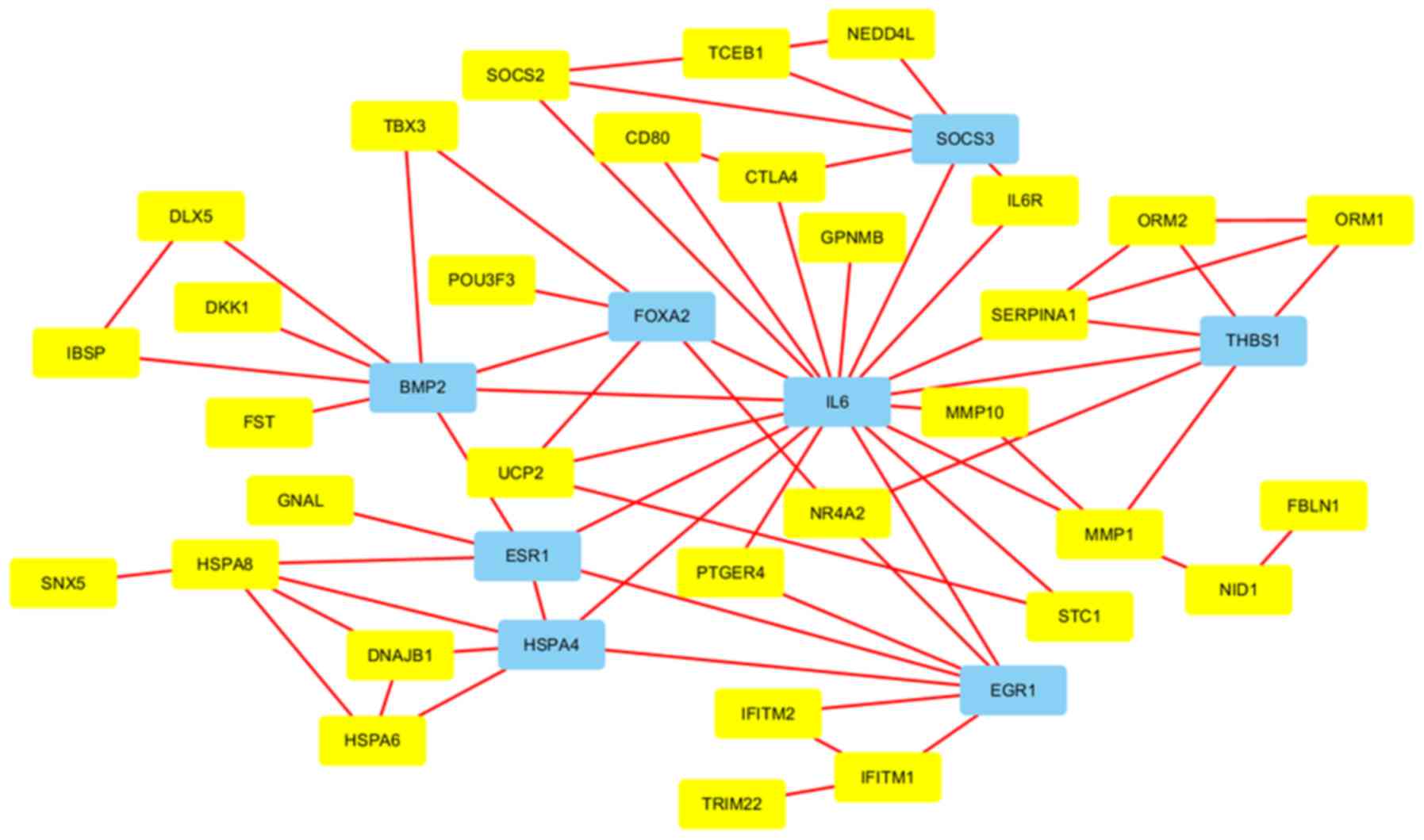
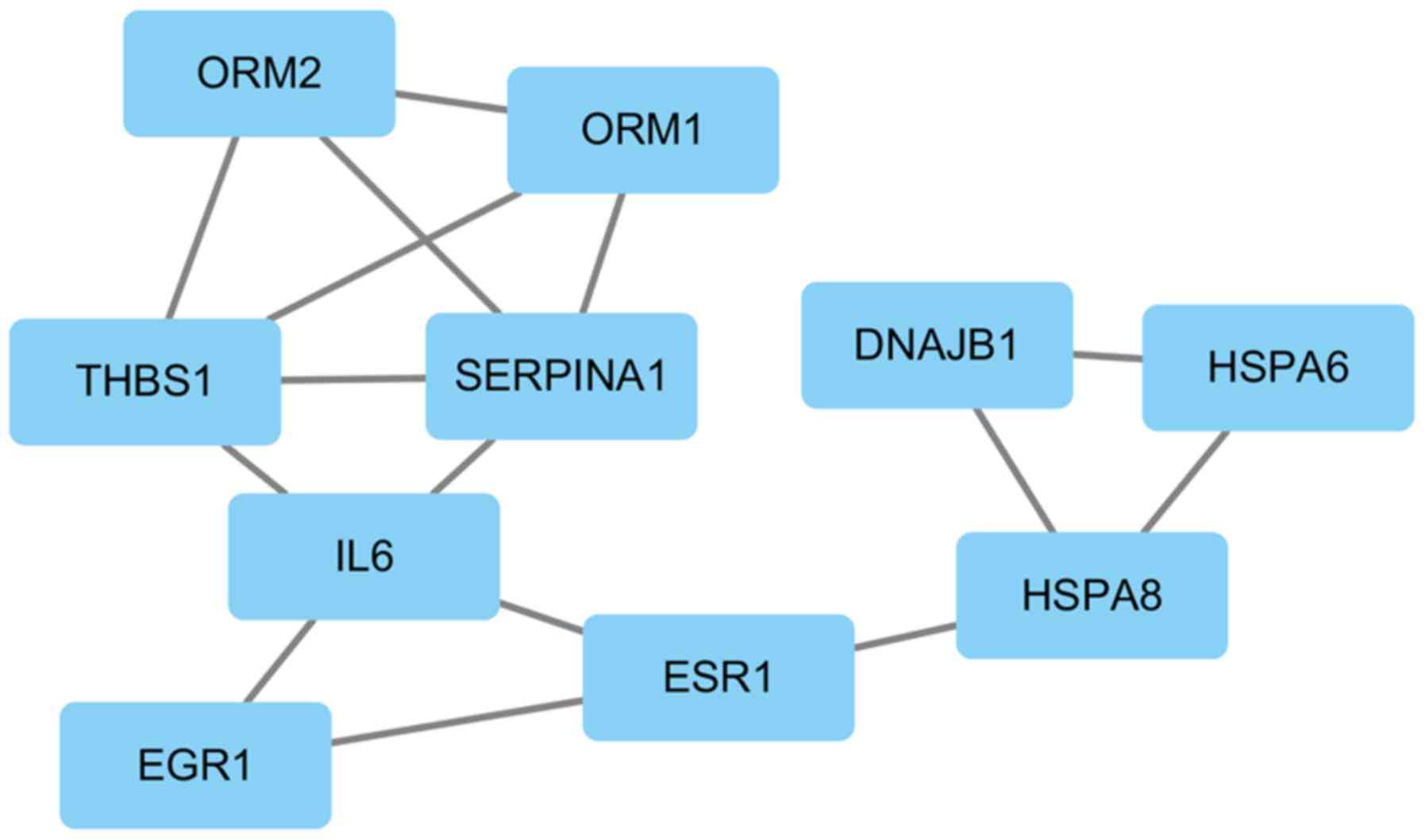
|
1
|
Luoma K, Riihimäki H, Luukkonen R,
Raininko R, Viikari-Juntura E and Lamminen A: Low back pain in
relation to lumbar disc degeneration. Spine (Phila Pa 1976).
25:487–492. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Andersson GB: Epidemiological features of
chronic low-back pain. Lancet. 354:581–585. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Maniadakis N and Gray A: The economic
burden of back pain in the UK. Pain. 84:95–103. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Katz JN: Lumbar disc disorders and
low-back pain: Socioeconomic factors and consequences. J Bone Joint
Surg Am. 88 Suppl 2:S21–S24. 2006. View Article : Google Scholar
|
|
5
|
Martin BI, Deyo RA, Mirza SK, Turner JA,
Comstock BA, Hollingworth W and Sullivan SD: Expenditures and
health status among adults with back and neck problems. JAMA.
299:656–664. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yorimitsu E, Chiba K, Toyama Y and
Hirabayashi K: Long-term outcomes of standard discectomy for lumbar
disc herniation: A follow-up study of more than 10 years. Spine
(Phila Pa 1976). 26:652–657. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li X, Han Y, Di Z, Cui J, Pan J, Yang M,
Sun G, Tan J and Li L: Percutaneous endoscopic lumbar discectomy
for lumbar disc herniation. J Clin Neurosci. 33:19–27. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Karasek M and Bogduk N: Twelve-month
follow-up of a controlled trial of intradiscal thermal anuloplasty
for back pain due to internal disc disruption. Spine (Phila Pa
1976). 25:2601–2607. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Uschold TD, Fusco D, Germain R, Tumialan
LM and Chang SW: Cervical and lumbar spinal arthroplasty: Clinical
review. AJNR Am J Neuroradiol. 33:1631–1641. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hilibrand AS and Robbins M: Adjacent
segment degeneration and adjacent segment disease: The consequences
of spinal fusion? Spine J. 4 (6 Suppl):190S–194S. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kepler CK, Ponnappan RK, Tannoury CA,
Risbud MV and Anderson DG: The molecular basis of intervertebral
disc degeneration. Spine J. 13:318–330. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Martin MD, Boxell CM and Malone DG:
Pathophysiology of lumbar disc degeneration: A review of the
literature. Neurosurg Focus. 13:E12002. View Article : Google Scholar
|
|
13
|
Lyons G, Eisenstein SM and Sweet MB:
Biochemical changes in intervertebral disc degeneration. Biochim
Biophys Acta. 673:443–453. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Antoniou J, Steffen T, Nelson F,
Winterbottom N, Hollander AP, Poole RA, Aebi M and Alini M: The
human lumbar intervertebral disc: Evidence for changes in the
biosynthesis and denaturation of the extracellular matrix with
growth, maturation, ageing, and degeneration. J Clin Invest.
98:996–1003. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Eyre DR and Muir H: Types I and II
collagens in intervertebral disc. Interchanging radial
distributions in annulus fibrosus. Biochem J. 157:267–270. 1976.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Roberts S, Menage J, Duance V and Wotton
SF: Type III collagen in the intervertebral disc. Histochem J.
23:503–508. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schollmeier G, Lahr-Eigen R and
Lewandrowski KU: Observations on fiber-forming collagens in the
anulus fibrosus. Spine (Phila Pa 1976). 25:2736–2741. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kazezian Z, Gawri R, Haglund L, Ouellet J,
Mwale F, Tarrant F, O'Gaora P, Pandit A, Alini M and Grad S: Gene
expression profiling identifies interferon signalling molecules and
IGFBP3 in human degenerative annulus fibrosus. Sci Rep.
5:156622015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Thompson JP, Pearce RH, Schechter MT,
Adams ME, Tsang IK and Bishop PB: Preliminary evaluation of a
scheme for grading the gross morphology of the human intervertebral
disc. Spine (Phila Pa 1976). 15:411–415. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gruber HE, Hoelscher GL, Ingram JA and
Hanley EN Jr: Genome-wide analysis of pain-, nerve- and
neurotrophin-related gene expression in the degenerating human
annulus. Mol Pain. 8:632012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gruber HE, Ingram JA, Hoelscher GL,
Zinchenko N, Hanley EN Jr and Sun Y: Asporin, a susceptibility gene
in osteoarthritis, is expressed at higher levels in the more
degenerate human intervertebral disc. Arthritis Res Ther.
11:R472009. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tsai TT, Lai PL, Liao JC, Fu TS, Niu CC,
Chen LH, Lee MS, Chen WJ and Fang HC: Increased periostin gene
expression in degenerative intervertebral disc cells. Spine J.
13:289–298. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gruber HE, Hoelscher GL and Hanley EN Jr:
Annulus cells from more degenerated human discs show modified gene
expression in 3D culture compared with expression in cells from
healthier discs. Spine J. 10:721–727. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
He J, Xue R, Li S, Lv J, Zhang Y, Fan L,
Teng Y and Wei H: Identification of the potential molecular targets
for human intervertebral disc degeneration based on bioinformatic
methods. Int J Mol Med. 36:1593–1600. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tang Y, Wang S, Liu Y and Wang X:
Microarray analysis of genes and gene functions in disc
degeneration. Exp Ther Med. 7:343–348. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guo W, Zhang B, Li Y, Duan HQ, Sun C, Xu
YQ and Feng SQ: Gene expression profile identifies potential
biomarkers for human intervertebral disc degeneration. Mol Med.
16:8665–8672. 2017.
|
|
27
|
Schubert AK, Smink JJ, Arp M, Ringe J,
Hegewald AA and Sittinger M: Quality assessment of surgical disc
samples discriminates human annulus fibrosus and nucleus pulposus
on tissue and molecular level. Int J Mol Sci. 19(pii): E17612018.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Roberts S, Caterson B, Menage J, Evans EH,
Jaffray DC and Eisenstein SM: Matrix metalloproteinases and
aggrecanase: Their role in disorders of the human intervertebral
disc. Spine (Phila Pa 1976). 25:3005–3013. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Deng B, Ren JZ, Meng XQ, Pang CG, Duan GQ,
Zhang JX, Zou H, Yang HZ and Ji JJ: Expression profiles of MMP-1
and TIMP-1 in lumbar intervertebral disc degeneration. Genet Mol
Res. 14:19080–19086. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Weiler C, Nerlich AG, Zipperer J,
Bachmeier BE and Boos N: 2002 SSE Award Competition in Basic
Science: Expression of major matrix metalloproteinases is
associated with intervertebral disc degradation and resorption. Eur
Spine J. 11:308–320. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Le Maitre CL, Freemont AJ and Hoyland JA:
Localization of degradative enzymes and their inhibitors in the
degenerate human intervertebral disc. J Pathol. 204:47–54. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Matsui Y, Maeda M, Nakagami W and Iwata H:
The involvement of matrix metalloproteinases and inflammation in
lumbar disc herniation. Spine (Phila Pa 1976). 23:863–869. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Visse R and Nagase H: Matrix
metalloproteinases and tissue inhibitors of metalloproteinases:
Structure, function, and biochemistry. Circ Res. 92:827–839. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Welgus HG, Jeffrey JJ and Eisen AZ: The
collagen substrate specificity of human skin fibroblast
collagenase. J Biol Chem. 256:9511–9515. 1981.PubMed/NCBI
|
|
35
|
Roberts S, Menage J, Duance V, Wotton S
and Ayad S: 1991 Volvo Award in basic sciences. Collagen types
around the cells of the intervertebral disc and cartilage end
plate: An immunolocalization study. Spine (Phila Pa 1976).
16:1030–1038. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ye S, Ju B, Wang H and Lee KB: Bone
morphogenetic protein-2 provokes interleukin-18-induced human
intervertebral disc degeneration. Bone Joint Res. 5:412–418. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kim DJ, Moon SH, Kim H, Kwon UH, Park MS,
Han KJ, Hahn SB and Lee HM: Bone morphogenetic protein-2
facilitates expression of chondrogenic, not osteogenic, phenotype
of human intervertebral disc cells. Spine (Phila Pa 1976).
28:2679–2684. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kim H, Lee JU, Moon SH, Kim HC, Kwon UH,
Seol NH, Kim HJ, Park JO, Chun HJ, Kwon IK and Lee HM: Zonal
responsiveness of the human intervertebral disc to bone
morphogenetic protein-2. Spine (Phila Pa 1976). 34:1834–1838. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hammes SR and Levin ER: Extranuclear
steroid receptors: Nature and actions. Endocr Rev. 28:726–741.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bay-Jensen AC, Slagboom E, Chen-An P,
Alexandersen P, Qvist P, Christiansen C, Meulenbelt I and Karsdal
MA: Role of hormones in cartilage and joint metabolism:
Understanding an unhealthy metabolic phenotype in osteoarthritis.
Menopause. 20:578–586. 2013.PubMed/NCBI
|
|
41
|
Song XX, Yu YJ, Li XF, Liu ZD, Yu BW and
Guo Z: Estrogen receptor expression in lumbar intervertebral disc
of the elderly: Gender- and degeneration degree-related variations.
Joint Bone Spine. 81:250–253. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen H, Herndon ME and Lawler J: The cell
biology of thrombospondin-1. Matrix Biol. 19:597–614. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gruber HE, Ingram JA and Hanley EN Jr:
Immunolocalization of thrombospondin in the human and sand rat
intervertebral disc. Spine (Phila Pa 1976). 31:2556–2561. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Risbud MV and Shapiro IM: Role of
cytokines in intervertebral disc degeneration: Pain and disc
content. Nat Rev Rheumatol. 10:44–56. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Dagistan Y, Cukur S, Dagistan E and Gezici
AR: Importance of IL-6, MMP-1, IGF-1, and BAX levels in lumbar
herniated disks and posterior longitudinal ligament in patients
with sciatic pain. World Neurosurg. 84:1739–1746. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kang JD, Georgescu HI, McIntyre-Larkin L,
Stefanovic-Racic M, Donaldson WF III and Evans CH: Herniated lumbar
intervertebral discs spontaneously produce matrix
metalloproteinases, nitric oxide, interleukin-6, and prostaglandin
E2. Spine (Phila Pa 1976). 21:271–277. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Studer RK, Vo N, Sowa G, Ondeck C and Kang
J: Human nucleus pulposus cells react to IL-6: Independent actions
and amplification of response to IL-1 and TNF-α. Spine (Phila Pa
1976). 36:593–599. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Takahashi H, Suguro T, Okazima Y, Motegi
M, Okada Y and Kakiuchi T: Inflammatory cytokines in the herniated
disc of the lumbar spine. Spine (Phila Pa 1976). 21:218–224. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Specchia N, Pagnotta A, Toesca A and Greco
F: Cytokines and growth factors in the protruded intervertebral
disc of the lumbar spine. Eur Spine J. 11:145–151. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Shamji MF, Setton LA, Jarvis W, So S, Chen
J, Jing L, Bullock R, Isaacs RE, Brown C and Richardson WJ:
Proinflammatory cytokine expression profile in degenerated and
herniated human intervertebral disc tissues. Arthritis Rheum.
62:1974–1982. 2010.PubMed/NCBI
|
|
51
|
Lee S, Moon CS, Sul D, Lee J, Bae M, Hong
Y, Lee M, Choi S, Derby R, Kim BJ, et al: Comparison of growth
factor and cytokine expression in patients with degenerated disc
disease and herniated nucleus pulposus. Clin Biochem. 42:1504–1511.
2009. View Article : Google Scholar : PubMed/NCBI
|