Introduction
Diabetes mellitus is a chronic disease that affects
~500 million people worldwide (1).
Functional β-cell apoptosis and dysfunctional insulin secretion are
the two primary mechanisms that are thought to be involved in the
pathogenesis of type 2 diabetes (T2D) (2–6).
Collectively, these mechanisms result in the inability to
compensate for insulin resistance. Therefore, enhancing β-cell
proliferation and increasing insulin secretion may be novel
strategies for the treatment of diabetes.
Sirtuins (SIRTs) are proteins that possess
nicotinamide-adenine dinucleotide-dependent deacetylase activity
and belong to the histone deacetylase (HDAC) family, which consist
of seven different subtypes in mammals (SIRT1-7) (7). SIRTs were originally identified to be
involved in lifespan prolongation; however, research has revealed
that they serve major roles in a number of additional biological
processes, including cell metabolism, aging and tumorigenesis
(7).
Although SIRT5 possesses weak deacetylase activity,
it catalyzes the modification of acidic lysine residues by
glutarylation, succinylation and malonylation (8–12).
Previous studies have identified thousands of potential SIRT5
targets, including metabolic enzymes involved in ketone body
synthesis, the tricarboxylic acid cycle, amino acid catabolism,
fatty-acid β-oxidation, glycolysis and ATP synthesis (13–15).
However, the functional role of SIRT5 in T2D remains unknown. SIRT5
regulates the expression of various metabolic enzymes and may also
serve a key function in T2D. The present study aimed to determine
the roles and underlying mechanism of SIRT5 in T2D.
Materials and methods
Cell culture
MIN6 and INS-1 cells were purchased from the
American Type Culture Collection (Manassas, VA, USA) and cultured
in low-glucose Dulbecco's modified Eagle's medium (5 mmol/l
glucose; HyClone; GE Healthcare Life Sciences, Logan, UT, USA),
supplemented with 100 U/ml penicillin, 0.1 mg/ml streptomycin
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 5.5
mM 2-mercaptoethanol and 10% horse serum (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C with 5% CO2.
Blood sample collection
The present study was approved by the Ethics
Committee of Taizhou People's Hospital (Taizhou, China), and
written informed consent was provided from patients and healthy
volunteers prior to enrolment. Blood samples were collected from
130 patients, including 65 patients with T2D patients (that had not
yet received treatment) and 65 healthy volunteers that were
admitted to Taizhou People's Hospital between 2015 to 2016, and
stored at −80°C prior to use. Healthy control subjects were
recruited from age- and gender-matched healthy blood donors.
Patients were diagnosed according to the criteria of the American
Diabetes Association (16). T2D
patients contained 30 males and 36 females, with age range of 11-87
years and an average age of 40±0.55 years. Blood glucose levels
were determined using ACCU-CHEK (Roche Diagnostics GmbH, Mannheim,
Germany).
Plasmid and transfection
pcDNA3.1-SIRT5 and pcDNA3.1-pancreatic and duodenal
homeobox 1 (PDX1) were purchased from Vigene Biosciences
(Rockville, MD, USA). The inactive form of the SIRT5 deacetylase
(T69A) was mutated using a Fast Mutagenesis System (Beijing
Transgen Biotech Co., Ltd., Beijing, China) according to the
manufacturer's protocol. RNAi-resistant SIRT5 (SIRT5R) was mutated
using Fast Mutagenesis System according to the sequence of SIRT5
siRNA. For overexpressing SIRT5, a total of 2.5 µg vector
(pcDNA3.1) and indicated plasmid were transfected into MIN6 and
INS-1 cells using lipo 2000 (Thermo Fisher Scientific Inc.).
Following transfection for 48 h, cells were collected and used for
subsequent experiments.
RNA interference
A total of 4×105 INS-1 or MIN6 cells were
transfected with 50 nM SIRT5 small interfering RNA (siSIRT5) or
scrambled siRNA (SCR, Sigma-Aldrich, St. Louis, MO, USA) using the
DharmaFECT 1 kit (Thermo Fisher Scientific Inc.) according to the
manufacturers' protocol. Following transfection for 48 h, the
expression of SIRT5 was determined using reverse transcription
quantitative polymerase chain reaction (RT-qPCR) and western
blotting. Each experiment was repeated three times. SIRT5 siRNA was
synthesized form Sigma-Aldrich (Darmstadt, Germany). The sequences
for SIRT5 were as follows: siRNA#1, GAGAAUUACAAGAGUCCAAUU and
siRNA#2, GGAGAUCCAUGGUAGCUUAUU.
RT-qPCR
Total RNA from blood samples and INS-1 or MIN6 cells
was extracted using the TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. A total
of 2 µg RNA was reverse transcribed into cDNA using the EasyScript
First-Strand cDNA Synthesis SuperMix kit (TransGen Biotech Co.,
Ltd., Beijing, China) according to the manufacturer's protocol. A
SYBR green Mix (Roche Diagnostics GmbH) was used to perform RT-qPCR
assay. The thermal cycling parameters were as follows: 33 cycles of
denaturation at 95°C for 4 min, annealing at 55°C for 30 sec
followed by extension at 72°C for 8 min. The following primers were
used: SIRT5, forward, 5′-AAATAACTAAAGCCCGCCTC-3′, and reverse,
5′-CCACTTTCTGCACTAACACC-3′; PDX1, forward,
5′-CCTTTCCCATGGATGAAGTC-3′, and reverse,
5′-GTGAGATGTACTTGTTGAATAGGA-3′; glyceraldehyde-3-phosphate
dehydrogenase (GAPDH), forward, 5′-ATGATGACATCAAGAAGGTGG-3′, and
reverse, 5′-TTGTCATACCAGGAAATGAGC-3′. The relative mRNA expression
was normalized to that of GAPDH and quantified using the
2−ΔΔCt method (17). Each
experiment was repeated three times.
Western blotting
Cells were harvested and lysed in
radioimmunoprecipitation assay (RIPA, Beyotime Institute of
Biotechnology, Jiangsu, China) buffer, and the protein
concentration of sample extracts was determined using a BCA protein
assay kit (Pierce; Thermo Fisher Scientific, Inc.). A total of 45
µg protein was loaded per lane and electrophoresed by 8% SDS-PAGE.
Samples were then transferred to a polyvinylidene fluoride
membrane. Membranes were blocked in 5% non-fat milk at room
temperature for 1 h. Membranes were then incubated with primary
antibodies against SIRT5 (1:1,000; cat. no. ab108968), PDX1
(1:1,000; cat. no. ab134150) or β-actin (1:7,000; cat. no. ab8226)
overnight at 4°C (All Abcam, Cambridge, UK). Following washing
three times with tris-buffered saline Tween 20, membranes were
incubated with secondary horseradish peroxidase-conjugated
antibodies (goat anti mouse, 1:3,000; ab6789; Abcam, Cambridge, UK;
goat anti rabbit, 1:5,000, ab205718; Abcam, Cambridge, UK) at room
temperature for 1 h. Specific protein bands were visualized using a
BeyoECL Plus ECL kit (Beyotime Institute of Biotechnology). Image J
software (version 1.48; National Institutes of Health, Besthesda,
MD, USA) was used for densitometry.
CCK-8 assay
A CCK-8 assay was used to investigate the effect of
SIRT5 expression on cell proliferation. INS-1 and MIN6 cells were
transfected with 2.5 µg pcDNA 3.1 vector, SIRT5 plasmid (Vigene
Biosciences, Rockville, MD, USA) or SIRT5 and PDX1 plasmid, 50 nM
scramble siRNA (SCR) or SIRT5 siRNA (siSIRT5, Sigma-Aldrich, St.
Louis, MO, USA), respectively. Following transfection for 48 h, a
total of 3×103 cells were seeded into 96-well plates
with 200 µl low-glucose Dulbecco's Modified Eagle's medium (5
mmol/l glucose; HyClone; GE Healthcare Life Sciences) supplemented
with 10% horse serum (Invitrogen; Thermo Fisher Scientific, Inc.).
Subsequently, 20 µl CCK-8 reagent (Beyotime Institute of
Biotechnology) was added to each well and cells were incubated for
a further 1 h at 37°C. Finally, the absorbance at 450 nm was read
using a microplate reader (BioTek Instruments Inc., Winooski, VT,
USA). Each experiment was repeated three times.
Colony formation assay
SIRT5 was overexpressed or knocked down in INS-1 and
MIN6 cells. Following transfection for 48 h, ~2×104
transfected INS-1 and MIN6 cells were seeded into 6-well plates.
Cells were cultured with serum-free low-glucose Dulbecco's modified
Eagle's medium for 14 days. Subsequently, cells were fixed with
methanol at room temperature for 10 min and stained with 0.5%
crystal violet at room temperature for 10 min. The colonies (cells
>50) were counted under a light microscope (magnification, 20×).
Each experiment was repeated three times. The Image J software
(v1.04; National Institutes of Health, Bethesda, MD, USA) was used
to assess colony formation
ChIP and qChIP assay
A ChIP assay was performed with 2.5 µg anti-H4K16ac
(cat. no. ab109463; Abcam) and goat anti-rabbit IgG (cat. no.
ab171870; Abcam) using a ChIP assay kit (Beyotime Institute of
Biotechnology) according to the manufacturer's protocol. The
antibody-bound DNA was detected in 2% agarose gel. Subsequently,
antibody-bound DNA was used to perform qChIP. SYBR green Mix (Roche
Diagnostics GmbH) was used to perform qPCR. The thermocycling
conditions were as follows: 30 cycles of denaturation at 95°C for 5
min, annealing at 56°C for 30 sec followed by extension at 72°C for
5 min. The following primers were used: PDX1, forward,
5′-AGGGTCTCATTCTGTCGTTC-3′; PDX1, reverse,
5′-GCCTGTAATCCCAGCTACTC-3′. The fold of enrichment was normalized
to that of IgG and quantified using the 2−ΔΔCq method
(17). Each experiment was repeated
three times.
Insulin secretion and insulin content
measurements
A total of ~4×105 cells were seeded into
a 24-well plate and transfected with a pcDNA 3.1 vector, SIRT5
plasmid or SIRT5 and PDX1 plasmid, scramble siRNA (SCR) or SIRT5
siRNA (siSIRT5). Following transfection for 24 h, MIN6 and INS-1
cells were also incubated with 1 or 20 mM glucose for 90 min and
the rate of insulin secretion was detected in each group. Cells
were washed with a secretion assay buffer [sub arachnoid block
(SAB); 20 mM HEPES (pH 7.2), 2.5 mM CaCl2, 1.16 mM
MgSO4, 1.2 mM KH2PO4, 4.7 mM KCl,
25.5 mM NaHCO3, 114 mM NaCl and 0.2% bovine serum
albumin (Invitrogen; Thermo Fisher Scientific, Inc.) with 2.8 mM
glucose three times. Subsequently, cells were incubated with 1.5 ml
SAB at 37°C and 5% CO2 for 2 h. Immediately following
incubation, insulin levels were analyzed using the Coat-A-Count
insulin RIA kit (Diagnostic Products Corporation, Los Angeles, CA,
USA)] according to the manufacturers' protocol. Cells were lysed
with RIPA buffer [1% NP-40/Triton X, 0.5% sodium deoxycholate, 0.1%
SDS, 150 mM NaCl, 50 mM Tris HCl (pH 8.0), 50 mM NaF and 2 mM
EDTA]. The cell lysates were centrifuged at 1×104 g for
5 min at 4°C, and shaken on ice for 30 min. The supernatant was
collected and used for the analysis of insulin content. Total
protein concentration was measured using a BCA Protein assay kit
(Pierce; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. The insulin content of each group was
normalized to protein concentration. Each experiment was repeated
three times.
Incubation of cells with valproic acid
(VPA)
INS-1 cells were transfected with vector (pcDNA3.1)
or SIRT5 plasmid and cultured with or without 20 mM glucose. A
total of 0.1, 0.5 or 1 mM VPA (cat. no. S1168; Selleck Chemicals,
Shanghai, China) was added to cells immediately following
transfection. The low-glucose Dulbecco's modified Eagle's medium (5
mmol/l glucose) supplemented with 10% horse serum was subsequently
removed and VPA-containing medium was refreshed 1 day following
transfection. Insulin secretion tests were also performed at 48 h
following transfection and treatment with VPA. Each experiment was
repeated three times.
Statistical analysis
Each experiment was repeated three times and the
results are expressed as the mean ± standard deviation. SPSS 18.0
software (SPSS, Inc., Chicago, IL, USA) was used for statistical
analysis. The Student's t-test was used to analyze the differences
between two groups and a one-way analysis of variance followed by a
Tukey's post hoc test, was used to analyze differences among
multiple groups. Associations between SIRT5 expression and blood
glucose levels were determined using the Pearson's correlation
coefficient. P<0.05 was considered to indicate a statistically
significant difference.
Results
Expression of SIRT5 is markedly
upregulated in patients with T2D
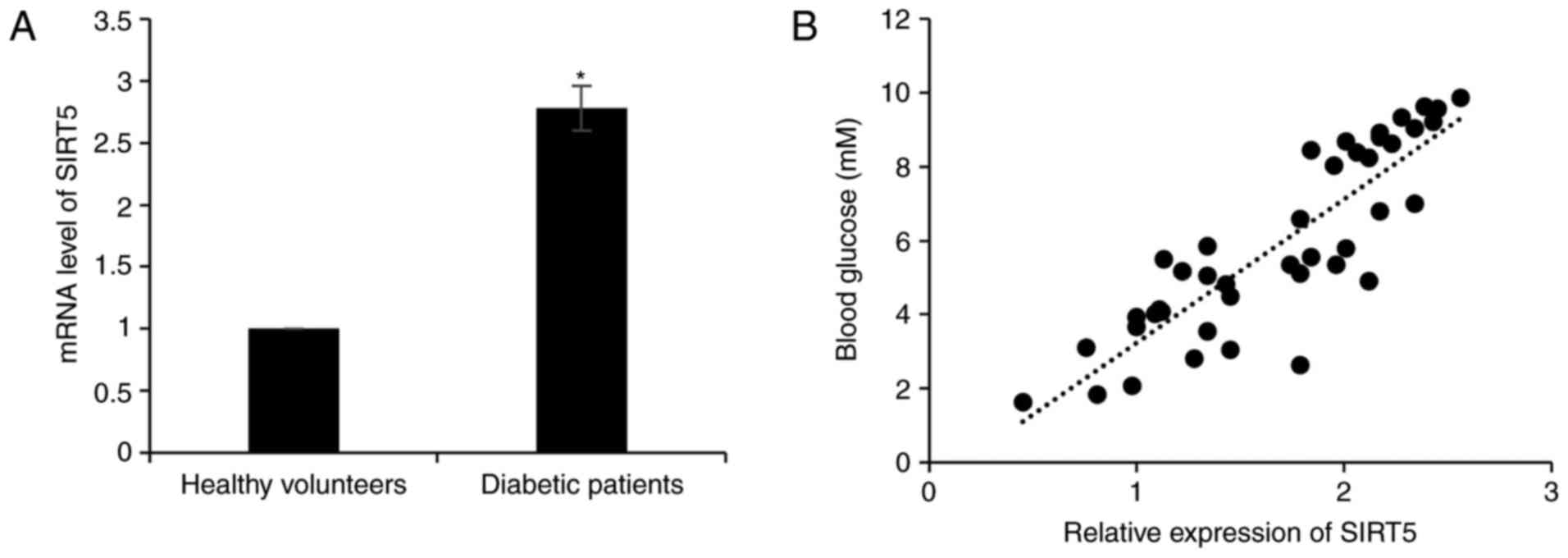
In order to investigate the role of SIRT5 in
diabetes, the expression of SIRT5 in plasma samples obtained from
65 patients with T2D and 65 healthy volunteers was assessed. The
result of RT-qPCR analysis revealed that SIRT5 mRNA levels were
significantly upregulated in patients with diabetes compared with
healthy volunteers (Fig. 1A). It was
also determined that the expression of SIRT5 was positively
correlated with blood glucose levels (r=0.87; Fig. 1B). In addition, the association
between patient clinicopathological features and SIRT5 expression
was analyzed, and it was revealed that a high SIRT5 expression was
significantly associated with age, but not with gender (Table I).
 | Table I.Association between SIRT5 expression
and the clinicopathologic features of 65 patients with type 2
diabetes. |
Table I.
Association between SIRT5 expression
and the clinicopathologic features of 65 patients with type 2
diabetes.
|
|
| SIRT5 protein
expression |
|---|
|
|
|
|
|---|
| Characteristic | Number | Low (n=23) | High (n=42) | P-value |
|---|
| Age |
|
|
| 0.011 |
|
<40 | 26 | 14 | 12 |
|
| ≥ 0 | 39 | 9 | 30 |
|
| Sex |
|
|
| 0.749 |
| Male | 30 | 10 | 20 |
|
|
Female | 35 | 13 | 22 |
|
| Blood glucose
level |
|
|
| <0.001 |
| <7.0
mM | 25 | 15 | 10 |
|
| >7.0
mM | 40 | 8 | 32 |
|
| PDX1
expression |
|
|
| <0.001 |
|
Low | 38 | 7 | 31 |
|
|
High | 27 | 16 | 11 |
|
SIRT5 suppresses insulin secretion and
the proliferation of pancreatic β-cell lines
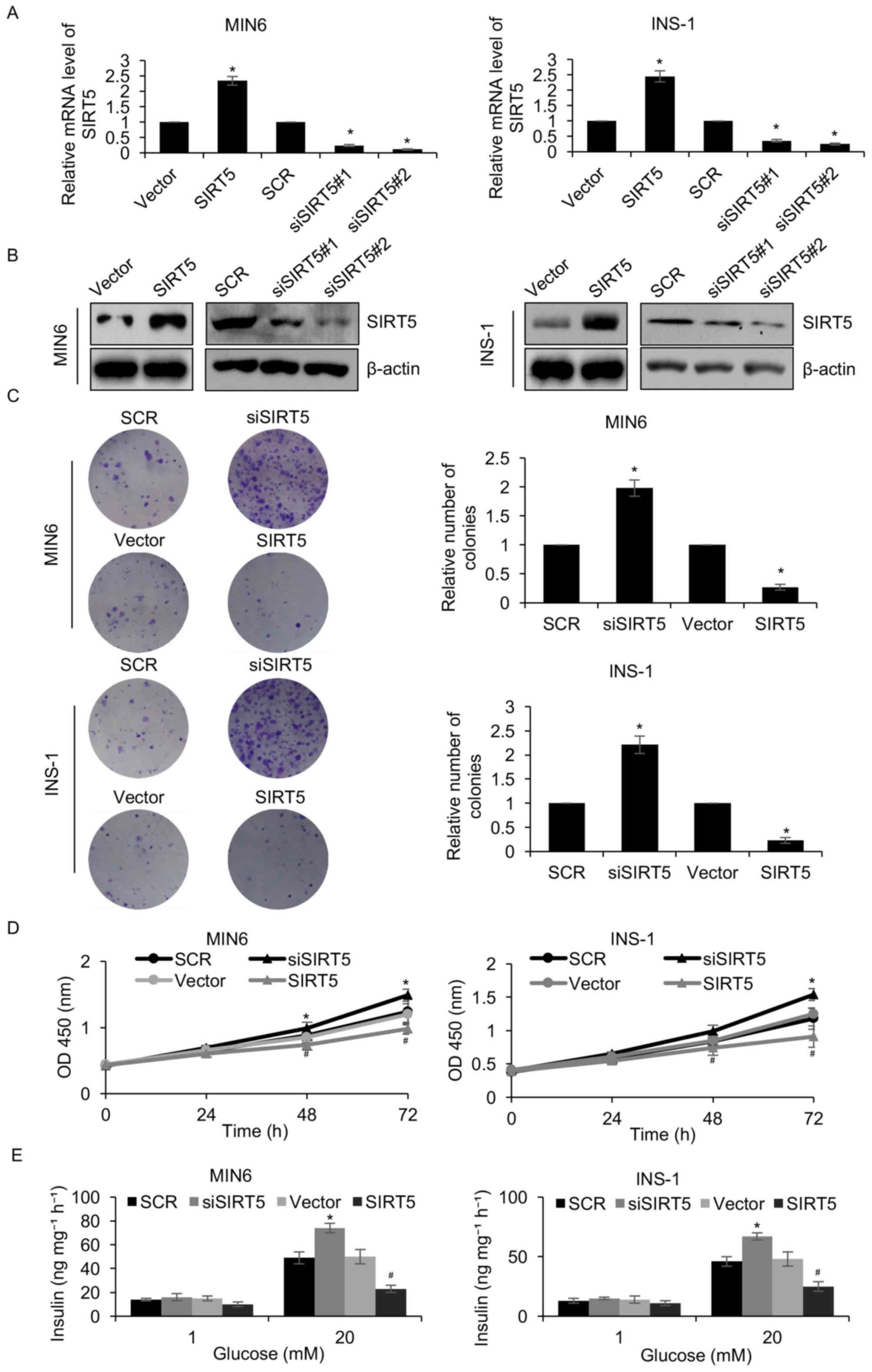
To elucidate the function of SIRT5 in diabetes, a
pcDNA3.1-SIRT5 plasmid was constructed and SIRT5 was overexpressed
or knocked down in MIN6 and INS-1 pancreatic β-cell lines.
Following a 48 h transfection, the expression of SIRT5 was
determined by RT-qPCR and western blotting analyses. SIRT5 was
significantly upregulated and downregulated in MIN6 and INS-1 cells
transfected with the pcDNA3.1-SIRT5 plasmid and siSIRT5,
respectively, when compared with their respective controls
(Fig. 2A and B). siSIRT5#2 was
demonstrated to be more efficient at repressing SIRT5 than
siSIRT5#1, and was therefore utilized in subsequent experiments. A
colony formation assay was performed to assess the effect of SIRT5
expression on the proliferation of pancreatic β-cell lines. The
results demonstrated that ectopic SIRT5 expression significantly
suppressed MIN6 and INS-1 cell colony formation when compared with
controls, whereas inhibition of SIRT5 expression had the opposite
effect (Fig. 2C). In addition, the
results of the CCK-8 assay indicated that upregulation of SIRT5
significantly inhibited MIN6 and INS-1 cell proliferation when
compared with controls, whereas SIRT5 inhibition demonstrated the
opposite effect (Fig. 2D). It was
also demonstrated that overexpression of SIRT5 significantly
decreased the level of insulin secretion following exposure to
glucose when compared with controls, whereas inhibition of SIRT5
expression significantly increased insulin secretion (Fig. 2E). Therefore, the results
demonstrated that SIRT5 downregulation facilitates insulin
secretion and the proliferation of pancreatic β-cell lines.
PDX1 is transcriptionally regulated by
SIRT5
As a HDAC enzyme, SIRT5 has been demonstrated to
regulate the expression of genes (7). The protein PDX1 serves a number of key
roles in the pathogenesis of diabetes (18). The authors of the present study
therefore hypothesized that SIRT5 may regulate the expression of
PDX1 in patients with diabetes. To verify this hypothesis, SIRT5
was overexpressed or knocked down in MIN6 and INS-1 cells and the
expression of PDX1 was determined using RT-qPCR and western
blotting analyses. The results demonstrated that PDX1 mRNA and
protein levels were regulated by SIRT5 (Fig. 3A and B). SIRT5 ectopic expression and
inhibition significantly reduced and increased the expression of
PDX1, respectively (Fig. 3A and B).
To investigate whether the effects of SIRT5 on the expression of
PDX1 were dependent on its deacetylase activity, an inactive form
of the SIRT5 deacetylase (T69A) was overexpressed. It was
demonstrated that ectopic expression of SIRT5 (T69A) had no
significant effect on the expression of PDX1 (Fig. 3A and B). Subsequently, a ChIP assay
was performed to determine whether SIRT5 regulates the
transcription of PDX1 expression through H4K16 deacetylation. The
results revealed that occupation of H4K16ac in the PDX1 promoter
region was significantly increased following knock down of SIRT5
(Fig. 3C). However, reconstituting
the expression of SIRT5R in MIN6 cells could partially offset the
effect of SIRT5-depletion (Fig. 3C).
Similar results were also observed in INS-1 cells (Fig. 3C). In addition, the expression of
PDX1 in plasma samples obtained from 65 patients with diabetes were
assessed. The results revealed that SIRT5 expression was negatively
associated with PDX1 expression (Table
I). Together, these results suggest that the HDAC activity of
SIRT5 may regulate the expression of PDX1 via H4K16
deacetylation.
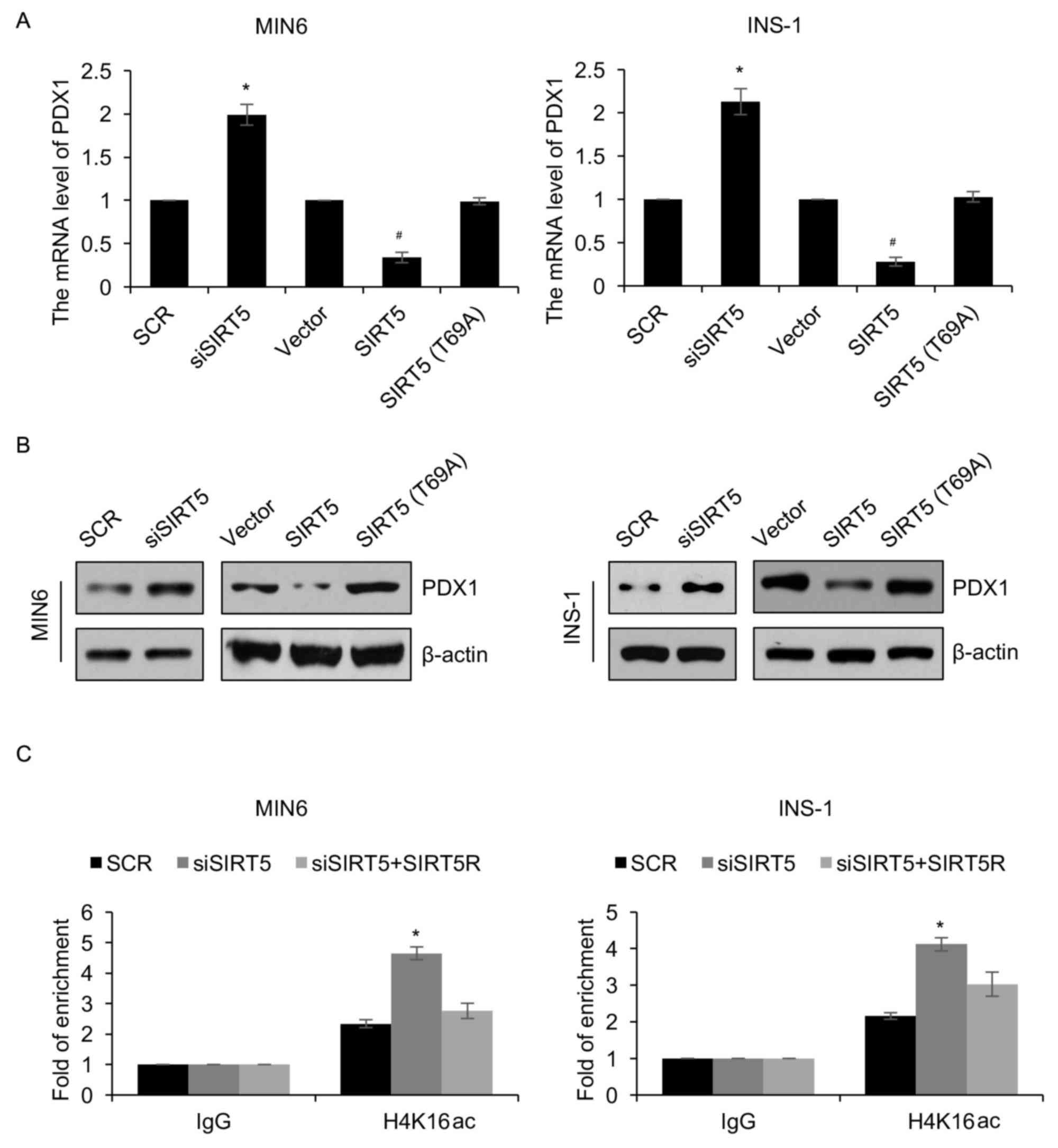 | Figure 3.PDX1 is transcriptionally regulated by
SIRT5. MIN6 and INS-1 cells were an empty plasmid vector, a plasmid
vector containing SIRT5, SCR, siSIRT5 or SIRT5 (T69A). Following 48
h transfection, the (A) mRNA and (B) protein levels of PDX1 was
detected using reverse transcription quantitative polymerase chain
reaction and western blotting analyses, respectively. (C) SIRT5 was
knocked down in treated MIN6 and INS-1 cells, and anti-H4K16ac
antibodies were used to perform qChIP assays. Each experiment was
repeated three times. The results are presented as the mean ±
standard deviation. *P<0.05 vs. SCR and #P<0.05
vs. vector. SIRT5, sirtuin 5; Vector, pcDNA3.1 plasmid vector; SCR,
scrambled small interfering RNA; siSIRT5, small interfering RNA
targeting sirtuin 5; PDX1, pancreatic and duodenal homeobox 1;
qChIP, quantitative chromatin immunoprecipitation. |
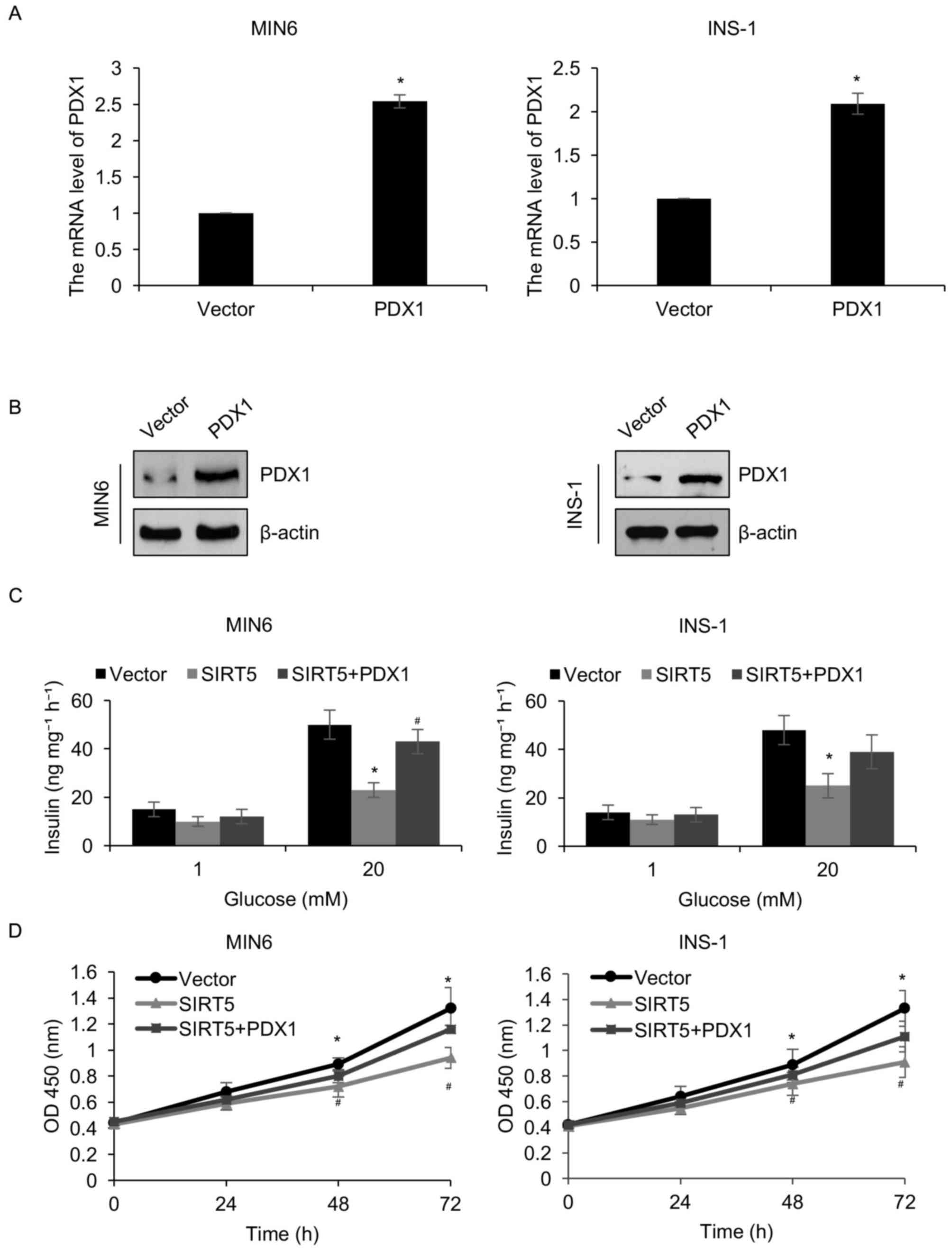
PDX1 is involved in SIRT5-mediated
insulin secretion and pancreatic β-cell proliferation
The results presented thus far have demonstrated
that SIRT5 may regulate the transcription of PDX1. Therefore, it
was hypothesized that PDX1 may serve as a downstream effector of
SIRT5. To verify this hypothesis, the pcDNA3.1-PDX1 plasmid was
constructed to increase the expression of PDX1 in MIN6 and INS-1
cells. Western blotting and RT-qPCR analyses were employed to
analyze PDX1 protein and mRNA levels, respectively. It was
demonstrated that the protein and mRNA levels of PDX1 were
significantly increased in MIN6 and INS-1 cells transfected with a
PDX1 when compared with controls (Fig.
4A and B). The involvement of PDX1 in SIRT5-mediated insulin
secretion and pancreatic β-cell proliferation was then determined.
It was demonstrated that the simultaneous overexpression of SIRT5
and PDX1 offset the effect of SIRT5 overexpression in INS-1 and
MIN6 cells (Fig. 4C). When cells
were simultaneously overexpressed with SIRT5 and PDX1, the
glucose-stimulated secretion of insulin was increased compared with
the cells overexpressing SIRT5 (Fig.
4C). Similarly, the CCK-8 assay results demonstrated that
combined SIRT5 and PDX1 overexpression significantly reduced the
effect of the SIRT5 overexpression on cell proliferation (Fig. 4D). These results indicate that PDX1
may function as a downstream target of SIRT5 and mediate pancreatic
β-cell proliferation and insulin secretion.
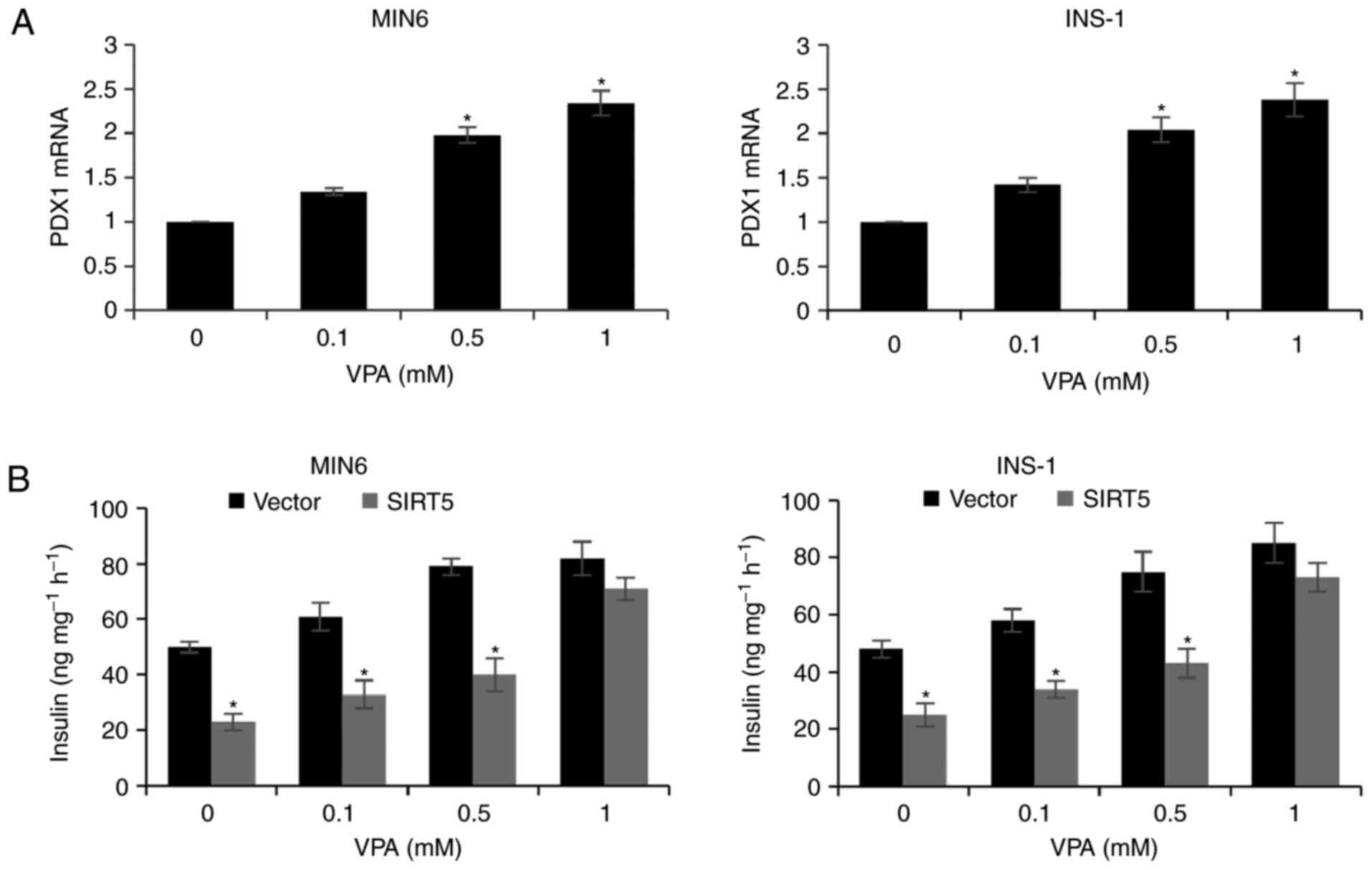
VPA increases PDX1 expression and
insulin secretion
Due to the HDAC activity of SIRT, the effect of VPA
treatment (a HDAC inhibitor) on pancreatic β-cells was assessed.
MIN6 and INS-1 cells incubated with 0.1, 0.5 and 1 mM VPA for 48 h
exhibited a significant increase in the expression of PDX1
(Fig. 5A). In addition, treatment
with increasing concentrations of VPA was associated with a
dose-dependent increase in insulin secretion following exposure to
glucose and SIRT5 overexpression decreased insulin secretion
compared with the control group (Fig.
5B). The results therefore suggest that VPA may be a novel
therapy for T2D, which is consistent with the results of previous
studies (19,20).
Discussion
The dysfunction of β-cells is a primary cause of
human T2D and results from the abnormal expression of specific
genes involved in regulating metabolic signaling pathways (21,22).
Chronically high carbohydrate diets may also contribute to β-cell
dysfunction (21). SIRT5 has been
demonstrated to serve key roles in various metabolic processes
(10,23,24),
including deacetylation and desuccinylation of CPS1 by SIRT5,
resulting in its increased enzymatic activity, thereby promoting
the urea cycle (25). However, the
role of SIRT5 in T2D has not yet been elucidated.
The present study demonstrated that SIRT5 was
upregulated in patients with T2D and that the expression of SIRT5
was associated with high blood glucose levels. It was also revealed
that the expression of SIRT5 was associated with patient age. This
may be due to alterations in the expression levels of specific
proteins as the body ages, such as SIRT1 (26), resulting in a high expression of
SIRT5. To investigate the effect of SIRT5 in T2D in the present
study, two pancreatic β-cell lines (MIN6 and INS-1) were utilized.
Functional experiments, including colony formation and CCK-8
assays, were performed. The results suggested that SIRT5 inhibition
facilitated pancreatic β-cell proliferation and insulin secretion.
A previous study demonstrated that SIRT5 facilitates cancer cell
growth and drug resistance in non-small cell lung cancer (27). This contradicts the results of the
present study; however, it should be considered that diabetes and
cancer are two different diseases. The present study hypothesized
that there are also many factors that may contribute to the
different functions of SIRT5, including cell microenvironment,
metabolism and oncogene activation.
PDX1 is the earliest tissue-selective transcription
factor expressed in the developing primordium, and is essential to
formation of all pancreatic cell types and the activity of adult
islet β cells (28). Thus, mice and
humans that completely lack Pdx1 function are apancreatic, while
haploinsufficiency primarily affects islet β cells following birth
(28). Moreover, β cell-specific
inactivation of Pdx1 in the adult mouse causes severe hyperglycemia
and loss of cell identity, with these cells transdifferentiating to
an islet α-like cell capable of secreting the glucagon hormone
(18). Previous studies have
demonstrated that PDX1 serves a primary role in diabetes and that
the downregulation of PDX1 expression aggravates diabetes mellitus
(29,30). The results of the present study
demonstrated that SIRT5 may regulate the transcription of PDX1
through its deacetylase activity. Despite SIRT5 only possessing
weak deacetylase activity, it was demonstrated that PDX1 is
negatively regulated by SIRT5. A previous study suggested that
SIRT5 may function through its de-succinylase activity (31). However, the role of succinylation in
cancer and metabolic diseases remains unknown. It is therefore
important to assess alterations in the level of specific histone or
protein succinylation in patients with diabetes mellitus using mass
spectrometry in future studies.
Previous studies have demonstrated that the HDAC
inhibitor, VPA, increases insulin secretion in vivo
(19,20). Consistent with these results, the
present study revealed that VPA facilitated the secretion of
insulin in two pancreatic β-cell lines. In addition, it was
determined that VPA may promote insulin secretion by enhancing the
expression of PDX1.
In conclusion, the current study investigated the
function of SIRT5 in T2D and the mechanisms underlying it role in
insulin secretion and pancreatic β-cell proliferation. The results
revealed that SIRT5 may be associated with the pathogenesis of T2D
and may serve as a novel biomarker for the diagnosis of T2D.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request
Authors' contributions
YM and XF conceived and designed the study, analyzed
the data, and wrote the paper. YM constructed the expression
plasmids, prepared the proteins, and performed experiments. All
authors have read and approved this manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Taizhou People's Hospital (Taizhou, China) and written
informed consent was provided from patients and healthy volunteers
prior to enrolment.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Matzinger M, Fischhuber K and Heiss EH:
Activation of Nrf2 signaling by natural products-can it alleviate
diabetes? Biotechnol Adv. Dec 28–2017.(Epub ahead of print).
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stoffers DA: The development of beta-cell
mass: recent progress and potential role of GLP-1. Horm Metab Res.
36:811–821. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tourrel C, Bailbe D, Lacorne M, Meile MJ,
Kergoat M and Portha B: Persistent improvement of type 2 diabetes
in the Goto-Kakizaki rat model by expansion of the beta-cell mass
during the prediabetic period with glucagon-like peptide-1 or
exendin-4. Diabetes. 51:1443–1452. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sakuraba H, Mizukami H, Yagihashi N, Wada
R, Hanyu C and Yagihashi S: Reduced beta-cell mass and expression
of oxidative stress-related DNA damage in the islet of Japanese
Type II diabetic patients. Diabetologia. 45:85–96. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Marchetti P, Del Guerra S, Marselli L,
Lupi R, Masini M, Pollera M, Bugliani M, Boggi U, Vistoli F, Mosca
F and Del Prato S: Pancreatic islets from type 2 diabetic patients
have functional defects and increased apoptosis that are
ameliorated by metformin. J Clin Endocrinol Metab. 89:5535–5541.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cozar-Castellano I, Fiaschi-Taesch N,
Bigatel TA, Takane KK, Garcia-Ocaña A, Vasavada R and Stewart AF:
Molecular control of cell cycle progression in the pancreatic
beta-cell. Endocr Rev. 27:356–370. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hirschey MD: Old enzymes, new tricks:
Sirtuins are NAD(+)-dependent de-acylases. Cell Metab. 14:718–719.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Colak G, Xie Z, Zhu AY, Dai L, Lu Z, Zhang
Y, Wan X, Chen Y, Cha YH, Lin H, et al: Identification of lysine
succinylation substrates and the succinylation regulatory enzyme
CobB in Escherichia coli. Mol Cell Proteomics. 12:3509–3520. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Park J, Chen Y, Tishkoff DX, Peng C, Tan
M, Dai L, Xie Z, Zhang Y, Zwaans BM, Skinner ME, et al:
SIRT5-mediated lysine desuccinylation impacts diverse metabolic
pathways. Mol Cell. 50:919–930. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rardin MJ, He W, Nishida Y, Newman JC,
Carrico C, Danielson SR, Guo A, Gut P, Sahu AK, Li B, et al: SIRT5
regulates the mitochondrial lysine succinylome and metabolic
networks. Cell Metab. 18:920–933. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Weinert BT, Scholz C, Wagner SA,
Iesmantavicius V, Su D, Daniel JA and Choudhary C: Lysine
succinylation is a frequently occurring modification in prokaryotes
and eukaryotes and extensively overlaps with acetylation. Cell Rep.
4:842–851. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Z, Tan M, Xie Z, Dai L, Chen Y and
Zhao Y: Identification of lysine succinylation as a new
post-translational modification. Nat Chem Biol. 7:58–63. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hirschey MD and Zhao Y: Metabolic
regulation by lysine malonylation, succinylation, and
glutarylation. Mol Cell Proteomics. 14:2308–2315. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tan M, Peng C, Anderson KA, Chhoy P, Xie
Z, Dai L, Park J, Chen Y, Huang H, Zhang Y, et al: Lysine
glutarylation is a protein posttranslational modification regulated
by SIRT5. Cell Metab. 19:605–617. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xie Z, Dai J, Dai L, Tan M, Cheng Z, Wu Y,
Boeke JD and Zhao Y: Lysine succinylation and lysine malonylation
in histones. Mol Cell Proteomics. 11:100–107. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
American Diabetes Association. Diagnosis
and classification of diabetes mellitus. Diabetes Care. 35 Suppl
1:S64–S71. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fujimoto K and Polonsky KS: Pdx1 and other
factors that regulate pancreatic beta-cell survival. Diabetes Obes
Metab. 11 Suppl 4:S30–S37. 2009. View Article : Google Scholar
|
|
19
|
Manaka K, Nakata M, Shimomura K, Rita RS,
Maejima Y, Yoshida M, Dezaki K, Kakei M and Yada T: Chronic
exposure to valproic acid promotes insulin release, reduces KATP
channel current and does not affect Ca (2+) signaling in mouse
islets. J Physiol Sci. 64:77–83. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Axelsson AS, Mahdi T, Nenonen HA, Singh T,
Hänzelmann S, Wendt A, Bagge A, Reinbothe TM, Millstein J, Yang X,
et al: Sox5 regulates beta-cell phenotype and is reduced in type 2
diabetes. Nat Commun. 8:156522017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Weir GC and Bonner-Weir S: Five stages of
evolving beta-cell dysfunction during progression to diabetes.
Diabetes. 53 Suppl 3:S16–S21. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ashcroft FM and Rorsman P: Diabetes
mellitus and the β cell: The last ten years. Cell. 148:1160–1171.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kumar S and Lombard DB: Mitochondrial
sirtuins and their relationships with metabolic disease and cancer.
Antioxid Redox Signal. 22:1060–1077. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu J, Sadhukhan S, Noriega LG, Moullan N,
He B, Weiss RS, Lin H, Schoonjans K and Auwerx J: Metabolic
characterization of a Sirt5 deficient mouse model. Sci Rep.
3:28062013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nakagawa T, Lomb DJ, Haigis MC and
Guarente L: SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and
regulates the urea cycle. Cell. 137:560–570. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Qadir MI and Anwar S: Sirtuins in brain
aging and neurological disorders. Crit Rev Eukaryot Gene Expr.
27:321–329. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lu W, Zuo Y, Feng Y and Zhang M: SIRT5
facilitates cancer cell growth and drug resistance in non-small
cell lung cancer. Tumour Biol. 35:10699–10705. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gannon M, Gamer LW and Wright CV:
Regulatory regions driving developmental and tissue-specific
expression of the essential pancreatic gene pdx1. Dev Biol.
238:185–201. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Spaeth JM, Gupte M, Perelis M, Yang YP,
Cyphert H, Guo S, Liu JH, Guo M, Bass J, Magnuson MA, et al:
Defining a novel role for the Pdx1 transcription factor in islet
β-cell maturation and proliferation during weaning. Diabetes.
66:2830–2839. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fujitani Y: Transcriptional regulation of
pancreas development and β-cell function [Review]. Endocr J.
64:477–486. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang
H, Kim J, Woo J, Kim JH, Choi BH, et al: Sirt5 is a NAD-dependent
protein lysine demalonylase and desuccinylase. Science.
334:806–809. 2011. View Article : Google Scholar : PubMed/NCBI
|