Introduction
Syphilis is a sexually transmitted disease caused by
infection with the spirochete Treponema pallidum (1,2).
Previous studies have indicated that the occurrence of syphilis may
result in severe complications, including dermatological diseases,
neurosyphilis and cardiovascular syphilis, which remain key causes
of morbidity worldwide (3,4). In addition, syphilis facilitates the
infectivity and susceptibility to HIV infection in the clinic
(5). Currently, the incidence of
syphilis is increasing in multiple developed countries (6,7).
Although various treatments have been explored, including
antibiotic therapy and antiviral therapy, these regimens are
unsatisfactory, particularly in specific populations (8,9).
Therefore, early diagnosis and effective treatment of patients at
the early stage of syphilis are essential for inhibition of T.
pallidum infection.
Previous reports have indicated that antibiotic
desensitization protocols may facilitate optimal and safe
antibiotic therapy in the appropriate clinical setting for patients
with T. pallidum infection (10).
Alternative antibiotics treatments are widely used for patients
with syphilis who are allergic to penicillin, including
doxycycline, tetracycline, ceftriaxone, erythromycin and
azithromycin (11,12). Clinical trials have indicated that
the efficacy of oral administration of azithromycin (2.0 g) is
equivalent to that of benzathine penicillin G for the treatment of
early syphilis in patients without HIV infection (13). However, drug resistance of T.
pallidum for azithromycin attenuates its therapeutic efficacy
for patients with syphilis (14).
Molecular analysis has indicated that the acidic repeat protein
(arp) gene, T. pallidum repeat (tpr) gene and
tp0548 gene are associated with the drug resistance of T.
pallidum (15,16). Therefore, it is critical to analyze
the association between gene subtypes of T. pallidum and
drug resistance of azithromycin.
The objectives of this study were to analyze the
association between gene subtype of T. pallidum and drug
resistance to azithromycin. In this study, tpr-positive
specimens were analyzed for the presence of A2058G and A2059G
mutations and the relative drug resistance to azithromycin was
determined among T. pallidum strains.
Materials and methods
Sample collection and molecular strain
typing
A total of 132 blood samples were collected from
female patients from the Yongkang region with primary syphilis in
The First People's Hospital of Yongkang (Zhejiang, China) between
May 2016 and June 2017. Patients with HIV or T. pallidum
history were excluded from the present study. The protocols were
approved by the Human Subjects Division of The First People's
Hospital of Yongkang. All patients were required to provide written
informed consent. T. pallidum strains were isolated from
patient blood samples using sequencing-based typing or enhanced
Centers for Disease Control and Prevention typing as described
previously (17). DNA samples were
tested for T. pallidum using quantitative polymerase chain
reaction (PCR) targeting the polA gene (18).
DNA extraction
T. pallidum DNA was isolated as previously
described (19). Briefly, DNA was
extracted from 200 µl of whole blood and DNAzol (Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) was used to extract DNA
from specimens, according to the manufacturer's protocols. Strain
typing was classified based on the analysis of three DNA target
regions: (1) Restriction fragment
length polymorphism analysis of sequence differences in the
tpr gene; (2) the number of
60 bp repeats in the arp gene; (3) sequence analysis of a short region of
the tp0548 gene (20).
PCR methods
The PCR assay for T. pallidum analyzed the
gene target polA as described previously (21). The primers used were as follows: F:
5′-CGTGTGGTATCAACTATGG-3′, R: 5′-TCAACCGTGTACTCAGTGC-3′. All PCR
products were analyzed using an ABI9700 GeneAmp PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). PCR was performed in a
25 µl reaction volume with 0.1 µg DNA, 1.5 mM MgCl2, 0.2
mM dNTP, 0.5 µM primers and 0.5 µl Taq polymerase (Takara Bio,
Inc., Otsu, Japan). PCR conditions were as follows: 96°C for 5 min,
followed by 40 cycles at 95°C for 30 sec, 56°C for 56 sec and 72°C
for 60 sec, followed by 72°C for 600 sec. PCR products were
analyzed on a 1.5% agarose gel. Then, PCR products were visualized
by staining with ethidium bromide and comparing with the molecular
size markers of a 100- or 2,000-bp ladder (New England BioLabs,
Inc., Ipswich, MA, USA).
Detection of drug resistance to
azithromycin
The polA gene was amplified to detect
resistance to azithromycin (22).
Briefly, PCR amplification of the 23S rRNA gene of T.
pallidum was treated by restriction enzyme digestion
(MboII) as described previously (23,24). PCR
were performed in a 50 µl reaction volume with 0.2 µg rRNA, 1.5 mM
MgCl2, 0.2 mM dNTP, 0.5 µM primers
(5′-GTGCCAGCMGCCGCGG-3′) and 1.0 µl Taq polymerase. PCR conditions
were as follows: 95°C for 5 min, followed by 35 cycles at 94°C for
30 sec, 54°C for 57 sec and 72°C for 60 sec, followed by 72°C for
600 sec. The azithromycin resistance genotypes were analyzed by DNA
sequencing of the PCR products after purification with a QIAquick
PCR purification kit (Qiagen, Inc., Valencia, CA, USA). T.
pallidum DNA was also evaluated using restriction enzyme
digestion for A2058G and A2059G mutations (MboII and
BsaI, respectively; New England BioLabs, Inc.) (25). The DNA sequences were obtained using
the DNA analyzer 3730×l (Applied Biosystems; Thermo Fisher
Scientific, Inc.) and analyzed using DNASTAR® software
version 3.0 (DNASTAR Inc., Madison, WI, USA).
Antimicrobial susceptibility
testing
All antibiotic disks (azithromycin) were purchased
from Abtek Biologicals Ltd. (Liverpool, UK). An antibiogram was
performed on Mueller-Hinton agar for 12 h at 37°C to determine the
antimicrobial agents resistance profiles to azithromycin (10 µg) or
PBS (10 µg). Antimicrobial susceptibility was performed according
to the European Committee on Antimicrobial Susceptibility
guidelines (26).
Statistical analysis
Data are expressed as mean ± standard deviation of
triplicate experiments. All data were analyzed using SPSS 17.0
software (SPSS, Inc., Chicago, IL, USA). Differences among groups
were analyzed by one-way analysis of variance with Tukey's multiple
comparisons test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Comparison of the gene types of T.
pallidum
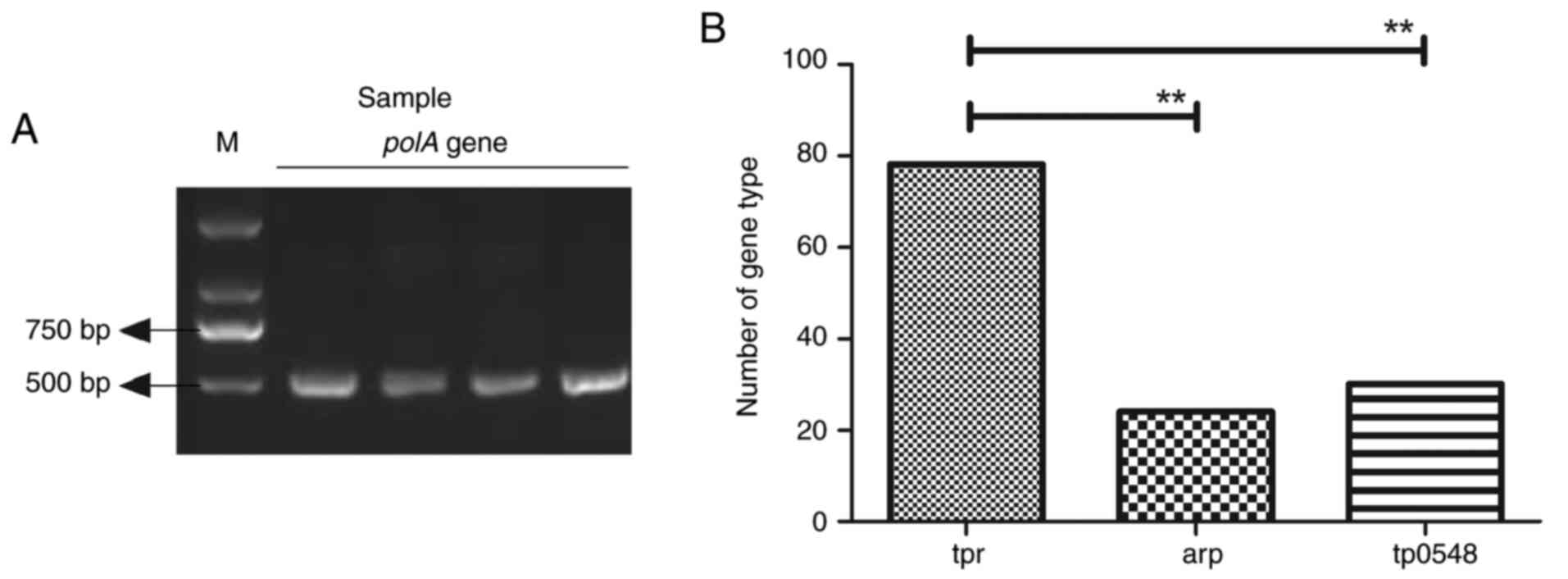
DNA samples from 132 genital ulcer specimens were
identified as T. pallidum positive, determined by PCR
targeting the polA gene (Fig.
1A). Three gene types were observed among the 132 T.
pallidum positive specimens (Fig.
1B). The restriction digestion assay indicated that 78 samples
were tpr gene type, 24 were arp gene type and 30 were
tp0548 gene type. These results suggest that tpr is
the most common gene type of T. pallidum.
Analysis of T. pallidum drug
resistance to azithromycin
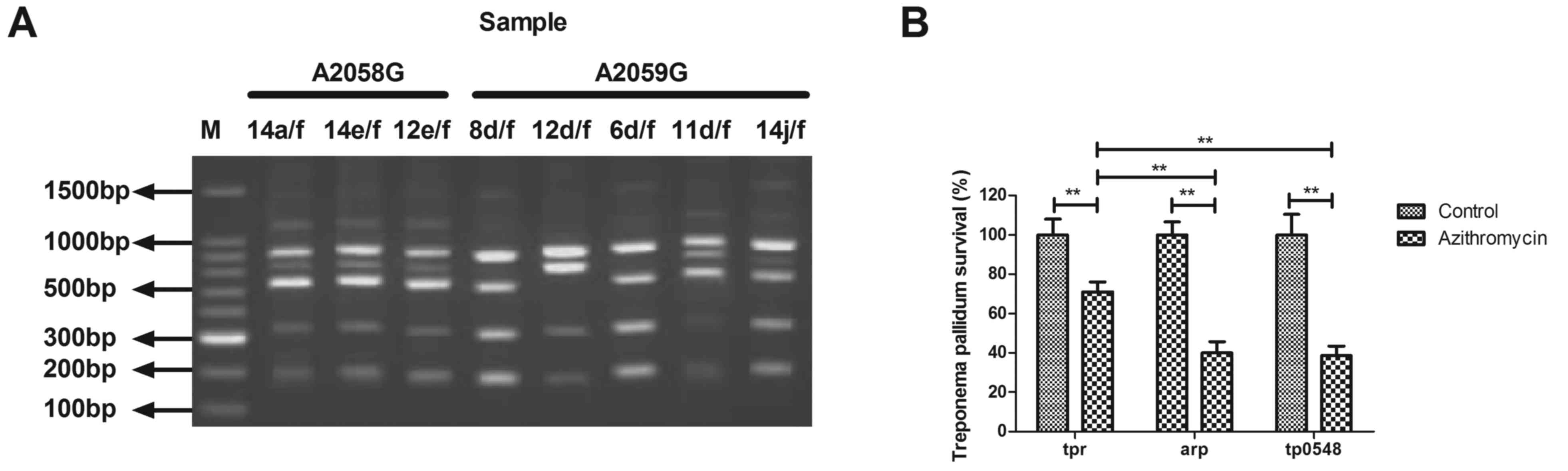
The antibiotic resistance of T. pallidum to
azithromycin was analyzed in this study. As indicated in Fig. 2A, the restriction fragment length
polymorphism analysis of the PCR amplicons from the representative
samples (A2058G: 14a/f, 14e/f and 12e/f; A2059G: 8d/f, 12d/f, 6d/f,
11d/f, 14j/f.) revealed that 94 of the T. pallidum specimens
were resistant to azithromycin. Gene type analysis indicated that
the tpr gene type presented significantly higher drug
resistance compared with the arp and tp0548 gene
types (Fig. 2B). These results
suggest that tpr gene type may be associated with drug
resistance of T. pallidum to azithromycin.
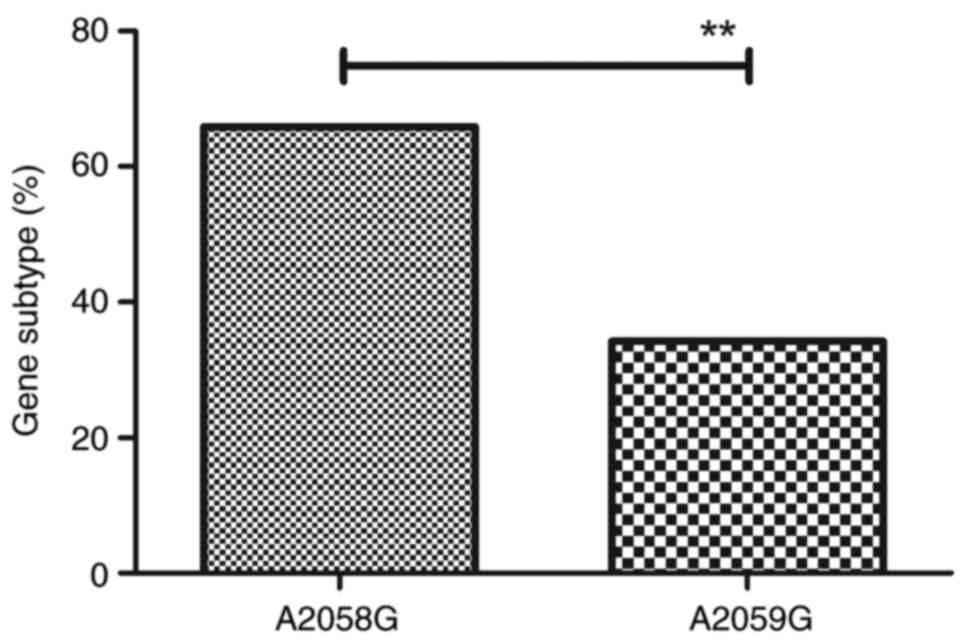
Comparison of the gene subtypes tpr of
T. pallidum
The association between gene subtype tpr of
T. pallidum and drug resistance was evaluated further. Eight
subtypes of tpr (14a/f, 14e/f, 12e/f, 12d/f, 6d/f, 11d/f,
14j/f, 8d/f) were identified among the 132 cases (Table I). It was identified that 23S rRNA
A2058G mutation was observed in gene subtypes 14a/f, 14e/f and
12e/f. A2059G mutation occurred in gene subtypes 8d/f, 12d/f, 6d/f,
11d/f and 14j/f. The proportion of azithromycin-resistant genotypes
harboring either the A2058G or the A2059G mutation among T.
pallidum was 65.8 and 34.2%, respectively (Fig. 3). These results indicate that
tpr gene subtypes of T. pallidum may be associated
with drug resistance to azithromycin.
 | Table I.Overview of tpr gene subtypes
of Treponema pallidum. |
Table I.
Overview of tpr gene subtypes
of Treponema pallidum.
| Gene type | A2058G | A2059G |
|---|
| 14a/f | + | − |
| 14e/f | + | − |
| 12e/f | + | − |
| 8d/f | − | + |
| 12d/f | − | + |
| 6d/f | − | + |
| 11d/f | − | + |
| 14j/f | − | + |
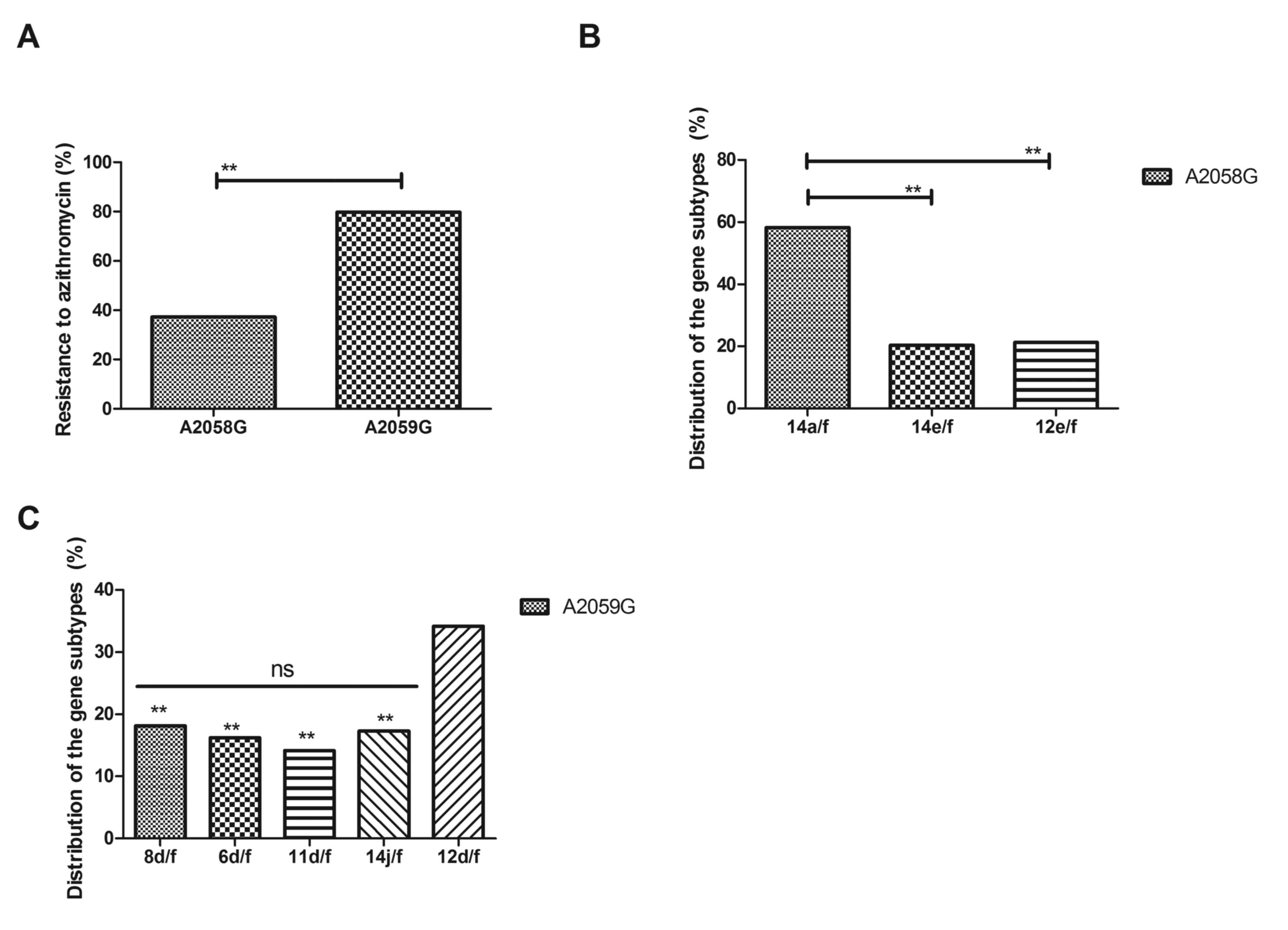
Analysis of drug resistance of A2058G
and A2059G mutations in gene subtype tpr of T. pallidum
The drug resistance of A2058G and A2059G mutations
in gene subtype tpr of T. pallidum was investigated
further. As indicated in Fig. 4A,
A2059G mutation demonstrated a higher drug resistance for
azithromycin compared with A2058G mutation, as determined by
antimicrobial susceptibility test. The distribution of gene
subtypes of T. pallidum with A2059G and A2058G mutation is
presented in Fig. 4B and C. These
results demonstrated that 14a/f of A2058G mutation and 12d/f of
A2059G mutation frequently occurred in tpr of T.
pallidum. These results indicate that A2058G and A2059G
mutations in gene subtype tpr of T. pallidum are
associated with drug resistance to azithromycin.
Discussion
Molecular subtyping for T. pallidum has
previously been explored in the tpr gene and arp gene
(27). A previous study also
indicated that azithromycin treatment failures are associated with
resistance in T. pallidum (28). However, the associations between
molecular subtyping of T. pallidum and azithromycin
resistance remain unclear. In the current study, the gene type of
T. pallidum evaluated and azithromycin resistance was
investigated in different gene types of T. pallidum. The
results indicate that out of 132 samples, 78 were tpr gene
type, 24 were arp gene type and 30 were tp0548 gene
type specimens of T. pallidum. The findings suggested that
tpr gene type presented higher azithromycin resistance
compared with arp and tp0548 gene type specimens of
T. pallidum.
Currently, azithromycin is widely used for the
treatment of T. pallidum in patients that are allergic to
penicillin (29). Although the
recommended azithromycin treatment for syphilis is effective,
azithromycin resistance in T. pallidum has emerged and is
increasing globally (30). It was
observed that T. pallidum has developed azithromycin
resistance to a varying extent (29). It has been identified that prevalence
of azithromycin resistance is substantial in China and consequently
that macrolides should not be used as a treatment option for early
or incubating syphilis in China (31). The results of the current study
identified three gene types, tpr, arp and tp0548, in
132 T. pallidum positive specimens from the Yongkang region.
Reports have also indicated that restriction fragment length
polymorphisms are associated with the drug resistance of T.
pallidum (32,33). In the current study, it was
identified that 94 specimens of T. pallidum presented
restriction fragment length polymorphisms, which were associated
with drug resistance of T. pallidum for azithromycin.
Notably, gene type analysis suggested that the tpr gene type
presents higher drug resistance compared with the arp and
tp0548 gene types of T. pallidum, which may be a
potential target for addressing drug resistance to
azithromycin.
A previous study indicated that tpr genes in
T. pallidum are likely to be relevant to the pathogenesis of
syphilis and drug resistance (34).
A previous report described A2058G and A2059G mutations, which are
associated with syphilis drug resistance to azithromycin (35). In the present study, the drug
resistance to azithromycin of A2058G and A2059G mutations in
tpr gene types of T. pallidum were compared. It was
indicated that A2059G mutation demonstrated higher drug resistance
for azithromycin compared with A2058G mutation. The findings also
revealed that the gene subtypes of T. pallidum 14a/f of
A2058G mutation and 12d/f of A2059G frequently occurred in
tpr of T. pallidum, which may be a potential target
for the treatment of syphilis. A limitation of the present study
was that it did not explore the clinical treatment of azithromycin
for patients with T. pallidum.
In conclusion, the current study identified that
12d/f tpr gene type of T. pallidum is the most common
gene type for drug resistance to azithromycin. The findings suggest
that A2059G mutation is associated with azithromycin resistance.
Further investigation is required into the molecular mechanism of
drug resistance of T. pallidum.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YL designed and performed the experiments. JL, WH,
HL, JZ, CL and CC analyzed the experimental data and constructed
the figures. The final version of the manuscript has been read and
approved by all authors.
Ethics approval and consent to
participate
The study protocol was approved by the Human
Subjects Division of The First People's Hospital of Yongkang
(Zhejiang, China). All patients provided written informed
consent.
Consent for publication
All patients provided written informed consent for
the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Stoltey JE and Cohen SE: Syphilis
transmission: A review of the current evidence. Sex Health.
12:103–109. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bristow CC, Larson E, Javanbakht M, Huang
E, Causer L and Klausner JD: A review of recent advances in rapid
point-of-care tests for syphilis. Sex Health. 12:119–125. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hawkes SJ, Gomez GB and Broutet N: Early
antenatal care: Does it make a difference to outcomes of pregnancy
associated with syphilis? A systematic review and meta-analysis.
PLoS One. 8:e567132013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gomez GB, Kamb ML, Newman LM, Mark J,
Broutet N and Hawkes SJ: Untreated maternal syphilis and adverse
outcomes of pregnancy: A systematic review and meta-analysis. Bull
World Health Organ. 91:217–226. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Campos LN, Guimaraes MD, Carmo RA, Melo
AP, Oliveira HN, Elkington K and McKinnon K: HIV, syphilis, and
hepatitis B and C prevalence among patients with mental illness: A
review of the literature. Cad Saude Publica. 24(Suppl 4):
S607–S620. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Naesens R, Vermeiren S, van Schaeren J and
Jeurissen A: False positive Lyme serology due to syphilis: Report
of 6 cases and review of the literature. Acta Clin Belg. 66:58–59.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Marinkovic Z and Dukic S: Historical and
medical review of syphilis-afflicted army leaders, rulers and
statesmen. Med Pregl. 64:423–427. 2011.PubMed/NCBI
|
|
8
|
Seppings L and Hamill M: A review of an
early syphilis outbreak in West Berkshire and Reading 2014–2015.
Sex Transm Infect. 92:3642016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kenyon CR, Osbak K and Tsoumanis A: The
Global Epidemiology of Syphilis in the Past Century-A systematic
review based on antenatal syphilis prevalence. PLoS Negl Trop Dis.
10:e00047112016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Magpantay G, Cardile AP, Madar CS, Hsue G
and Belnap C: Antibiotic desensitization therapy in secondary
syphilis and Listeria infection: Case reports and review of
desensitization therapy. Hawaii Med J. 70:266–268. 2011.PubMed/NCBI
|
|
11
|
Balaskas K, Spencer S and D'Souza Y:
Peripapillary choroidal neovascularisation in the context of ocular
syphilis is sensitive to combination antibiotic and corticosteroid
treatment. Int Ophthalmol. 33:159–162. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kahn RH, Moseley KE, Johnson G and Farley
TA: Potential for community-based screening, treatment, and
antibiotic prophylaxis for syphilis prevention. Sex Transm Dis.
27:188–192. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hook EW III, Behets F, van Damme K,
Ravelomanana N, Leone P, Sena AC, Martin D, Langley C, McNeil L and
Wolff M: A phase III equivalence trial of azithromycin versus
benzathine penicillin for treatment of early syphilis. J Infect
Dis. 201:1729–1735. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Read P, Jeoffreys N, Tagg K, Guy RJ,
Gilbert GL and Donovan B: Azithromycin-resistant syphilis-causing
strains in Sydney, Australia: Prevalence and risk factors. J Clin
Microbiol. 52:2776–2781. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Castro R, Prieto E, Aguas MJ, Manata MJ,
Botas J and Pereira FM: Molecular subtyping of treponema pallidum
subsp. pallidum in lisbon, portugal. J Clin Microbiol.
47:2510–2512. 2009. View Article : Google Scholar
|
|
16
|
Flasarova M, Smajs D, Matejkova P,
Woznicova V, Heroldova-Dvorakova M and Votava M: Molecular
detection and subtyping of Treponema pallidum subsp. Pallidum in
clinical specimens. Epidemiol Mikrobiol Imunol. 55:105–111.
2006.(In Czech).
|
|
17
|
Mikalova L, Strouhal M, Cejkova D,
Zobaníková M, Pospíšilová P, Norris SJ, Sodergren E, Weinstock GM
and Šmajs D: Genome analysis of Treponema pallidum subsp. Pallidum
and subsp. pertenue strains: Most of the genetic differences are
localized in six regions. PLoS One. 5:e157132010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen CY, Chi KH, George RW, Cox DL,
Srivastava A, Rui Silva M, Carneiro F, Lauwers GY and Ballard RC:
Diagnosis of gastric syphilis by direct immunofluorescence staining
and real-time PCR testing. J Clin Microbiol. 44:3452–3456. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Castro R, Aguas MJ, Batista T, Araujo C,
Mansinho K and Pereira Fda L: Detection of Treponema pallidum Sp.
Pallidum DNA in Cerebrospinal Fluid (CSF) by Two PCR Techniques. J
Clin Lab Anal. 30:628–632. 2016.
|
|
20
|
Kondratiev NV, Alfimova MV and Golimbet
VE: A search of target regions for association studies between DNA
methylation and cognitive impairment in schizophrenia. Zh Nevrol
Psikhiatr Im S S Korsakova. 117:72–75. 2017.(In Russian).
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pillay A, Liu H, Ebrahim S, Chen CY, Lai
W, Fehler G, Ballard RC, Steiner B, Sturm AW and Morse SA:
Molecular typing of Treponema pallidum in South Africa:
Cross-sectional studies. J Clin Microbiol. 40:256–258. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu H, Rodes B, Chen CY and Steiner B: New
tests for syphilis: Rational design of a PCR method for detection
of Treponema pallidum in clinical specimens using unique regions of
the DNA polymerase I gene. J Clin Microbiol. 39:1941–1946. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jia S, Li J, Park SR, Ryu Y, Park IH, Park
JH, Hong SS, Kwon SW and Lee J: Combined application of dispersive
liquid-liquid microextraction based on the solidification of
floating organic droplets and charged aerosol detection for the
simple and sensitive quantification of macrolide antibiotics in
human urine. J Pharm Biomed Anal. 86:204–213. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Haanpera M, Huovinen P and Jalava J:
Detection and quantification of macrolide resistance mutations at
positions 2058 and 2059 of the 23S rRNA gene by pyrosequencing.
Antimicrob Agents Chemother. 49:457–460. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lukehart SA, Godornes C, Molini BJ,
Sonnett P, Hopkins S, Mulcahy F, Engelman J, Mitchell SJ, Rompalo
AM, Marra CM and Klausner JD: Macrolide resistance in Treponema
pallidum in the United States and Ireland. N Engl J Med.
351:154–158. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kassim A, Omuse G, Premji Z and Revathi G:
Comparison of clinical laboratory standards institute and european
committee on antimicrobial susceptibility Testing guidelines for
the interpretation of antibiotic susceptibility at a University
teaching hospital in Nairobi, Kenya: A cross-sectional study. Ann
Clin Microbiol Antimicrob. 15:212016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tian H, Li Z, Li Z, Hou J, Zheng R, Li F,
Liu R, Liu B, Wang C and Zhang F: Molecular typing of Treponema
pallidum: Identification of a new sequence of tp0548 gene in
Shandong, China. Sex Transm Dis. 41:5512014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Prevalence of the 23S rRNA A2058G point
mutation and molecular subtypes in Treponema pallidum in the United
States, 2007 to 2009. A2058G. Prevalence Workgroup. 39:794-798,
2012.
|
|
29
|
Pandori MW, Gordones C, Castro L, Engelman
J, Siedner M, Lukehart S and Klausner J: Detection of azithromycin
resistance in Treponema pallidum by real-time PCR. Antimicrob
Agents Chemother. 51:3425–3430. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Katz KA and Klausner JD: Azithromycin
resistance in Treponema pallidum. Curr Opin Infect Dis. 21:83–91.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen XS, Yin YP, Wei WH, Wang HC, Peng RR,
Zheng HP, Zhang JP, Zhu BY, Liu QZ and Huang SJ: High prevalence of
azithromycin resistance to Treponema pallidum in geographically
different areas in China. Clin Microbiol Infect. 19:975–979. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen CY, Chi KH, Pillay A, Nachamkin E, Su
JR and Ballard RC: Detection of the A2058G and A2059G 23S rRNA gene
point mutations associated with azithromycin resistance in
Treponema pallidum by use of a TaqMan real-time multiplex PCR
assay. J Clin Microbiol. 51:908–913. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu BR, Yang CJ, Tsai MS, Lee KY, Lee NY,
Huang WC, Wu H, Lee CH, Chen TC, Ko WC, et al: Multicentre
surveillance of prevalence of the 23S rRNA A2058G and A2059G point
mutations and molecular subtypes of Treponema pallidum in Taiwan,
2009–2013. Clin Microbiol Infect. 20:802–807. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Giacani L, Molini B, Godornes C, Barrett
L, van Voorhis W, Centurion-Lara A and Lukehart SA: Quantitative
analysis of tpr gene expression in Treponema pallidum isolates:
Differences among isolates and correlation with T-cell
responsiveness in experimental syphilis. Infect Immun. 75:104–112.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Matejkova P, Flasarova M, Zakoucka H,
Borek M, Kremenová S, Arenberger P, Woznicová V, Weinstock GM and
Smajs D: Macrolide treatment failure in a case of secondary
syphilis: A novel A2059G mutation in the 23S rRNA gene of Treponema
pallidum subsp. pallidum. J Med Microbiol. 58:832–836. 2009.
View Article : Google Scholar : PubMed/NCBI
|