Introduction
Hepatic fibrosis is a wound healing response to a
variety of chronic stimuli that results in the excessive production
and deposition of extracellular matrix (ECM) in the liver (1). Hepatic stellate cells (HSCs) are the
central link and the cytological basis of liver fibrosis. The
activation of HSCs is a well-accepted critical event in hepatic
fibrosis and an attractive target for treatment. Activated HSCs are
able to secrete various cytokines, including transforming growth
factor (TGF)-β and platelet-derived growth factor (PDGF), which act
as regulators of various different signaling transduction pathways
in the formation of liver fibrosis (2). PDGF, which is one of the most effective
mitogens in terms of exerting functions on HSCs, regulates liver
fibrosis via autocrine and paracrine signaling throughout the
entire HSC activation process (3). A
previous study demonstrated that PDGF was able to promote
insulin-like growth factor (IGF)-1, secretion and the release of
IGF binding proteins in HSC cells and therefore induce liver
fibrosis via PDGF self-interaction (4). HSCs are also the primary cell type
responsible for the progression of liver fibrosis and serve a
central role in the pathogenesis of liver fibrosis (5). The HSC-T6 immortal cell line is
established by transfecting rat HSCs with the SV40 virus and has
the phenotype of activated HSC and can virtually be pass aged
infinitely (5). In the present
study, HSC-T6 was used as a cell model to investigate the role of
amygdalin in regulating the mRNA expression levels of PDGF and IGF
and the protein expression levels of PDGF and its receptor
(PDGFR).
Materials and methods
Experimental cells
The HSC-T6 cell line was cryopreserved and
resuscitated by the Institute of Tropical Medicine, Guangzhou
University of Chinese Medicine (Guangzhou, China).
Experimental drugs
Amygdalin (molecular weight, 457.4; cat. no.
110820-200403; 20 mg/tube) was purchased from the National
Institute for the Control of Pharmaceutical and Biological Products
(Beijing, China). The storage solution was prepared by diluting
amygdalin in water to a concentration of 10−5,
10−4 and 10−3 mol/l. The solution then under
intermittent sterilization at a low-temperature (4°C) and was
preserved at −20°C for future use. Prior to experiments, the
stocking solution was prepared by diluting the storage solution
with Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) to a concentration of 20
mmol/l, followed by sterilization with microporous membranes and
preservation at −20°C.
Chemical reagents
The following chemical reagents were used in the
present study: High-glucose DMEM (Gibco; Thermo Fisher Scientific,
Inc.); fetal bovine serum (FBS; Hangzhou Sijiqing Biological
Engineering Materials Co., Ltd., Hangzhou, China); L-glutamine,
dimethyl sulfoxide, trypsin acrylamide, pancreatic enzymes,
methanol and Tris-base (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany); EDTA (Shanghai Shenggong Biology Engineering Technology
Service, Ltd., Shanghai, China); penicillin and streptomycin (North
China Pharmaceutical Co., Ltd., Shijiazhuang, China); RNA
extraction kit, RNA reverse transcription kit, dNTP, RNase
inhibitor and oligo-dT (Tiangen Biotech Co., Ltd., Beijing, China);
DNA marker, polymerase chain reaction (PCR) kit and optimized PCR
kit (Tiangen Biotech Co., Ltd.); protein extraction kit (cat. no.
DB1011; Bioscience Co., Ltd., Shanghai, China); bovine serum
albumin (Shanghai Pufei Biotech Co., Ltd., Shanghai, China);
nitrocellulose membrane and filter paper (Whatman; GE Healthcare
Life Sciences, Shanghai, China); horseradish peroxidase-conjugated
secondary antibody (cat. no. 365802; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA); antibodies against PDGF(cat. no. 360511;
Bioworld Technology, Inc., St. Louis Park, MN, USA); fluorescein
isothiocyanate secondary antibody (1:100; cat. no. 375702; Santa
Cruz Biotechnology, Inc.) and antibodies against PDGFR-α (cat. no.
361162) and PDGFR-β (cat. no. 360924; Bioworld Technology,
Inc.).
Equipment
The following instruments were used in the present
study: Clean bench (cat. no. YJ-875; Suzhou Purification
Engineering Installation Co., Ltd., Suzhou, China,); cell culture
flask (Corning Incorporated, Corning, NY, USA); cell counting slice
and millipore filter (0.45 and 0.2 µm, respectively; Shanghai
Peninsula Industrial Co., Ltd, Shanghai, China); fluorescence
microscope (Leica Microsystems, Inc., Buffalo Grove, IL, USA);
inverted microscope (BX600; Olympus Corporation, Tokyo, Japan);
−86°C cryogenic refrigerator (Revco; Thermo Fisher Scientific,
Inc.); incubator (2300–2E; Sheldon Manufacturing, Inc., Cornelius,
OR, USA); microplate reader (ELX800; BioTek Instruments, Inc.,
Winooski, VT, USA); PCR amplifier (Revco; Thermo Fisher Scientific,
Inc.); homogenizer (Ningbo Scientz Biotechnology Co., Ltd., Ningbo,
China); pipette (Eppendorf, Hamburg, Germany); film analysis system
(Tanon Science and Technology Co., Ltd., Shanghai, China);
ultraviolet spectrophotometer (Hach Company, Loveland, CO, USA);
electrophoresis system (Bio-Rad Laboratories, Inc., Hercules, CA,
USA); and flow cytometer (Beckman Coulter, Inc., Brea, CA,
USA).
Cell treatment
The HSC-T6 immortal cell line, which has unlimited
passing characteristics, was generated by transfecting SV40 virus
into rat HSC cells. The transfected HSC-T6 cell line was purchased
from the Type Culture Collection of the Chinese Academy of Sciences
(Shanghai, China). HSC-T6 cells (1×106 cells/well) were
planted in a 6-well plate and incubated at 37°C with 5%
CO2 for 24 h and DMEM containing 5% FBS. The cells were
subsequently allowed to attach and following growth to 50%
confluence, the medium was replenished. HSC-T6 cells were then
randomly divided into four groups and incubated with different
amounts of amygdalin as follows: Control (DMEM containing 2% FBS),
low dose (10−5 mol/l), mid dose (10−4 mol/l)
and high dose (10−3 mol/l) at 37°C and 5% CO2
for 48 h. Amygdalin in each treatment group was dissolved in DMEM
containing 2% FBS.
RNA extraction
Amygdalin treatment was administered as
aforementioned and three repeats were performed for each group.
Following 48 h drug treatment, cells were digested and the total
RNA was extracted using a TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
Briefly, 1 ml lysis buffer was added to cells, mixed and allowed to
stand for 5 min. Subsequently 200 µl CHCl3 was added,
the mixture was vortexed for 30 sec, allowed to stand for 10 min
and centrifuged at a speed of 14,800 × g for 10 min at 4°C. The
supernatant was collected and added to 1/2 (vol/vol) ethanol,
allowed to stand for 2 min and centrifuged at a speed of 14,800 × g
at 4°C for 30 sec. Subsequently, 0.5 ml deproteinized buffer was
added and the mixture was allowed to stand for 2 min, at which
point the mixture was again centrifuged at a speed of 14,800 × g at
4°C for 30 sec. The precipitate was added to 0.7 ml wash buffer,
allowed to stand for 2 min, followed by centrifuging at 14,800 × g
at 4°C for 30 sec. This washing step was repeated twice. RNA was
collected following centrifuging the isolation column at a speed of
14,800 × g at 4°C for 2 min and air-drying for 10 min. Then, 50 µl
RNase-free water was added to the collected RNA, allowed to stand
for 2 min and centrifuged again at 14,800 × g at 4°C. The
supernatant was stored at −70°C prior to further use.
Reverse transcription-quantitative PCR
(RT-qPCR)
The expression profiles of PDGF and IGF were
measured using a one-step SYBR PrimeScript RT-PCR kit (Takara Bio,
Inc., Otsu, Japan) in an ABI Prism 7500 (Applied Biosystems; Thermo
Fisher Scientific, Inc.) following the manufacturer's protocol. An
RNA reverse transcription kit, dNTP, RNase inhibitor and oligo-dT
(all Tiangen Biotech Co., Ltd., Beijing, China) were used for
RT-qPCR. The 20 µl reaction mixture contained 4 µl cDNA, 2 µl 10X
buffer, 0.4 µl dNTP (10 mol/l), 0.4 µl primer mix and 0.5 U Taq
polymerase and ddH2O. The qPCR procedure was as follows:
Initial denaturation at 94°C for 2 min, followed by 30 cycles of
denaturation at 94°C for 30 sec, annealing at 58°C for 30 sec and
elongation at 72°C for 45 sec, with a final elongation at 72°C for
10 min and short storage at 4°C. The primer sequences utilized in
RT-qPCR were as follows: PDGF, forward, 5′-GATCCGCTCCTTTGATGATC-3′
and reverse, 5′-GTCTCACACTTGCATGCCAG-3′; IGF, forward
5′-AAGCCTACAAAGTCAGCTCC-3′ and reverse, 5′-CGTCTTGTTTCCTGCACTTC-3′;
and β-actin, forward, 5′-TGGTGGGTATGGGTCAGAAGGACTC-3′ and reverse,
5′-CATGGCTGGGGTGTTGAAGGTCTCA-3′. All primers were synthesized by
Invitrogen; Thermo Fisher Scientific, Inc. The relative expression
was analyzed using the 2−ΔΔCq method (6) and the expression of all transcripts
were normalized to that of the housekeeping gene β-actin.
Western blotting
Whole-cell lysates were isolated using
radioimmunoprecipitation assay lysis buffer (Thermo Fisher
Scientific, Inc.) and proteins were extracted using a BCA protein
assay kit (Thermo Fisher Scientific, Inc.) following the
manufacturer's protocol. The PDGF protein was analyzed using
western blotting as previously described (7). For electrophoresis, 15% SDS-PAGE was
prepared by mounting glass slides with 0.75-mm comb and injecting
the solution from the side. The gel was allowed to solidify at room
temperature for 30–45 min and protein samples were loaded following
boiling for 3–5 min. Electrophoresis was performed under 110 V for
60 min and the gel was subsequently removed from the rack and
balanced with the transfer buffer at room temperature for 30 min.
Also, a nitrocellulose membrane was placed (at a 45° angle) into
the transfer buffer and balanced for 10–15 min. The membrane was
placed on the gel, bubbles were removed and the transfer device was
locked and placed into the electrophoresis rack with each electrode
connected properly. Transmembrane electrophoresis was performed
under 100 V for 60 min. Following protein transfer, the membrane
was incubated at 4°C with the 5 ml blocking buffer and the PDGF
monoclonal antibody (1:100; cat. no. 360511; Bioworld Technology,
Inc.) overnight and then incubated with horseradish
peroxidase-conjugated secondary antibodies (1:100) at room
temperature for 1 h. Immunoreactive bands were detected by enhanced
chemiluminescence (ECL) reagent (Pierce; Thermo Fisher Scientific,
Inc.), visualized by autoradiography and quantified using the
Quantity One analysis system (version 4.4.02; Bio-Rad Laboratories,
Inc.). β-actin (1:100; cat. no. 390521; Bioworld Technology, Inc.)
was utilized as the internal control.
Flow cytometry
Cells were routinely cultured, digested using 0.25%
pancreatic enzymes and 0.25% EDTA and washed with PBS prior to
centrifugation at 200 × g at 4°C for 5 min. The cells were blocked
using 1% BSA (Pierce, Thermo Fisher Scientific, Inc.) at 4°C for 3
h. The supernatant was discarded and cells were incubated with a
primary antibodies (1:100; 0.5 ml); antibodies against PDGFR-α
(cat. no. 361162); antibodies against PDGFR-β (cat. no. 360924;
Bioworld Technology, Inc.) at room temperature for 30 min, washed
three times with PBS (3 min each) and centrifuged at 200 × g at 4°C
for 2 min for collection. Subsequently, the fluorescein
isothiocyanate secondary antibody (1:100; 0.5 ml; cat. no. 375702;
Santa Cruz Biotechnology, Inc.) was added and the cells were
incubated for 30 min at room temperature. Cells were then washed
three times with PBS (3 min each). Cells were then fixed in 1%
paraformaldehyde at room temperature for 30 min and adjusted to a
concentration of 5×105 cells/ml prior to analysis. PBS
was utilized as a negative control for PDGFR-α. However, PDGFR-β
has no wild-type group, so incubation using secondary antibodies
alone served as the control. Flow cytometry was then performed
using a Beckman Coulter flow cytometer. Results were analyzed using
Win MDI 2.9 software (Bio-Rad Laboratories, Inc.).
Statistical analysis
Data were presented as the mean ± standard deviation
and three repeats were performed for each group. Statistical
analysis was performed using one-way analysis of variance followed
by a Student-Newman-Keuls test. The differences between the two
groups were compared using a Student's t-test. Statistical analysis
was performed using SPSS 13.0 software (SPSS, Inc., Chicago, IL,
USA). P<0.05 indicated that the difference between groups was
statistically significant.
Results
PT-qPCR detection of PDGF mRNA
A high dose of amygdalin significantly suppressed
the mRNA expression of PDGF compared with the control group
(P<0.05; Fig. 1A). However, low
or mid doses of amygdalin had no significant effect on suppressing
PDGF mRNA expression compared with the control group. Furthermore,
the mid and low dose treatments led to an increase of PDGF mRNA
compared with the control group (P<0.05; Fig. 1A). These results suggest that effect
of amygdalin in suppressing the mRNA expression of PDGF is
dose-dependent.
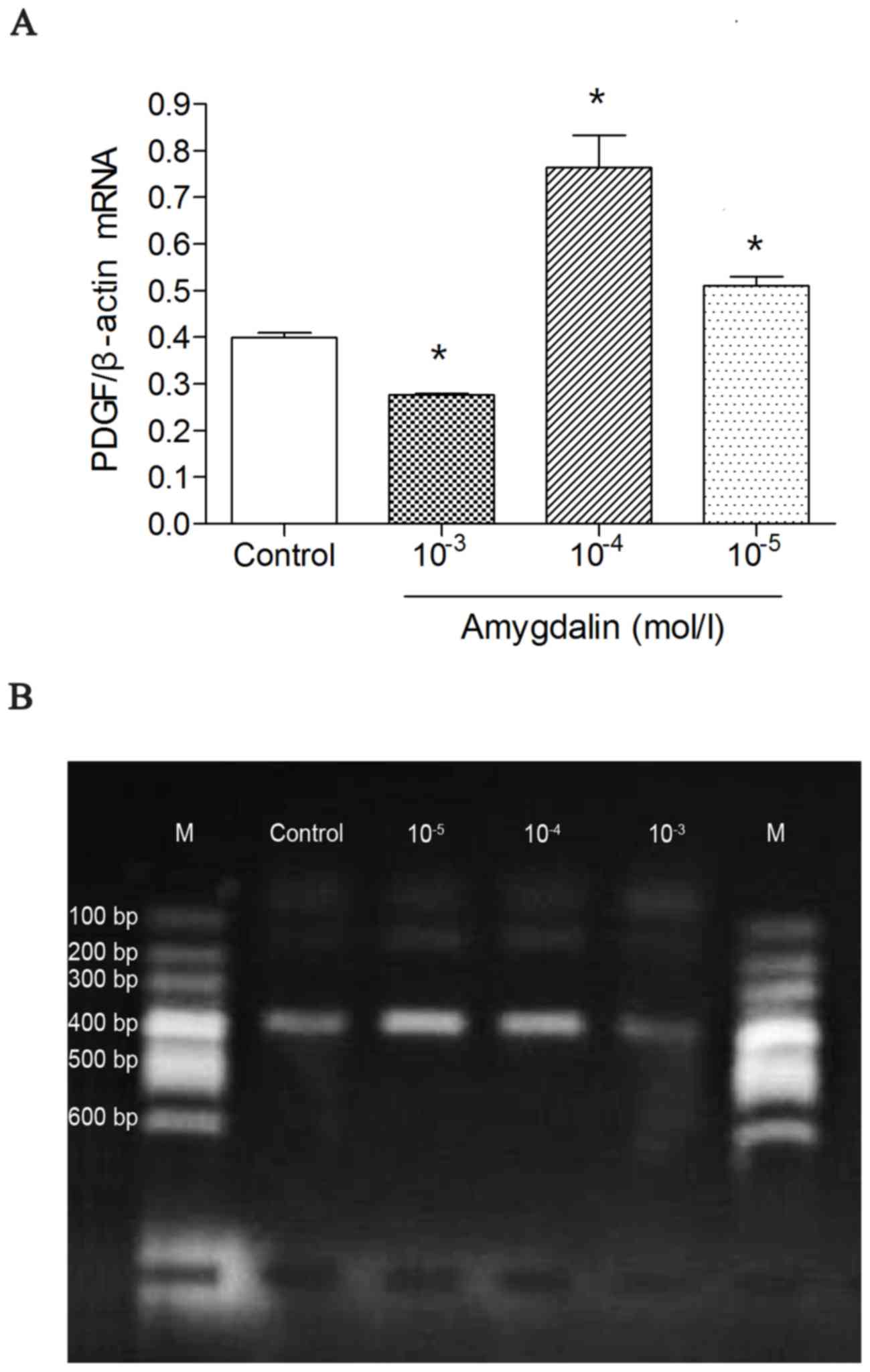 | Figure 1.RT-qPCR results showing PDGF mRNA
levels in the four groups. (A) RT-qPCR was used to measure the
expression of PDGF mRNA. (B) HSC-T6 cells were incubated with
amygdalin for 48 h and the PDGF mRNA levels were assessed using
RT-qPCR. *P<0.05 vs. control. The high dose group
(10−3 mol/l amygdalin) exhibited a significant decrease
compared with the control group. In addition, the low- and mid-dose
groups (10−5 mol/l amygdalin; 10−4 mol/l
amygdalin, respectively) exhibited a significant increase in PDGF
mRNA, compared with the control group. RT-qPCR, reverse
transcription quantitative polymerase chain reaction; PDGF,
platelet-derived growth factor; Con, control; 10−5, low
dose, 10−5 mol/l amygdalin; 10−4, mid dose,
10−4 mol/l amygdalin; 10−3, high dose,
10−3 mol/l amygdalin; M, DNA marker. |
RT-qPCR detection of IGF mRNA
A high dose of amygdalin significantly suppressed
the mRNA expression of IGF compared with the other experimental
groups and the control group (P<0.05; Fig. 2A). Furthermore, low or mid-doses of
amygdalin had no significant effect on suppressing the mRNA
expression of IGF, compared with the control group. However, the
mid dose treatment did lead to an increase of IGF mRNA compared
with the control group (Fig. 2A).
This result suggested a dose-dependent effect of amygdalin in
suppressing the mRNA expression of IGF.
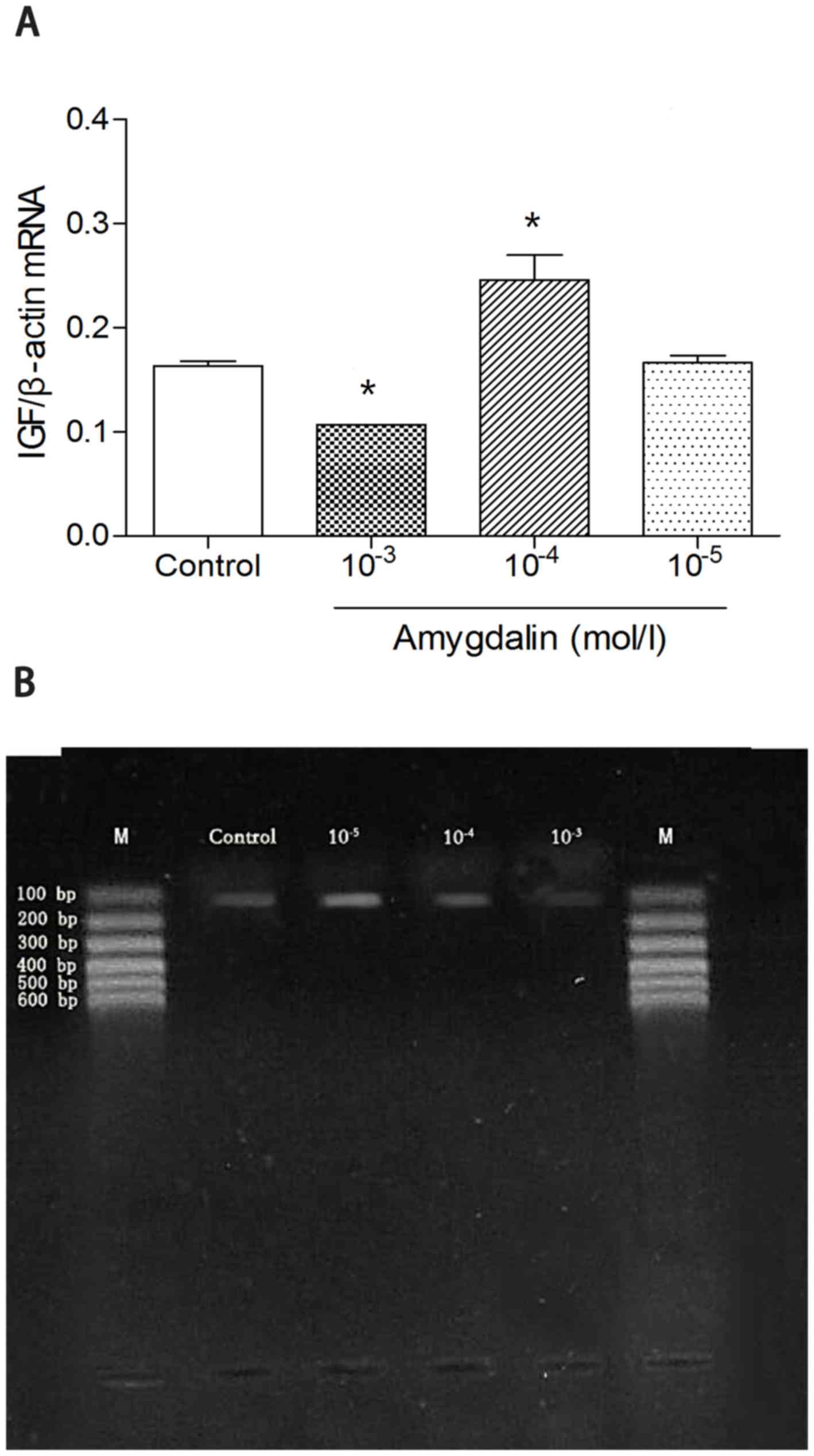 | Figure 2.RT-qPCR results presenting IGF mRNA
levels in the four groups. (A) RT-qPCR analysis was used to measure
the expression of IGF mRNA. (B) HSC-T6 cells were incubated with
amygdalin for 48 h and the level of IGF mRNA was assessed using
RT-qPCR. *P<0.05 vs. Control. The high dose group
(10−3 mol/l amygdalin) exhibited a significant decrease
compared with the control group. In addition, the mid dose group
(10−4 mol/l amygdalin) exhibited a significant increase
in IGF mRNA, compared with the control group. RT-qPCR, reverse
transcription quantitative polymerase chain reaction; IGF,
insulin-like growth factor; Con, control; 10−5, low
dose, 10−5 mol/l amygdalin; 10−4, mid dose,
10−4 mol/l amygdalin; 10−3, high dose,
10−3 mol/l amygdalin; M, DNA marker. |
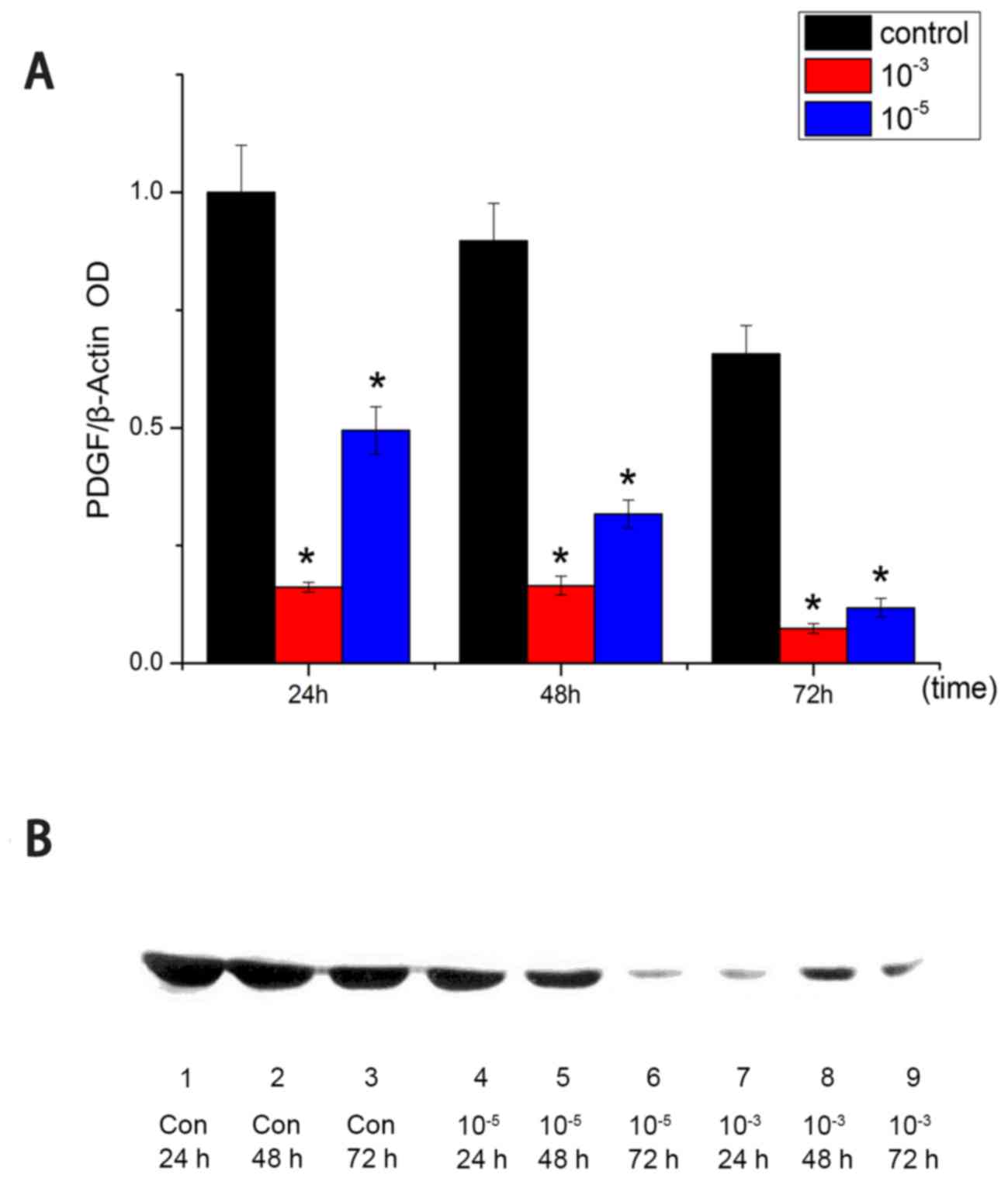
Western blot analysis of PDGF
protein
Following 24, 48 and 72 h of treatment, high dose
and low dose amygdalin groups demonstrated decreased PDGF protein
expression, compared with the control group (P<0.05; Fig. 3A). For all time points, the high-dose
group had the least amount of PDGF protein, followed by the
low-dose groups. This indicated a dose-dependent effect of
amygdalin on suppressing PDGF expression.
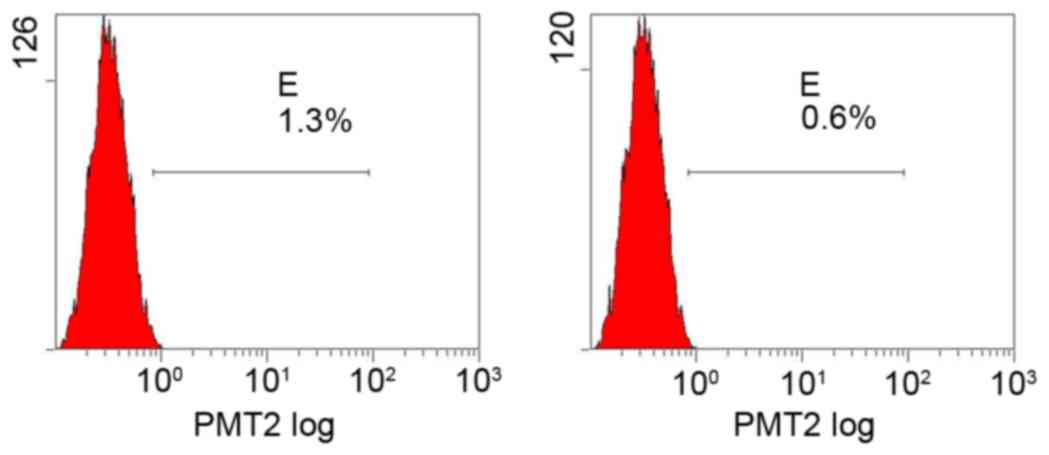
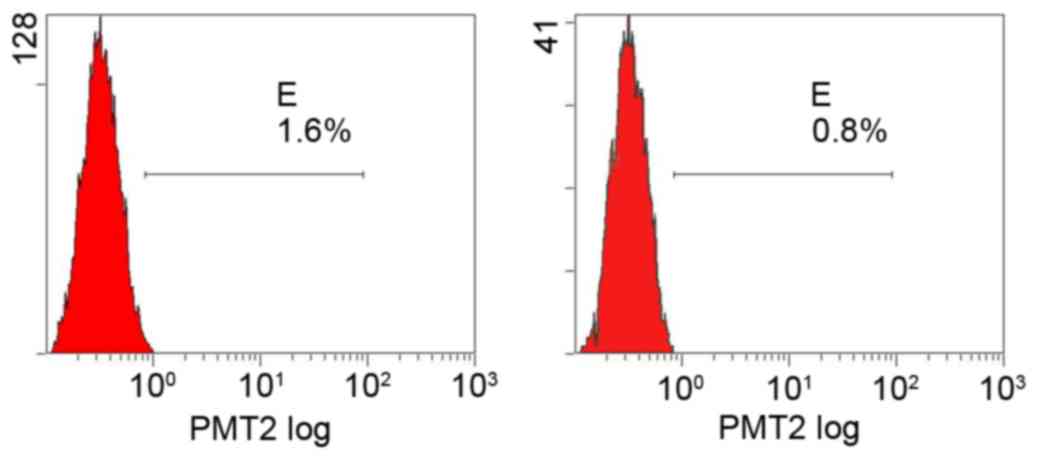
Flow cytometry determination of PDGFR
levels
Flow cytometry detected only a trace expression of
PDGFRs in all groups and the effect of amygdalin on PDGFR levels
was not significant compared with the control group (Figs. 4–7;
Table I).
 | Table I.Comparison of PDGFR levels between the
four groups (%). |
Table I.
Comparison of PDGFR levels between the
four groups (%).
| Group | PDGFR-α | PDGFR-β |
|---|
| High dose | 1.3 | 0.6 |
| Mid dose | 1.0 | 0.7 |
| Low dose | 1.3 | 0.7 |
| Control | 1.6 | 0.8 |
Discussion
PDGF, which is released from platelets, is a serum
mitogen that serves a primary role in various processes of cell
biology, including the enhancement of cell division and the control
of maturation. It also affects glial cell growth and
differentiation. PDGF is generated by a variety of cell types and
when liver failure occurs it is predominantly secreted from
nonparenchymal cells, including Kupffer and hepatic sinusoidal
endothelial cells, or from activated HSCs through autocrine and
paracrine mechanisms to regulate downstream pathways (8,9). PDGF
serves an important role in stimulating HSC proliferation and
altering its cellular skeleton distribution, thereby promoting HSCs
to differentiate into myofibroblasts (MFBs) (10). In chemical terms, PDGF is a dimeric
glycoprotein consisting of two A(-AA) or B(-BB) chains, or a
combination of the two(-AB); therefore, PDGF has three dimeric
isoforms: PDGF-AA, PDGF-BB and PDGF-AB (11). The PDGF signaling network is
activated when PDGF binds to its receptor on the membrane. PDGFR is
a single-strand transmembrane protein of 170–180 kDa containing two
subunits, PDGFR-α and PDGFR-β, which make three different
combinations, PDGFR-αα, PDGFR-ββ and PDGFR-αβ (12). As the α subunit is highly affinitive
to the A chain whereas the β subunit is affinitive to the B chain,
PDGFR-αα binds only to PDGF-A, PDGFR-ββ to PDGF-BB and PDGFR-αβ to
both PDGF-BB and PDGF-AB (13).
PDGF-BB and PDGFR-ββ have important roles in regulating liver
fibrosis as PDGFR is typically expressed in the β form and binds to
PDGF-BB during liver fibrosis (14).
PDGF binds to PDGFR and the dimerized receptor activates the
autophosphorylation of various sites within their cytosolic
domains, which serve to mediate the signal transduction (14). PDGF affects cells in various ways: i)
It stimulates HSC proliferation. PDGF has no effect on unactivated
HSCs, but for HSCs activated from spontaneous activation or
stimulated by culturing with TGF-β or Kupffer cell conditioned
medium, PDGF is able to promote their proliferation through a
dose-dependent regulation. ii) It stimulates collagen synthesis and
inhibits collagen degeneration. In liver fibrosis lesions, the PDGF
location is highly associated with the distribution of mononuclear
macrophages and collagen-producing cells and also with the
deposition of collagen types I and III (15). Furthermore, PDGF is positively
associated with the tissue inhibitor of metalloproteinase-1 level,
which serves an important role in degenerating ECM (15). iii) In addition to being a
chemotactic factor of mononuclear macrophages, neutrophils and
HSCs, PDGF also promotes HSC migration in a dose-dependent manner
(16). A previous study revealed
that the expression of PDGF and PDGFR are significantly higher in
liver fibrosis than in normal liver tissues and its activity
increased with the severity of liver fibrosis (17). Particularly, the expression of PDGF
in HSCs exhibited a significant positive correlation with the level
of fibrosis. This suggests that PDGF has an important role in
changing HSC to MFB cells.
IGF is a polypeptide mitogen that is similar to
proinsulin; therefore, it has an important role in regulating cell
proliferation, differentiation and apoptosis and mediates cell
growth, synthetic metabolism, glucose reduction and immune
regulation (18). Pinzani and Marra
(4) previously demonstrated that
PDGF enhanced the secretion and release of IGF and its binding
proteins in rat HSCs and also promoted HSC division and
proliferation by stimulating DNA synthesis in cells. Another study
revealed that exogenous IGF significantly promoted HSC
proliferation, indicating that IGF may be produced through
autocrine and paracrine functions of HSCs and are further
associated with the repair of liver damage by promoting liver
fibrosis (19).
In the present study, the mRNA expression of PDGF
and IGF was significantly suppressed by the high dose of amygdalin
compared with that observed in controls. Whereas the high-dose
group exhibited a significant decrease compared with the control
group, the low- and mid-dose groups did not suppress PDGF and IGF
mRNA expression. The mid-dose treatments led to a significant
increase of PDGF and IGF mRNA compared with the control group. This
difference may be due to the fact that relatively low doses of
amygdalin are not sufficient to transfer signals to its
receptor.
In the present study, amygdalin exhibited a
long-term suppressive effect on PDGF protein levels, from 24 to 72
h. However, amygdalin treatment exhibited no significant effect on
PDGFR. First, this may be associated with limitations of the
experiment performed. As the fluorescence of PDGFR may be quenched
easily, improper behaviors during sample handling and loading may
result in false-negative results. However, due to time limitations,
this could not be repeated in the present study and awaits further
investigation. Second, due to differences between HSC-T6 and in
vivo HSCs, HSC-T6 cells may fail to express PDGFR.
Therefore, the present study suggests that amygdalin
reduced the production of PDGF and IGF by down regulating their
transcription of genes. In this way, the influence of PDGF and IGF
on HSCs may be reduced, thereby realizing the function of
anti-fibrosis during the formation of liver fibrosis.
In conclusion, amygdalin is able to reduce the
transcription of PDGF and IGF mRNA and the expression of PDGF
protein. Amygdalin also decreased the synthesis and release of PDGF
and IGF, thereby reducing the influence of PDGF and IGF on HSCs,
thus protecting the liver from fibrosis. IGF and PDGF have been
identified as significant mitogens for liver myofibroblasts (LMFs),
a cell population that serves a role in liver fibrogenesis. A
previous study demonstrated that both IGF-I and PDGF are important
growth-promoting factors of LMFs in vitro (10). In the present study, amygdalin
reduced the transcription of PDGF and IGF mRNA and the expression
of PDGF protein. Furthermore, amygdalin also decreased the
synthesis and release of PDGF and IGF. The data presented herein
indicates that IGF and PDGF serve important roles in liver
fibrogenesis, such that these may be attractive targets for future
antifibrotic therapy.
Acknowledgements
Not applicable.
Funding
The present study was supported by Guangdong
Province Natural Science Foundation of China (grant no.
S2012010008917).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
Not applicable.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lee UE and Friedman SL: Mechanisms of
hepatic fibrogenesis. Best Pract Res Clin Gastroenterol.
25:195–206. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yoshiji H, Kuriyama S, Noguchi R, Ikenaka
Y, Yoshii J, Yanase K, Namisaki T, Kitade M, Yamazaki M, Asada K,
et al: Amelioration of liver fibrogenesis by dual inhibition of
PDGF and TGF-beta with a combination of imatinib mesylate and ACE
inhibitor in rats. Int J Mol Med. 17:899–904. 2006.PubMed/NCBI
|
|
3
|
Di Sario A, Bendia E, Svegliati-Baroni G,
Marzioni M, Ridolfi F, Trozzi L, Ugili L, Saccomanno S, Jezequel AM
and Benedetti A: Rearrangement of the cytoskeletal network induced
by platelet-derived growth factor in rat hepatic stellate cells:
Role of different intracellular signaling pathways. J Hepatol.
36:179–190. 2002. View Article : Google Scholar
|
|
4
|
Pinzani M and Marra F: Cytokine receptors
and signaling in hepatic stellate cells. Semin Liver Dis.
21:397–416. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Puche JE, Saiman Y and Friedman SL:
Hepatic stellate cells and liver fibrosisi. Compr Physiol.
3:1473–1492. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta DeltaC(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang W, Huang XR, Li AG, Liu F, Li JH,
Truong LD, Wang XJ and Lan HY: Signaling mechanism of TGF-beta1 in
prevention of renal inflammation: Role of Smad7. J Am Soc Nephrol.
16:1371–1383. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Burt AD: C L Oakley Lecture (1993)
Cellular and molecular aspects of hepatic fibrosis. J Pathol.
170:105–114. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wong L, Yamasaki G, Johnson RJ and
Friedman SL: Induction of beta-platelet-derived growth factor
receptor in rat hepatic lipocytes during cellular activation in
vivo and in culture. J Clin Invest. 94:1563–1569. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Di Sario A, Bendia E, Svegliati-Baroni G,
Marzioni M, Ridolfi F, Trozzi L, Ugili L, Saccomanno S, Jezequel AM
and Benedetti A: Rearrangement of the cytoskeletal network induced
by platelet-derived growth factor in rat hepatic stellate cells:
Role of different intracellular signalling pathways. J Hepatol.
36:179–190. 2002. View Article : Google Scholar
|
|
11
|
Ge X, Chen S, Liu M, Liang T and Liu C:
Evodiamine attenuates PDGF-BB-induced migration of rat vascular
smooth muscle cells through activating PPARγ. Int J Mol Sci.
16:28180–28093. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kuai J, Mosyak L, Brooks J, Cain M, Carven
GJ, Ogawa S, Ishino T, Tam M, Lavallie ER, Yang Z, et al:
Characterization of binding mode of action of a blocking
anti-platelet-derived growth factor (PDGF)-B monoclonal antibody,
MOR8457, reveals conformational flexibility and avidity needed for
PDGF-BB to bind PDGF receptor-β. Biochemistry. 54:1918–1929. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pinzani M, Milani S, Herbst H, DeFranco R,
Grappone C, Gentilini A, Caligiuri A, Pellegrini G, Ngo DV,
Romanelli RG and Gentilini P: Expression of platelet-derived growth
factor and its receptors in normal human liver and during active
hepatic fibrogenesis. Am J Pathol. 148:785–800. 1996.PubMed/NCBI
|
|
14
|
Rong R, Wang YC, Hu LQ, He QQ, Zhou XF,
Wang TH and Bu PL: Role of endogenous PDGF-BB in cultured
cardiomyocytes exposed to hypoxia. Neuropeptides. 50:43–49. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Borkham-Kamphorst E and Weiskirchen R: The
PDGF system and its antagonists in liver fibrosis. Cytokine Growth
Factor Rev. 28:53–61. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Carloni V, Romanelli RG, Pinzani M, Laffi
G and Gentilini P: Focal adhesion kinase and phospholipase C gamma
involvement in adhesion and migration of human hepatic stellate
cells. Gastroenterology. 112:522–531. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cao S, Yaqoob U, Das A, Shergill U,
Jagavelu K, Huebert RC, Routray C, Abdelmoneim S, Vasdev M, Leof E,
et al: Neuropilin-1 promotes cirrhosis of the rodent and human
liver by enhancing PDGF/TGF-beta signaling in hepatic stellate
cells. J Clin Invest. 120:2379–2394. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang G, Wang W, Cao Q, Gu J, Mi X, Wang
K, Chen G and Wang X: Insulin growth factor-1 (IGF-1) enhances
hippocampal excitatory and seizure activity through IGF-1
receptor-mediated mechanisms in the epileptic brain. Clin Sci
(Lond). 129:1047–1060. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gentilini A, Marra F, Gentilini P and
Pinzani M: Phosphatidylinositol-3 kinase and extracellular
signal-regulated kinase mediate the chemotactic and mitogenic
effects of insulin-like growth factor-I in human hepatic stellate
cells. J Hepatol. 32:227–234. 2000. View Article : Google Scholar : PubMed/NCBI
|