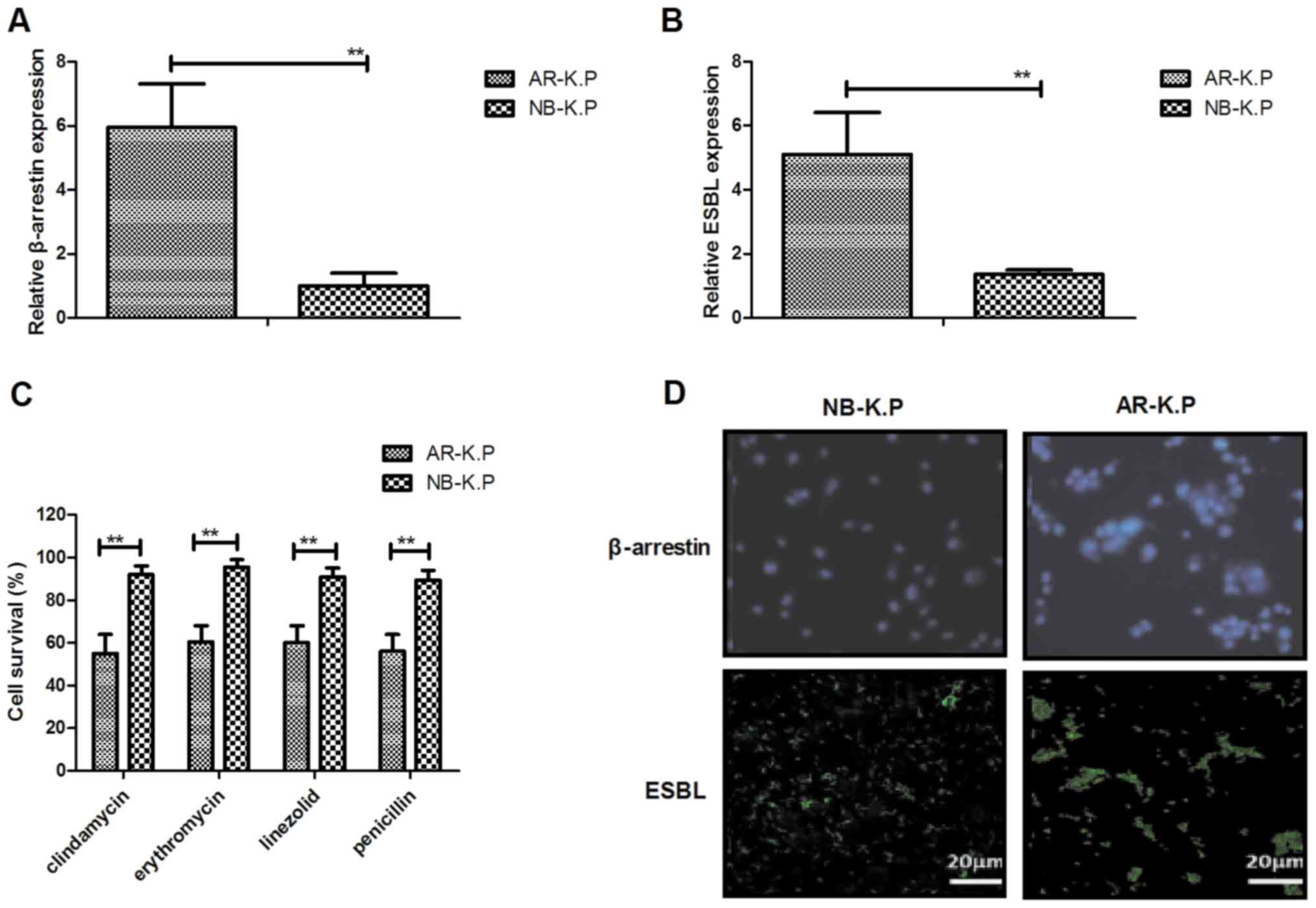
Fig. 1A and B, β-arrestin and ESBL
expression levels were downregulated in AR-K.P. compared to
those in NB-K.P. As displayed in Fig. 1C, AR-K.P. was resistant to the
antimicrobial drugs clindamycin, erythromycin, linezolid and
penicillin. In addition, the location of β-arrestin and ESBL
proteins in AR-K.P. was identified. The results indicated
that β-arrestin was located in the intracellular compartment and
that ESBL was expressed in the cytomembrane (Fig. 1D). These results suggested that
Klebsiella pneumoniae is resistant to antimicrobial drugs
and that β-arrestin and ESBL gene expression levels were
upregulated in AR-K.P.
Roles of β-arrestin and ESBL in
Klebsiella pneumoniae growth and ant-microbial drug resistance
In order to assess the role β-arrestin and ESBL in
Klebsiella pneumoniae growth, the activities of β-arrestin
and ESBL in Klebsiella pneumoniae were first evaluated. As
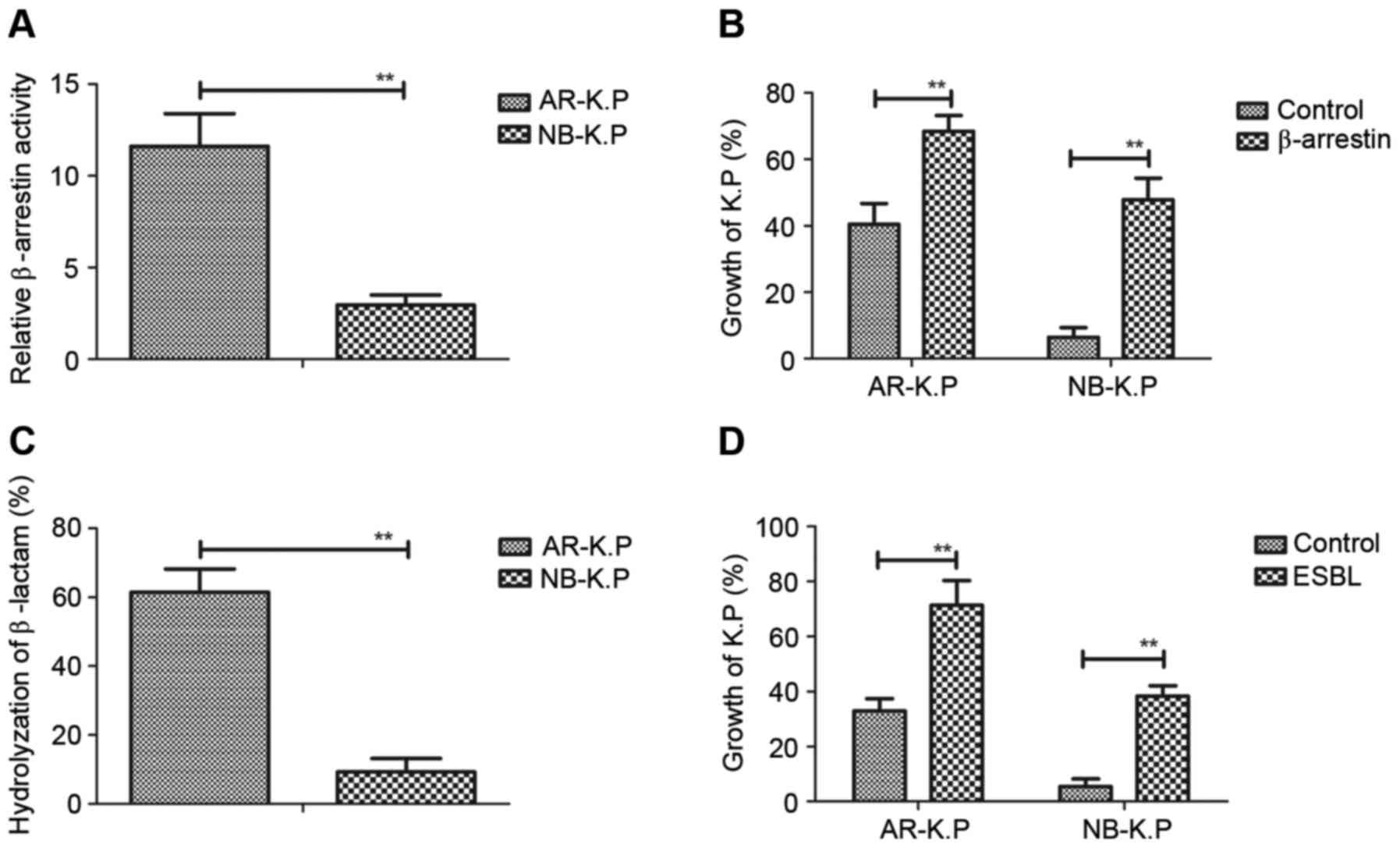
presented in Fig. 2A, the
β-arrestin-luciferase signal was higher in AR-K.P. compared
with that in NB-K.P. The growth of Klebsiella
pneumoniae in the presence of penicillin (10 mg/l) was prompted
after endogenous β-arrestin expression (Fig. 2B). As displayed in Fig. 2C, ESBL expressed in AR-K.P.
more efficiently hydrolyzed β-lactam compared with that in
NB-K.P. Of note, restoration of ESBL in Klebsiella
pneumoniae contributed to the capacity of antimicrobial drug
resistance, resulting in the promotion Klebsiella pneumoniae
growth in penicillin-containing medium (Fig. 2D). These results indicated that
β-arrestin and ESBL enhanced antimicrobial drug resistance and
promoted the growth of Klebsiella pneumoniae.
Knockdown of β-arrestin decreases ESBL
expression and growth of Klebsiella pneumoniae
To identify the association between β-arrestin, ESBL
and the antimicrobial drugs resistances of Klebsiella
pneumoniae, the β-arrestin-mediated signaling pathway was
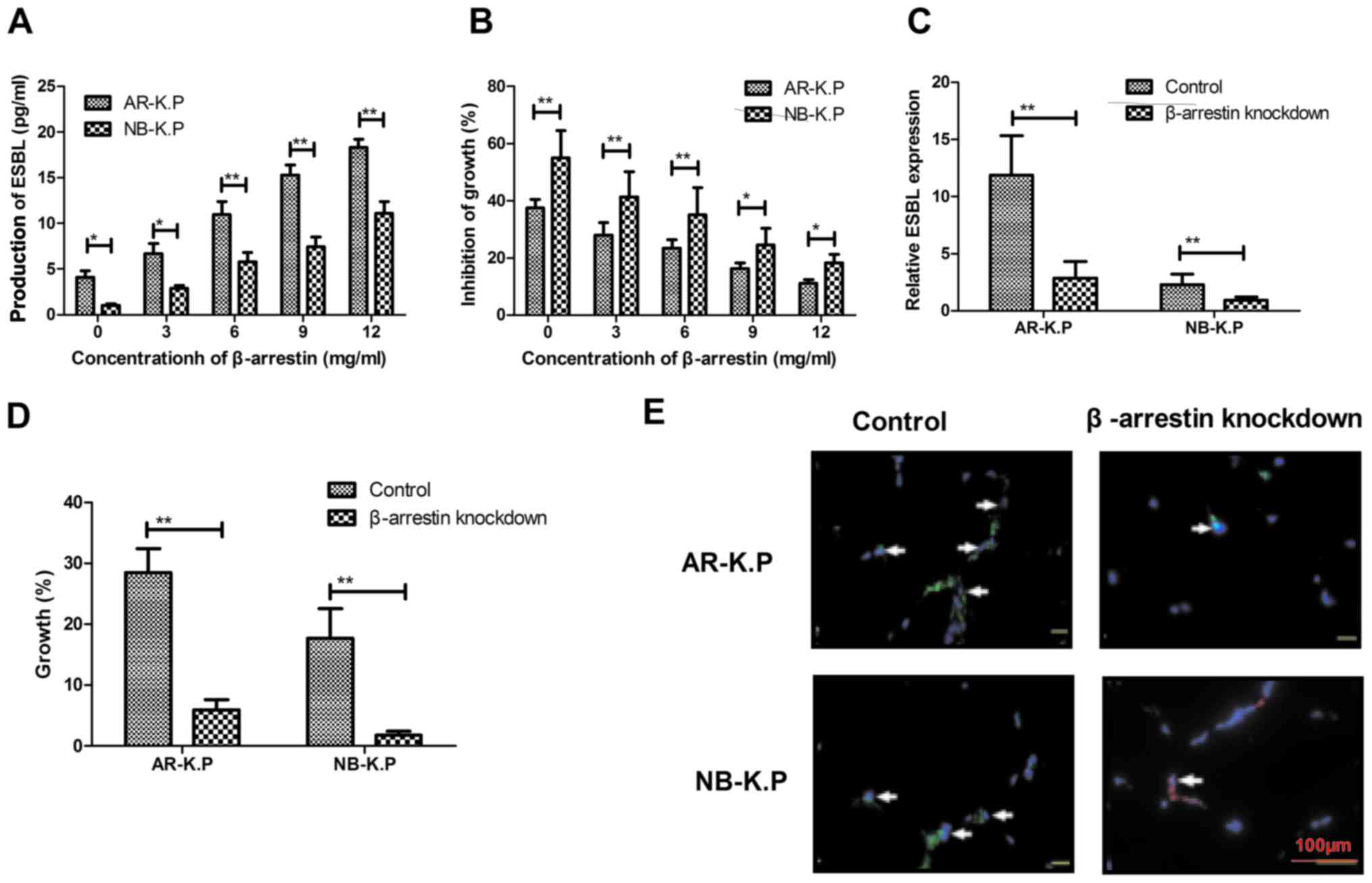
analyzed. As presented in Fig. 3A,
β-arrestin stimulated ESBL production in Klebsiella
pneumoniae and in a dose-dependent manner. The increasing
production of ESBL allowed Klebsiella pneumoniae to survive
in an environment containing penicillin antibiotics (Fig. 3B). Of note, β-arrestin knockdown led
to downregulation of ESBL expression and inhibited the growth of
Klebsiella pneumoniae (Fig. 3C
and D). β-arrestin knockdown also attenuated ESBL signal
intensity on the surface of Klebsiella pneumoniae (Fig. 3E). Collectively, these findings
suggested that β-arrestin knockdown induced the downregulation of
ESBL, which attenuated the antimicrobial drug resistance of
AR-K.P.
Knockdown of β-arrestin decreases the
virulence and antimicrobial drug resistance of Klebsiella
pneumoniae in vivo
To investigate the inhibitory effects of β-arrestin
knockdown on the biotic resistance capacity of Klebsiella
pneumoniae, mice were infected with Klebsiella
pneumoniae with or without β-arrestin knockdown and treated
with penicillin. In mice infected with Klebsiella pneumoniae
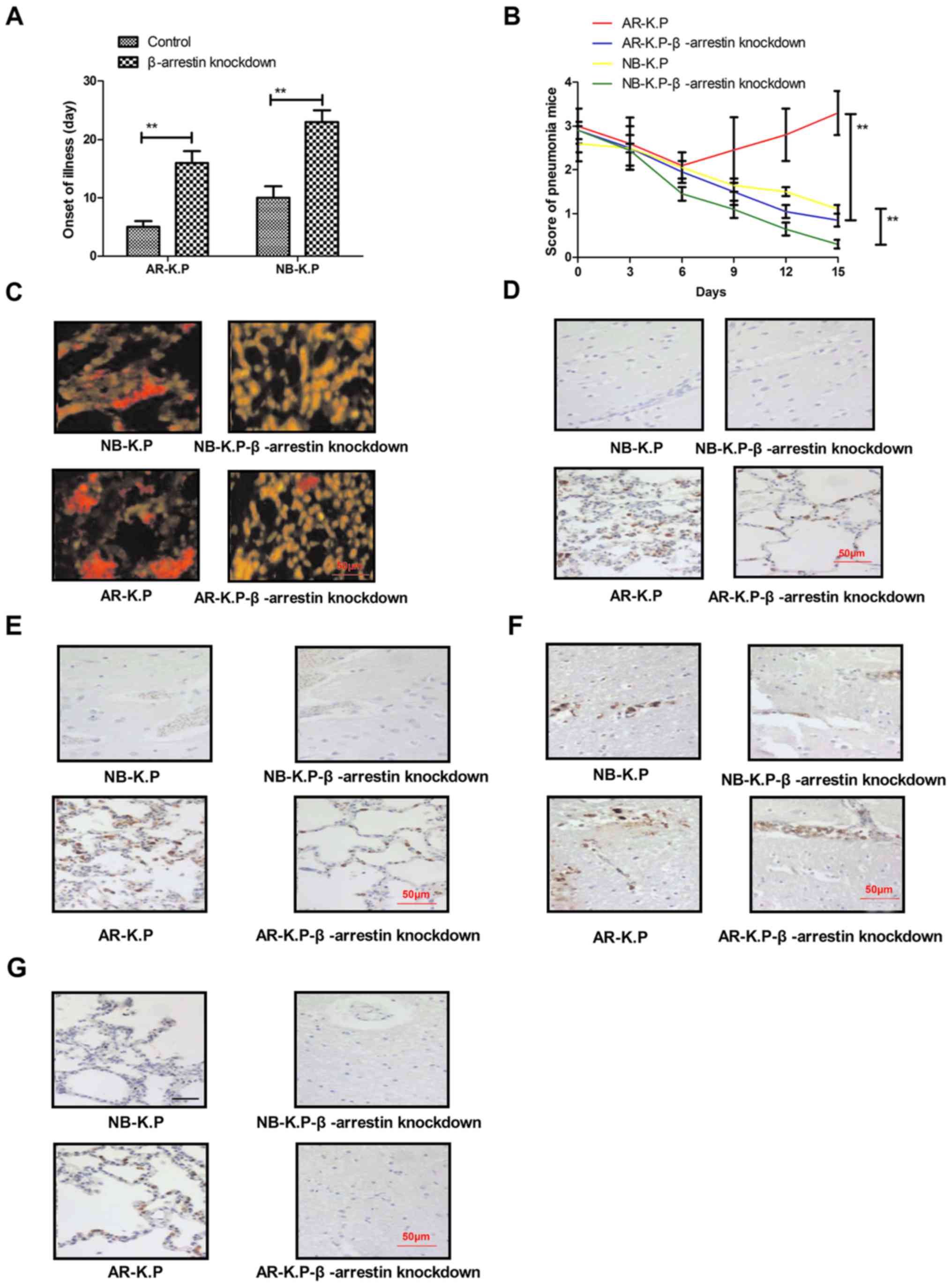
with β-arrestin knockdown, the onset of pneumonia was delayed
(Fig. 4A). Penicillin treatment was
also found to efficiently inhibit the proliferation of
Klebsiella pneumoniae with β-arrestin knockdown, resulting
in rapid recovery of pneumonia mice, as indicated by the decline in
pneumonia score (Fig. 4B).
Immunohistochemistry revealed that the Klebsiella pneumoniae
cells with β-arrestin knockdown were almost eradicated after a
15-day penicillin treatment compared to that in the control groups
(Fig. 4C). Of note, the expression
levels of two important members of the β-lactamase signal pathway
in Klebsiella pneumoniae, β-lactamase repressor (BlaR)1 and
Blal, were markedly decreased after penicillin treatment on day 7
(Fig. 4D and E). In addition, the
results demonstrated that β-arrestin and ESBL expression were
significantly downregulated in the lungs of mice infected with
Klebsiella pneumoniae with β-arrestin knockdown (Fig. 4F and G). Taken together, these
findings suggested that knockdown of β-arrestin decreased the
virulence and antimicrobial drug resistance of Klebsiella
pneumoniae in vivo.
Discussion
At present, treatment with β-lactam-based
antibiotics is the most common therapeutic strategy for
Klebsiella pneumoniae infection in the clinic (36). However, overuse and misuse of
antibiotics leads to rapid evolution of antibiotic-resistant
Klebsiella pneumoniae strains. In addition, clinical
evidence has suggested that Klebsiella pneumoniae has become
the most common pathogenic bacterium causing nosocomial infectious
due to the high virulence factors and general occurrence of
resistance to most antibiotics (37,38). The
association between antimicrobial drug resistance and biofilm
formation along with ESBL lactamase produced in Escherichia
coli has been assessed in a previous study (39). In line with this, the present study
indicated that antibiotic resistance of Klebsiella
pneumoniae depended on the expression of ESBL, which
efficiently hydrolyzed penicillin present in the medium. The
results demonstrated that β-arrestin recruitment significantly
regulated ESBL expression, resulting in a marked improvement of
antibiotic resistance of Klebsiella pneumoniae in vitro and
in vivo. These findings suggested that β-arrestin may be a
potential target to address the antibiotic resistance of
Klebsiella pneumoniae.
Based on previous studies, the increasing antibiotic
resistance of gram-negative bacilli enhances the rate of associated
morbidities and mortalities worldwide (40,41).
Klebsiella pneumoniae is a gram-negative bacterium and an
epidemiological study based on clinical data has revealed that
antibiotic resistance of Klebsiella pneumoniae is increasing
(42). However, antibiotic treatment
is the most common therapeutic regiment for pulmonary infections
with Klebsiella pneumoniae in patients with pneumonia
(43). Clinical experience has
demonstrated that antibiotic resistance of Klebsiella
pneumoniae has caused great problems in the treatment of
pneumonia, as only few antibiotic drugs may be applied to patients
infected with Klebsiella pneumoniae (44). Among these antibiotic drugs, β-lactam
antibiotics exhibit a higher efficiency in killing Klebsiella
pneumoniae. However, in recent years, drug resistance of
Klebsiella pneumoniae has led to insufficient efficacy of
β-lactam antibiotics (45). A study
has indicated that extended-spectrum β-lactamase produced by
Klebsiella pneumoniae may be associated with antibiotic
resistance (46). The mechanism of
β-lactamase hydrolyzing β-lactam antibiotics has been demonstrated
in Klebsiella pneumoniae (47). A previous study has identified
evolutionarily conserved genetic associations between β-lactamases
and antibiotic-producing bacteria (48). In addition, antibiotic resistance
genes originating from antibiotic-producing microorganisms may be
integrated into the genome of other pathogens through transduction
and/or transformation (49,50).
Theoretically, simultaneous and long-term analysis
of Klebsiella pneumoniae resistance to various
antimicrobials may contribute to an enhanced knowledge for
improving current and future perspectives regarding the therapeutic
use of these drugs (51). In a
previous study, β-arrestin recruitment was found to be closely
associated with β-lactamase enzyme fragments and with antibiotic
resistance of Klebsiella pneumoniae (52). The present study revealed that
β-arrestin knockdown regulated ESBL on the surface of Klebsiella
pneumoniae, which decreased antibiotic resistance in
vivo. Antibiotic-resistant bacteria and antibiotic resistance
genes can be passed on between different bacteria (7), which supports that clinical resistance
is intimately correlated with environmental resistance (8,9). The
present study found that β-arrestin may be a potential target for
inhibiting the antibiotic resistance of Klebsiella
pneumoniae.
In conclusion, despite the trend of the
antimicrobial susceptibility of Klebsiella pneumoniae
continuously decreasing due to the emergence of antibiotic
resistance, the molecular mechanisms of this resistance have
remained to be fully elucidated. The present study demonstrated
that regulation of β-arrestin modifies ESBL expression in
Klebsiella pneumoniae. Importantly, knockdown of β-arrestin
gene expression reduced the antibiotic-resistant features of
Klebsiella pneumoniae. In line with previously reported
pre-clinical findings (53), the
present study revealed that following β-arrestin knockdown, the
mean number of Klebsiella pneumoniae cells was significantly
decreased in the presence of penicillin, and that the sensitivity
to antibiotics was restored in vivo. Taken together, the
molecular mechanisms of Klebsiella pneumoniae resistance to
antibiotics proceed through the β-arrestin recruitment-induced ESBL
signaling pathway, suggesting that β-arrestin may be a potential
target to address antibiotic resistance of Klebsiella
pneumoniae.
Acknowledgements
This study was supported by Science and Technology
Fund of Tianjin Health and Family Planning Commission (grant no.
2014KZ029).
References
|
1
|
Li W, Dong K, Ren J and Qu X: A
β-Lactamase-imprinted responsive hydrogel for the treatment of
antibiotic-resistant bacteria. Angew Chem Int Ed Engl.
55:8049–8053. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yosef I, Manor M and Qimron U:
Counteracting selection for antibiotic-resistant bacteria.
Bacteriophage. 6:e10969962016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Singh N, Sit MT, Chung DM, Lopez AA,
Weerackoon R and Yeh PJ: How often are antibiotic-resistant
bacteria said to ‘evolve’ in the news? PLoS One. 11:e01503962016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sharma VK, Johnson N, Cizmas L, McDonald
TJ and Kim H: A review of the influence of treatment strategies on
antibiotic resistant bacteria and antibiotic resistance genes.
Chemosphere. 150:702–714. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lim MK, Lai PS, Ponnampalavanar SS, Syed
Omar SF, Taib NA, Yusof MY, Italiano CM, Kong DC and Kamarulzaman
A: Antibiotics in surgical wards: Use or misuse? A newly
industrialized country's perspective. J Infect Dev Ctries.
9:1264–1271. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zimmer BL: Combating antibiotic-resistant
bacteria: The Microbiology lab answers the challenge. MLO Med Lab
Obs. 47:30–33. 2015.PubMed/NCBI
|
|
7
|
Çöl A, Dedeić-Ljubović A, Salimović-Bešić
I and Hukic M: Antibiotic resistance profiles and genetic
similarities within a new generation of carbapenem-resistant
Acinetobacter calcoaceticus-A. Baumannii complex resistotypes in
bosnia and herzegovina. Microb Drug Resist. 22:655–661. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tahrani L, Soufi L, Mehri I, Najjari A,
Hassan A, Van Loco J, Reyns T, Cherif A and Ben Mansour H:
Isolation and characterization of antibiotic-resistant bacteria
from pharmaceutical industrial wastewaters. Microb Pathog.
89:54–61. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Conen A, Frei R, Adler H, Dangel M, Fux CA
and Widmer AF: Microbiological screening is necessary to
distinguish carriers of plasmid-mediated AmpC
beta-lactamase-producing enterobacteriaceae and extended-spectrum
beta-lactamase (ESBL)-producing Enterobacteriaceae because of
clinical similarity. PLoS One. 10:e01206882015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schauss T, Glaeser SP, Gütschow A, Dott W
and Kämpfer P: Improved detection of extended spectrum
beta-lactamase (ESBL)-producing Escherichia coli in input and
output samples of German biogas plants by a selective
pre-enrichment procedure. PLoS One. 10:e01197912015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bonnedahl J, Stedt J, Waldenström J,
Svensson L, Drobni M and Olsen B: Comparison of extended-spectrum
β-lactamase (ESBL) CTX-M genotypes in franklin gulls from Canada
and Chile. PLoS One. 10:e01413152015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Valenza G, Nickel S, Pfeifer Y, Pietsch M,
Voigtländer E, Lehner-Reindl V and Höller C: Prevalence and genetic
diversity of extended-spectrum β-lactamase (ESBL)-producing
Escherichia coli in nursing homes in Bavaria, Germany. Vet
Microbiol. 200:138–141. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gajul SV, Mohite ST, Mangalgi SS, Wavare
SM and Kakade SV: Klebsiella pneumoniae in septicemic neonates with
special reference to extended spectrum β-lactamase, AmpC, metallo
β-lactamase production and multiple drug resistance in tertiary
care hospital. J Lab Physicians. 7:32–37. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sudarwanto M, Akineden Ö, Odenthal S,
Gross M and Usleber E: Extended-spectrum β-lactamase
(ESBL)-producing Klebsiella pneumoniae in bulk tank milk from dairy
farms in Indonesia. Foodborne Pathog Dis. 12:585–590. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rastogi V, Nirwan PS, Jain S and Kapil A:
Nosocomial outbreak of septicaemia in neonatal intensive care unit
due to extended spectrum β-lactamase producing Klebsiella
pneumoniae showing multiple mechanisms of drug resistance. Indian J
Med Microbiol. 28:380–384. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bae D, Cheng CM and Khan AA:
Characterization of extended-spectrum β-lactamase (ESBL) producing
non-typhoidal Salmonella (NTS) from imported food products. Int J
Food Microbiol. 214:12–17. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Walther B, Lübke-Becker A, Stamm I, Gehlen
H, Barton AK, Janssen T, Wieler LH and Guenther S: Suspected
nosocomial infections with multi-drug resistant E. coli, including
extended-spectrum beta-lactamase (ESBL)-producing strains, in an
equine clinic. Berl Munch Tierarztl Wochenschr. 127:421–427.
2014.PubMed/NCBI
|
|
18
|
Schaufler K, Bethe A, Lübke-Becker A,
Ewers C, Kohn B, Wieler LH and Guenther S: Putative connection
between zoonotic multiresistant extended-spectrum beta-lactamase
(ESBL)-producing Escherichia coli in dog feces from a veterinary
campus and clinical isolates from dogs. Infect Ecol Epidemiol.
5:253342015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ibrahim DR, Dodd CE, Stekel DJ, Ramsden SJ
and Hobman JL: Multidrug resistant, extended spectrum β-lactamase
(ESBL)-producing Escherichia coli isolated from a dairy farm. FEMS
Microbiol Ecol. 92:fiw0132016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sanchez GV, Master RN, Clark RB, Fyyaz M,
Duvvuri P, Ekta G and Bordon J: Klebsiella pneumoniae antimicrobial
drug resistance, United States, 1998–2010. Emerg Infect Dis.
19:133–136. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Barger A, Schaade L, Krause G and Kramer
M: Strategies for recognition, prevention and control of
antimicrobial resistances in Germany-a draft from the Federal
German Ministry of Health. Gesundheitswesen. 70:631–635. 2008.(In
German). View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Barthet G, Framery B, Gaven F, Pellissier
L, Reiter E, Claeysen S, Bockaert J and Dumuis A:
5-hydroxytryptamine 4 receptor activation of the extracellular
signal-regulated kinase pathway depends on Src activation but not
on G protein or beta-arrestin signaling. Mol Biol Cell.
18:1979–1991. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lipsky R, Potts EM, Tarzami ST, Puckerin
AA, Stocks J, Schecter AD, Sobie EA, Akar FG and Diversé-Pierluissi
MA: beta-Adrenergic receptor activation induces internalization of
cardiac Cav1.2 channel complexes through a beta-arrestin 1-mediated
pathway. J Biol Chem. 283:17221–17226. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mittal N, Roberts K, Pal K, Bentolila LA,
Fultz E, Minasyan A, Cahill C, Pradhan A, Conner D, DeFea K, et al:
Select G-protein-coupled receptors modulate agonist-induced
signaling via a ROCK, LIMK, and β-arrestin 1 pathway. Cell Rep.
5:1010–1021. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Borroni EM, Cancellieri C, Vacchini A,
Benureau Y, Lagane B, Bachelerie F, Arenzana-Seisdedos F, Mizuno K,
Mantovani A, Bonecchi R and Locati M: β-arrestin-dependent
activation of the cofilin pathway is required for the scavenging
activity of the atypical chemokine receptor D6. Sci Signal.
6:ra30.1–11, S1-S3. 2013. View Article : Google Scholar
|
|
26
|
Zoudilova M, Kumar P, Ge L, Wang P, Bokoch
GM and DeFea KA: Beta-arrestin-dependent regulation of the cofilin
pathway downstream of protease-activated receptor-2. J Biol Chem.
282:20634–20646. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zscheppang K, Kurth I, Wachtel N,
Dubrovska A, Kunz-Schughart LA and Cordes N: Efficacy of beta1
integrin and EGFR targeting in sphere-forming human head and neck
cancer cells. J Cancer. 7:736–745. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hsueh PR, Ko WC, Wu JJ, Lu JJ, Wang FD, Wu
HY, Wu TL and Teng LJ: Consensus statement on the adherence to
Clinical and Laboratory Standards Institute (CLSI) Antimicrobial
Susceptibility Testing Guidelines (CLSI-2010 and CLSI-2010-update)
for Enterobacteriaceae in clinical microbiology laboratories in
Taiwan. J Microbiol Immunol Infect. 43:452–455. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Aguileta MA, Rojas-Rivera D, Goossens V,
Estornes Y, Van Isterdael G, Vandenabeele P and Bertrand MJ: A
siRNA screen reveals the prosurvival effect of protein kinase A
activation in conditions of unresolved endoplasmic reticulum
stress. Cell Death Differ. 23:1670–1680. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wirries A, Breyer S, Quint K, Schobert R
and Ocker M: Thymoquinone hydrazone derivatives cause cell cycle
arrest in p53-competent colorectal cancer cells. Exp Ther Med.
1:369–375. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
George TP and Thomas T: Discrete wavelet
transform de-noising in eukaryotic gene splicing. BMC
Bioinformatics. 11 Suppl 1:S502010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Weigel RM, Qiao B, Barber DA, Teferedegne
B, Kocherginskaya S, White BA and Isaacson RE: Identification of
patterns of transmission of Salmonella within swine production
systems using pulsed field gel electrophoresis (PFGE) and
repetitive sequence polymerase chain reaction (REP-PCR): A
quantitative analysis. Berl Munch Tierarztl Wochenschr.
114:397–400. 2001.PubMed/NCBI
|
|
33
|
Xiao S, Wang J and Xiao N: MicroRNAs as
noninvasive biomarkers in bladder cancer detection: A diagnostic
meta-analysis based on qRT-PCR data. Int J Biol Markers.
31:e276–e285. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kishimoto M, Yamana H, Inoue S, Noda T,
Myojin T, Matsui H, Yasunaga H, Kawaguchi M and Imamura T:
Sivelestat sodium and mortality in pneumonia patients requiring
mechanical ventilation: Propensity score analysis of a Japanese
nationwide database. J Anesth. 31:405–412. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ceccarelli G, Falcone M, Giordano A,
Mezzatesta ML, Caio C, Stefani S and Venditti M: Successful
ertapenem-doripenem combination treatment of bacteremic
ventilator-associated pneumonia due to colistin-resistant
KPC-producing Klebsiella pneumoniae. Antimicrob Agents Chemother.
57:2900–2901. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
McCullough AR, Parekh S, Rathbone J, Del
Mar CB and Hoffmann TC: A systematic review of the public's
knowledge and beliefs about antibiotic resistance-authors'
response. J Antimicrob Chemother. 71:23662016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bryce A, Hay AD, Lane IF, Thornton HV,
Wootton M and Costelloe C: Global prevalence of antibiotic
resistance in paediatric urinary tract infections caused by
Escherichia coli and association with routine use of antibiotics in
primary care: Systematic review and meta-analysis. BMJ.
352:i9392016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Neupane S, Pant ND, Khatiwada S, Chaudhary
R and Banjara MR: Correlation between biofilm formation and
resistance toward different commonly used antibiotics along with
extended spectrum beta lactamase production in uropathogenic
Escherichia coli isolated from the patients suspected of urinary
tract infections visiting Shree Birendra Hospital, Chhauni,
Kathmandu, Nepal. Antimicrob Resist Infect Control. 5:52016.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Moradi J, Hashemi FB and Bahador A:
Antibiotic resistance of Acinetobacter baumannii in Iran: A
systemic review of the published literature. Osong Public Health
Res Perspect. 6:79–86. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Khademi F, Poursina F, Hosseini E, Akbari
M and Safaei HG: Helicobacter pylori in Iran: A systematic review
on the antibiotic resistance. Iran J Basic Med Sci. 18:2–7.
2015.PubMed/NCBI
|
|
42
|
Lee CY, Tsai HC, Lee SS and Chen YS:
Concomitant emphysematous prostatic and periurethral abscesses due
to Klebsiella pneumoniae: A case report and review of the
literature. Southeast Asian J Trop Med Public Health. 45:1099–1106.
2014.PubMed/NCBI
|
|
43
|
Oliva A, Gizzi F, Mascellino MT, Cipolla
A, D'Abramo A, D'Agostino C, Trinchieri V, Russo G, Tierno F,
Iannetta M, et al: Bactericidal and synergistic activity of
double-carbapenem regimen for infections caused by
carbapenemase-producing Klebsiella pneumoniae. Clin Microbiol
Infect. 22:147–153. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rosen DA, Hilliard JK, Tiemann KM, Todd
EM, Morley SC and Hunstad DA: Klebsiella pneumoniae FimK promotes
virulence in murine pneumonia. J Infect Dis. 213:649–658. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Netikul T and Kiratisin P: Genetic
Characterization of carbapenem-resistant Enterobacteriaceae and the
spread of carbapenem-resistant Klebsiella pneumonia ST340 at a
university hospital in Thailand. PLoS One. 10:e01391162015.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Walker CD and Shankaran S: Extended
antibiotic resistance in carbapenemase-producing Klebsiella
pneumoniae: A case series. Am J Infect Control. 44:1050–1052. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Sekowska A, Janicka G, Kłyszejko C, Wojda
M, Wróblewski M and Szymankiewicz M: Resistance of Klebsiella
pneumoniae strains producing and not producing ESBL
(extended-spectrum beta-lactamase) type enzymes to selected
non-beta-lactam antibiotics. Med Sci Monit. 8:BR100–BR104.
2002.PubMed/NCBI
|
|
48
|
Sanguinetti CM, De Benedetto F and
Miragliotta G: Bacterial agents of lower respiratory tract
infections (LRTIs), beta-lactamase production, and resistance to
antibiotics in elderly people. DEDALO Study Group. Int J Antimicrob
Agents. 16:467–471. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Boggio SB and Roveri OA: Catalytic
properties of an endogenous beta-lactamase responsible for the
resistance of Azospirillum lipoferum to beta-lactam antibiotics.
Microbiology. 149:445–450. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Uraz G, Simsek H and Celik B:
beta-Lactamase activities and resistance to antibiotics of
Haemophilus influenzae, H. parainfluenzae and H. aphrophilus
strains identified in throat cultures from children. Drug Metabol
Drug Interact. 16:217–228. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Soge OO, Adeniyi BA and Roberts MC: New
antibiotic resistance genes associated with CTX-M plasmids from
uropathogenic Nigerian Klebsiella pneumoniae. J Antimicrob
Chemother. 58:1048–1053. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ikeda Y, Kumagai H, Okazaki H, Fujishiro
M, Motozawa Y, Nomura S, Takeda N, Toko H, Takimoto E, Akazawa H,
et al: Monitoring β-arrestin recruitment via β-lactamase enzyme
fragment complementation: Purification of peptide E as a
low-affinity ligand for mammalian bombesin receptors. PLoS One.
10:e01274452015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ramirez MS, Traglia GM, Lin DL, Tran T and
Tolmasky ME: Plasmid-Mediated antibiotic resistance and virulence
in Gram-begatives: The Klebsiella pneumoniae paradigm. Microbiol
Spectr. 2:2014. View Article : Google Scholar
|