Introduction
Breast cancer, one of the most common female
malignant tumors, is a leading cause of cancer mortality worldwide
(1). Genetic mutations have been
demonstrated to be causative of the tumorigenesis and maintenance
of breast cancer. Over the past decades, an increasing amount of
evidence has shown that microRNAs (miRNAs), a small class of
non-coding RNAs, play a pivotal role in breast cancer, in which
they may function as oncogenes or tumor suppressor genes (2–4).
miRNAs are derived from exons of protein-coding and
non-coding genes (5,6), and are then transcribed by polymerase
II as a long primary transcript (primiR). Pri-miRNAs are further
processed by Drosha, leading to the excision and release of
approximately 70 nucleotide hairpin precursors termed pre-miRNAs
(7). After being exported from the
nucleus by exportin-5, the pre-miRNAs are subsequently cleaved by
Dicer, releasing the 22 nt miRNA-miRNA duplex, one strand of which
in turn is incorporated into the RNA-induced silencing complex
(miRISC), and eventually functions as a mature miRNA. The ‘seed’
region of the mature miRNA (nucleotides 2–8 at the 5′ end of a
miRNA) binds partially or completely to mRNA 3′-untranslated
regions (3′-UTRs) of specific protein-coding genes (8). miRNAs regulate their targets by
directly cleaving mRNAs or inhibiting protein synthesis, which
depends on the degree of complementarity with the 3′-UTR of their
targets (6). Complete
complementarity between miRNAs and the 3′-UTR of their targets
leads to mRNA degradation, while partial complementarity results in
translation inhibition of the target protein; the latter commonly
occurs in human cells (9).
There has been a large body of evidence showing the
significant difference in expression of miRNAs between breast
cancer and normal breast tissues (10,11).
These differentially expressed miRNAs are critical to the
proliferation (12,13), apoptosis (14,15),
invasion (16–19) and therapy resistance (20,21)
of breast cancer. In this study, miRNAs (miRs)-497, -198, -373 and
-1289 were selected for expression validation in breast cancer
samples. Correlation analysis was conducted to characterize the
association of miR-497 expression abnormality with pathological
characteristics. miR-497 mimics were used to determine the impact
of miR-497 on the proliferation, apoptosis and cell cycle of breast
cancer cells, as well as to determine its downstream targets.
Materials and methods
Specimens
In this study, 48 pairs of breast cancer and normal
specimens were collected from the Department of Breast and Thyroid
Surgery of Shanghai Tenth People’s Hospital, Shanghai, China. All
the samples were confirmed as invasive ductal breast cancer, and no
patients had received any chemotherapy or radiotherapy prior to
surgery.
Cell culture
The MCF-7 breast cancer cell line used in this study
was obtained from the Chinese Science Institute and grown in DMEM
(Gibco, New York, NY, USA) supplemented with 10% fetal bovine serum
(FBS) (Gibco), as well as 100 units of penicillin/ml and 100 μg of
streptomycin/ml (Enpromise, Hangzhou, China). Cells were incubated
at 37°C in a humidified chamber supplemented with 5%
CO2.
Real-time polymerase chain reaction (PCR)
analyses
We followed the protocol of Chen et al for
primer design and real-time reverse transcription (RT)-PCR
(22). The analyzed miRNAs
included miR-497, miR-198, miR-373 and miR-1289. miRNAs were
harvested according to the instructions of the miRcute miRNA
isolation kit (Tiangen, Beijing, China). The following primers were
used for the U6 small nuclear RNA, which was used as an internal
control: 5′-GTCCTATCCAGT GCAGGGTCCGAGGTGCACTGGATACGACAAAATATGG
AAC-3′, 5′-TGCGGGTGCTCGCTTCGCAGC-3′ and 5′-CCA GTGCAGGGTCCGAGGT-3′.
cDNA was generated by reverse transcription according to the
instructions of the PrimeScript™ RT-PCR kit (Takara, Shiga, Japan).
PCR parameters for miRNA quantification were as follows: 2 min at
95°C, and then 40 cycles of 30 sec at 95°C, and 45 sec at 60°C.
Total RNA was isolated using TRIzol (Invitrogen,
Carlsbad, CA, USA), and cDNA was generated by reverse transcription
following the protocol of the PrimeScript RT-PCR kit (Takara).
Quantitative real-time PCR was performed on a 7900HT fast RT-PCR
instrument (Applied Biosystems, Singapore) using SYBR-Green as the
detection fluorophore. All the primers used were as follows: Bcl-w
sense, CACCCAGGT CTCCGATGAAC and antisense, TTGTTGACACTCTCA GCACAC;
p65 sense, GGGAAGGAACGCTGTCAGAG and antisense,
TAGCCTCAGGGTACTCCATCA; Bcl-xL sense, GGTCGCATTGTGGCCTTTTTC and
antisense, TGCTGCATTGTTCCCATAGAG; Bcl-2 sense, GAACTG
GGGGAGGATTGTGG and antisense, CCGGTTCAGGTA CTCAGTCA; caspase-3
sense, ATGGAAGCGAATCAATGG ACTC and antisense,
CTGTACCAGACCGAGATGTCA; caspase-8 sense, CCTGTCACTGTCTTGTACCCT and
antisense, CCCGCAGTATCTTGCCTCC; β-actin sense, AGC
GAGCATCCCCCAAAGTT and anti sense, GGGCACGAA GGCTCATCATT. The PCR
parameters for relative quantification were as follows: 2 min at
95°C, followed by 40 cycles of 15 sec at 95°C and 30 sec at 60°C.
Each sample was tested in triplicate. The mRNA level of β-actin was
used as the internal control, and gene-specific mRNA expression was
normalized against β-actin expression.
Transfection assay
Cells (1x106) were added into each well
of a 6-well plate and cultured with DMEM medium without serum and
antibiotics. As the confluency of MCF-7 breast cancer cells reached
80–90%, miR-497 mimics and lipofect at the ratio of 1 μg:3 μl were
diluted to 250 μl by DMEM medium, respectively, and incubated for 5
min at room temperature. miR-497 mimics and the lipofect dilution
were gently combined and incubated for 20 min. Subsequently, 500 μl
of the complexes were added to each well. After 4–5 h of
incubation, DMEM medium was replaced by DMEM with 10% FBS, and all
the cells were incubated at 37°C in a CO2 incubator for
48 h prior to further testing.
Western blot analysis
Protein samples were separated with 12%
SDS-polyacrylamide gel (SDS-PAGE) and transferred onto PVDF
membranes (Beyotime, Haimen, China). Immune complexes were formed
by incubation of the proteins with primary antibodies (R&D
Systems, Minneapolis, MN, USA) overnight at 4°C. Blots were washed
and incubated for 1 h with HRP-conjugated anti-mouse secondary
antibodies (R&D Systems). Immunoreactive protein bands were
detected with an Odyssey Scanning system.
Cell proliferation assay
Cell proliferation was assessed using an MTT assay
kit (Sigma, Santa Clara, CA, USA). A total of 4–5 h after miR-497
mimics transfection, cells with various concentrations of miR-497
mimics were trypsinized and counted, respectively. Cells (1,000)
were plated in each well of 96-well plates (BD Biosciences,
Corning, NY, USA) in triplicate and incubated at 37°C, and cell
proliferation was assessed at 24, 36 and 48 h, following the
instructions of the MTT proliferation assay kit. The growth
inhibition rate was calculated using the following equation: growth
inhibition rate = [the mean optical density (OD) of controls - the
mean OD of samples]/the mean OD of controls.
Apoptosis assay
Thirty-six hours after miR-497 mimics transfection,
adherent cells were trypsinized. Annexin V incubation reagent (100
μl) was prepared by combining: 10 μl 10X binding buffer, 10 μl
propidium iodide (PI), 1 μl Annexin V-FITC and 79 μl deionized,
distilled H2O. Cells were gently resuspended in the
prepared Annexin V incubation reagent at a concentration of
105 to 106 cells per 100 μl. Samples were
incubated in the dark for 15 min at room temperature. All the
samples were processed by flow cytometry (FACSCanto™ II, BD
Biosciences, USA). FACS analyses were performed at least 3 times
with reproducible results.
Cell cycle assay
Cells were harvested and centrifuged at 1,200 rpm
for 5 min and washed twice in PBS. Subsequently, 3.0 ml ice-cold
70% ethanol was added dropwise and cells were fixed in this final
70% ethanol solution for at least 30 min. A total of 250 μl 0.05
g/l PI staining solution was added into each sample and incubated
for 30 min at room temperature and then analyzed by a flow
cytometer (FACSCanto™ II, BD Biosciences). The proliferation index
was calculated according to the following equation: (S+G2/M)/(G0/
G1+S+G2/M). The mock was the control groups treated with lipofect,
the negative control (NC) was the control groups transfected with a
short RNA sequence. The controls were not treated with miR-497
mimics.
Statistical analysis
Data were expressed as the means ± standard
deviation from at least 3 separate experiments performed in
triplicate. Correlations with tumor characteristics were made with
Spearman analysis. For RT-PCR analyses, based on the
2−ΔΔCt method described by Livak and
Schmittgen (23),
semi-quantitative analysis was used for gene quantification. As
variances were not equal, the statistical differences were analyzed
using a two independent sample non-parametric test (Mann-Whitney U
test) and a K-independent non-parametric sample test
(Kruskal-Wallis test) in MCF-7 breast cancer cells, and the null
hypothesis was rejected at the 0.05 level.
Results
Underexpression of miR-497 in breast
cancer
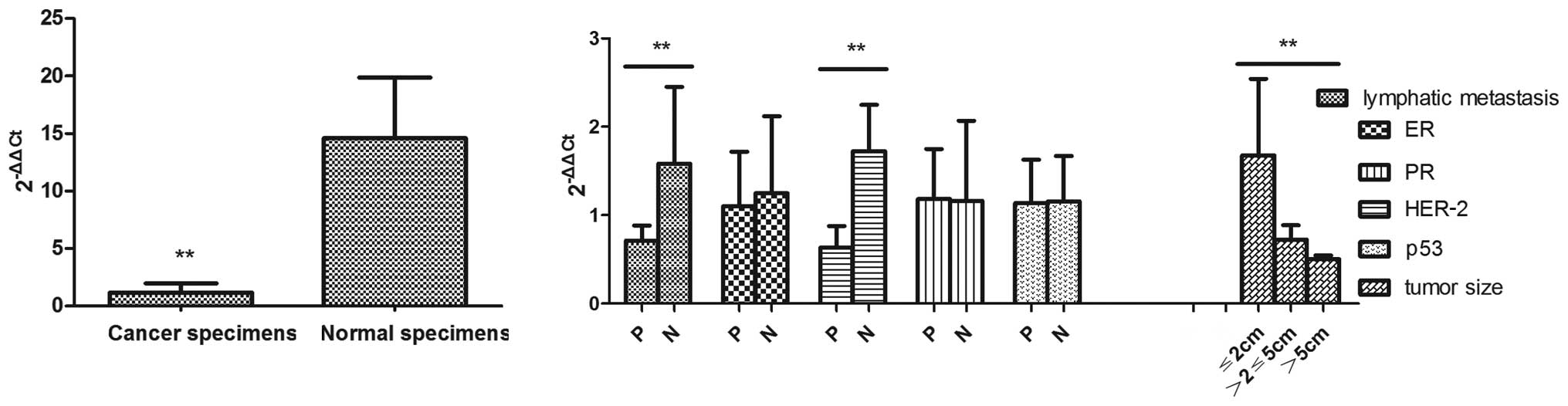
The RT-PCR data showed that miR-497 expression was
relatively reduced in the breast cancer specimens in comparison to
the normal tissues, the relative expression of which was
1.181±0.779 and 14.599±5.266 (P<0.01), respectively (Fig. 1A). Markedly, the abnormal
expression pattern of miR-497 was negatively correlated with
pathological stage, lymphatic metastasis, tumor size and human
epidermal growth factor receptor-2 (HER-2) (P<0.01). No
correlation was observed for miR-497 with estrogen receptor (ER),
progesterone receptor (PR) and p53 (P>0.05). No significant
expression alteration of miR-198, miR-373 and miR-1289 was found
between breast cancer and normal tissues (Fig. 1B).
Suppression of tumor proliferation by
miR-497
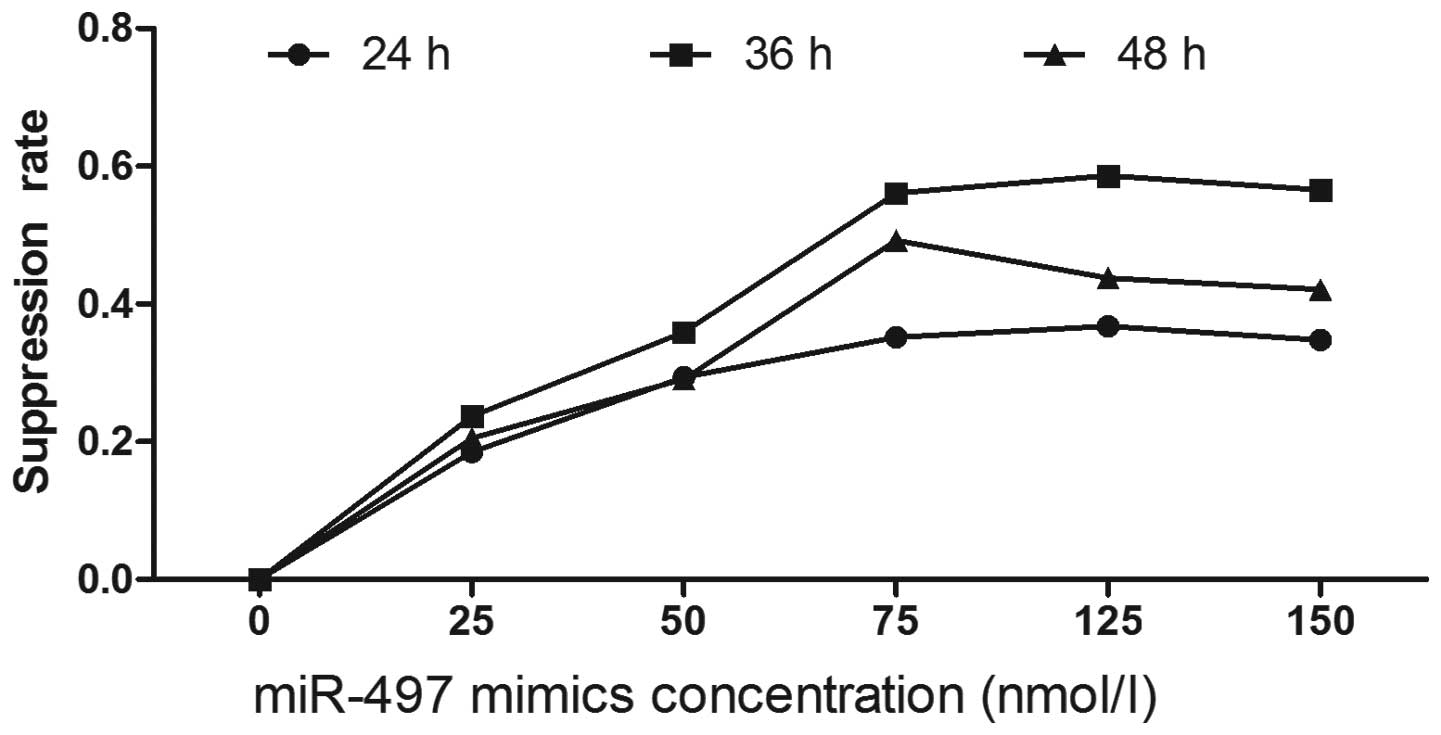
To explore the potential impact of miR-497 on the
proliferation of breast cancer cells, miR-497 mimics were used to
interfere with tumor cells at the concentrations of 25, 50, 75, 100
and 125 nmol/ l. Cellular viability and proliferation were measured
following the protocol of the MTT assay kit at 24, 36 and 48 h.
Compared to negative controls, miR-497 significantly repressed the
growth of breast cancer cells. Suppression of cell growth by
miR-497 was time- and dosage-dependent, and miR-497 at the
concentration of 100 nmol/l and at 36 h showed the greatest
inhibitory effect. As the concentration exceeded 100 nmol/ l, no
significant alteration of the inhibition rate was observed
(Fig. 2).
miR-497 induces the apoptosis of breast
cancer cells
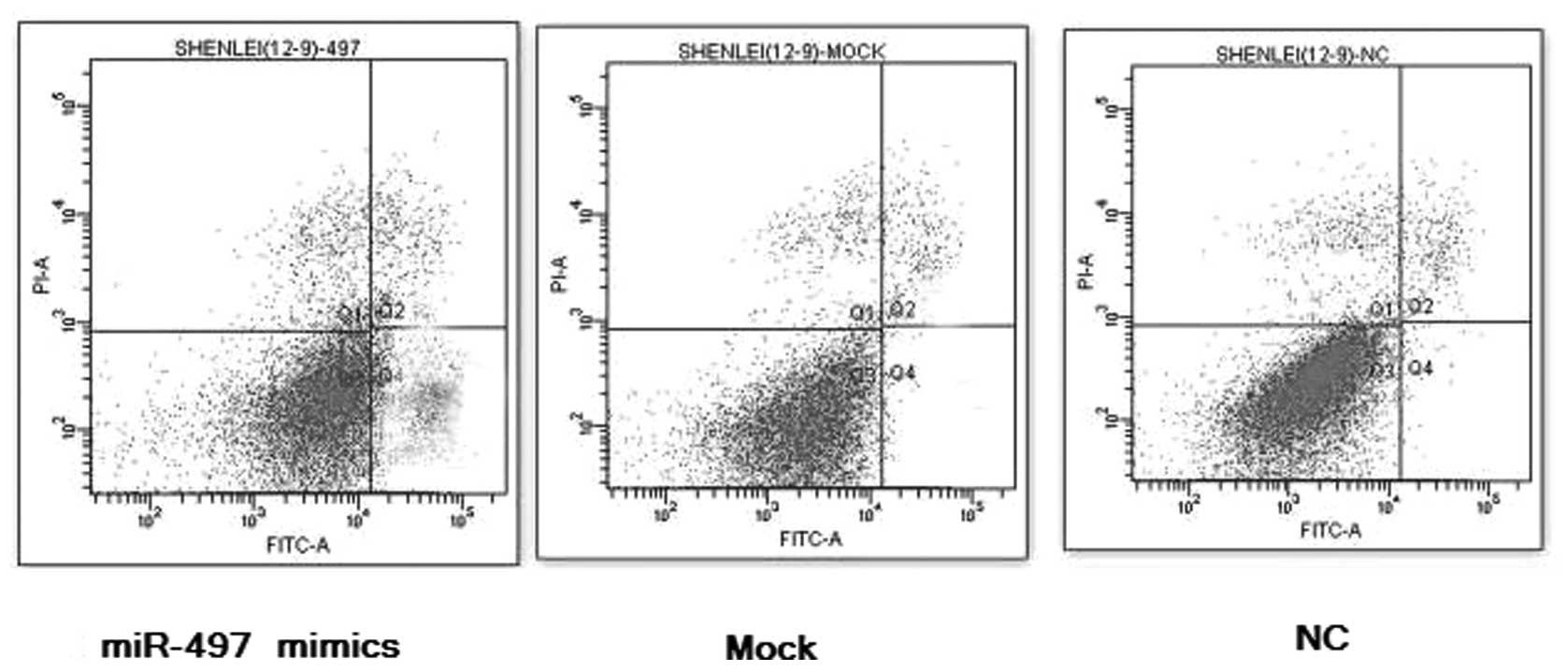
To examine whether miR-497 facilitates the apoptosis
of breast cancer cells, MCF-7 cancer cells were transfected with
100 nmol/l of miR-497 mimics for 36 h. Flow cytometry data
indicated that the elevated expression of miR-497 induced early
apoptosis, compared to the lipofect-treated controls and the
negative controls, and the percentage of early apoptotic cancer
cells of the miR-497 treatment groups was markedly increased, which
shows that miR-497 can act as an apoptosis inducer in breast cancer
in vitro, P<0.01, n=3 (Fig.
3).
miR-497 disrupts the cell cycle of breast
cancer cells
As thoroughly described previously, miR-497 is
capable of repressing the proliferation and promote the apoptosis
of breast cancer cells. After the transfection of miR-497 mimics at
the concentration of 100 nmol/l for 36 h, flow cytometry analysis
revealed that the percentage of G0/G1 phase cells (76.23±1.78%)
dramatically increased in the treatment groups, which was
statistically higher than those in the lipofect-treated groups and
negative controls (70.21±1.52%, 69.81±1.36%), and while the
proportion of S-phase cells decreased (12.79±0.91%), the percentage
of G2/M phase cells was not significantly altered. Furthermore, the
cellular proliferation index of the treatment groups dropped to
23.76±0.62, suggesting that miR-497 can initiate G0/G1 phase arrest
and that up-regulation of miR-497 expression leads to the reduction
of the S-phase cells as well as the proliferation index (Fig. 4).
miR-497 directly targets Bcl-w
To understand the molecular mechanisms by which
miR-497 inhibits tumor cell growth and induces cell apoptosis,
several members of the apoptotic pathway were selected, including
Bcl-w, p65, Bcl-2, Bcl-xL, caspase-3 and caspase-8 for further
determination of the possible downstream targets. Real-time PCR
analysis indicated that Bcl-w expression was significantly
different between the miR-497 mimics-transfected groups and
lipofect-treated and negative controls, the relative expression of
which were 1.004±0.109, 4.303±0.332 and 3.971±0.382, respectively,
while no significant alteration of p65, Bcl-2, Bcl-xL, caspase-3
and caspase-8 expression was observed (Fig. 5A). To validate the possibility that
miR-497 may target Bcl-w in breast cancer cells, we performed
Western blot analysis. In concordance with RT-PCR results, Bcl-w
protein expression was significantly decreased with the
overexpression of miR-497, suggesting that miR-497 suppresses Bcl-w
expression at the protein level. These data demonstrate that
miR-497 is capable of inhibiting the growth and facilitating the
apoptosis of breast cancer cells via targeting of the proapoptotic
gene, Bcl-w (Fig. 5B).
Discussion
The discovery of the first miRNA, lin-4, in
Caenorhabditis elegans symbolized a new era of miRNAs. Their
biogenesis, function and potential application have become active
areas of research, particularly in cancer. In this study, we
selected 4 miRNAs and employed RT-PCR to explore their expression
patterns in breast cancer. Compared to the normal breast tissues,
miR-497 expression was significantly down-regulated in the breast
cancer specimens. Markedly, the miR-497 expression pattern was
negatively correlated with pathological stage, lymphatic
metastasis, tumor size and HER-2, and no correlation was found
between miR-497 and ER, PR and p53. HER-2 is relatively
underexpressed in normal breast tissue and overexpressed in 20–30%
of breast cancer tissues. It has been shown that HER-2-positive
breast cancer is highly metastatic, proliferative and more likely
to relapse. In addition, lymphatic metastasis, higher pathological
stage and positive HER-2 are connected to worse prognosis,
suggesting that breast cancer patients with elevated expression of
miR-497 have better prognosis, and miR-497 may turn out to be a new
prognostic marker for breast cancer.
Functional assays have revealed that miR-497 greatly
inhibits the cellular growth, induces the apoptosis and disrupts
the cell cycle of breast cancer cells. Recently, there has been a
large body of evidence for the involvement of miRNAs in the
apoptosis of breast cancer. miR-9-3, down-regulated in breast
cancer, plays a role in the p53-related apoptotic pathway (24). miR-26b impairs the viability and
triggers the apoptosis of MCF7 human breast cancer cells by
targeting SLC7A11 (25). miR-145
exhibited a pro-apoptotic effect through a death-promoting loop
between miR-145 and TP53 (26).
The suppression of miR-21 by antisense oligonucleotides hindered
tumor cell growth both in vitro and in the xenograft mouse
model via the down-regulation of Bcl-2, which consequently
increased apoptosis and decreased cell proliferation (14).
Bcl-w, an anti-apoptotic member of the Bcl-2 family,
has been demonstrated to be closely associated to cancer formation
and progression (27). To date,
even though the mutation of Bcl-w has not been verified to be
causative of cancer formation, there is evidence that the elevated
expression of Bcl-w combined with other oncogenes contributes to
tumor occurrence. Target prediction obtained from the target scan
showed that the 3′-UTR of Bcl-w contained potential binding
sequences complementary to miR-497. There are 3 complementary
binding sites in the conserved region, and 1 in the non-conserved
region, providing a theoretical basis for the target prediction of
miR-497. Target validation by RT-PCR and Western blot analysis
further confirmed the prediction. Similar to this study, Lin et
al described that the up-regulation of miR-122 expression in
hepatic cancer led to the decrease of Bcl-w mRNA and protein
expression (28).
In conclusion, given the underexpression pattern of
miR-497 as well as its potential pro-apoptotic function, it may be
concluded that miR-497 acts as a tumor suppressor by targeting
Bcl-w in breast cancer. Therefore, the up-regulation of miR-497
artificially offers us a promising new direction for breast cancer
treatment in the future.
Acknowledgements
We give special thanks to all the
teachers at the Central Laboratory of Shanghai Tenth People’s
Hospital for their help and support.
References
|
1
|
WHO fact sheet No.297. World Health
Organization. Geneva, 2006
|
|
2
|
M ShiN GuoMicroRNA expression and its
implications for the diagnosis and therapeutic strategies of breast
cancerCancer Treat
Rev35328334200910.1016/j.ctrv.2008.12.00219171434
|
|
3
|
J Le QuesneC CaldasMicro-RNAs and breast
cancerMol Oncol4230241201020537965
|
|
4
|
MV IorioP CasaliniE TagliabueMicroRNA
profiling as a tool to understand prognosis, therapy response and
resistance in breast cancerEur J
Cancer4427532759200810.1016/j.ejca.2008.09.03719022662
|
|
5
|
S Griffiths-JonesAnnotating noncoding RNA
genesAnnu Rev Genomics Hum
Genet8279298200710.1146/annurev.genom.8.080706.092419
|
|
6
|
DP BartelMicroRNAs, genomics biogenesis
mechanism and functionCell116281297200414744438
|
|
7
|
K BrevingA Esquela-KerscherThe
complexities of microRNA regulation: mirandering around the
rulesInt J Biochem Cell
Biol4213161329201010.1016/j.biocel.2009.09.01619800023
|
|
8
|
RI GregoryR ShiekhattarMicroRNA biogenesis
and cancerCancer
Res6535093512200510.1158/0008-5472.CAN-05-029815867338
|
|
9
|
S VoliniaGA CalinCG LiuA microRNA
expression signature of human solid tumors defines cacer gene
targetsProc Natl Acad Sci
USA10322572261200610.1073/pnas.051056510316461460
|
|
10
|
MV IorioM FerracinCG LiuMicroRNA gene
expression deregulation in human breast cancerCancer
Res6570657070200510.1158/0008-5472.CAN-05-178316103053
|
|
11
|
LX YanXF HuangQ ShaoMicroRNA miR-21
overexpression in human breast cancer is associated with advanced
clinical stage, lymph node metastasis and patient poor
prognosisRNA1423482360200810.1261/rna.103480818812439
|
|
12
|
ZR YuCG WangM WangA cyclin D1/microRNA
17/20 regulatory feedback loop in control of breast cancer cell
proliferationJ Cell
Biol182509517200810.1083/jcb.20080107918695042
|
|
13
|
S WangC BianZ YangmiR-145 inhibits breast
cancer cell growth through RTKNInt J
Oncol3414611466200919360360
|
|
14
|
ML SiS ZhuH WumiR-21-mediated tumor
growthOncogene2627992803200710.1038/sj.onc.121008317072344
|
|
15
|
W KongLL HeM CoppolaMicroRNA-155 regulates
cell survival, growth, and chemosensitivity by targeting FOXO3a in
breast cancerJ Biol
Chem2851786917879201010.1074/jbc.M110.10105520371610
|
|
16
|
M SachdevaYY MoMicroRNA-145 suppresses
cell invasion and metastasis by directly targeting mucin 1Cancer
Res70378387201010.1158/0008-5472.CAN-09-202119996288
|
|
17
|
S ValastyanF ReinhardtN BenaichA
pleiotropically acting microRNA, miR-31, inhibits breast cancer
metastasisCell13710321046200910.1016/j.cell.2009.03.04719524507
|
|
18
|
HL WuSM ZhuYY MoSuppression of cell growth
and invasion by miR-205 in breast cancerCell
Res19439448200910.1038/cr.2009.1819238171
|
|
19
|
L MaJ Teruya-FeldsteinRA WeinbergTumour
invasion and metastasis initiated by microRNA-10b in breast
cancerNature449682688200710.1038/nature0617417898713
|
|
20
|
YZ PanME MorrisAM YuMicroRNA-328
negatively regulates the expression of breast cancer resistance
protein (BCRP/ABCG2) in human cancer cellsMol
Pharmacol7513741379200910.1124/mol.108.05416319270061
|
|
21
|
ZX LiangH WuJ XiaInvolvement of miR-326 in
chemotherapy resistance of breast cancer through modulating
expression of multidrug resistance-associated protein 1Biochem.
Pharmacol79817824201010.1016/j.bcp.2009.10.017
|
|
22
|
C ChenDA RidzonAJ BroomerReal-time
quantification of microRNAs by stem-loop RT-PCRNucleic Acids
Res33e179200510.1093/nar/gni17816314309
|
|
23
|
KJ LivakTD SchmittgenAnalysis of relative
gene expression data using real-time quantitative PCR and the
2−ΔΔCt
methodMethods25402408200110.1006/meth.2001.126211846609
|
|
24
|
PY HsuDE DeatherageBA
RodriguezXenoestrogen-induced epigenetic repression of microRNA-9-3
in breast epithelial cellsCancer
Res6959365945200910.1158/0008-5472.CAN-08-491419549897
|
|
25
|
XX LiuXJ LiB ZhangMicroRNA-26b is
underexpressed in human breast cancer and induces cell apoptosis by
targeting SLC7A11FEBS
Lett58513631367201110.1016/j.febslet.2011.04.01821510944
|
|
26
|
R SpizzoMS NicolosoL LupinimiR-145
participates with TP53 in a death-promoting regulatory loop and
targets estrogen receptor-alpha in human breast cancer cellsCell
Death Differ17246254201010.1038/cdd.2009.117
|
|
27
|
IH BaeMJ ParkSH YoonBcl-w promotes gastric
cancer cell invasion by inducing matrix metalloproteinase-2
expression via phosphoinositide 3-kinase, Akt, and Sp1Cancer
Res6649914995200610.1158/0008-5472.CAN-05-425416707418
|
|
28
|
CJ LinHY GongHC TsengMir-122 targets an
anti-apoptotic gene, Bcl-w, in human hepatocellular carcinoma cell
linesBiochem Biophys Res
Commun375315320200810.1016/j.bbrc.2008.07.15418692484
|