Introduction
Hepatocellular carcinoma is the third leading cause
of mortality from cancer, with an increasing incidence worldwide.
As a result, it is one of the most serious threats to health in the
global population (1).
Deregulation of oncogenes or tumor suppressors has been shown to be
closely associated with the development and progression of
hepatocellular carcinoma (2,3).
Accordingly, the development of effective therapeutic targets for
hepatocellular carcinoma is urgently required.
microRNAs (miRNAs) are non-coding RNAs formed of
18–25 nucleotides that can cause the inhibition of gene expression
at a post-transcriptional level by directly binding to the
3′-untranslational region (UTR) of mRNAs (4). Deregulation of miRNAs, such as
miR-204, miR-331, miR-125b, miR-148b and miR-148a, has been
demonstrated to play an important role in hepatocellular carcinoma
(5–9). Among these miRNAs, miR-148a has been
demonstrated to act as a tumor suppressor in several types of
cancer, including gastric, non-small cell lung and colorectal
cancer (10–12). For instance, the expression level
of miR-148a was reduced in gastric cancer tissues and cell lines,
and it could regulate various target genes and pathways involving
tumor proliferation, invasion and metastasis (10).
Deregulation of miR-148a has additionally been shown
to affect the poor prognosis of hepatocellular carcinoma,
associated with the overexpression of ubiquitin-specific protease
4, an identified target of miR-148a (9). miR-148a has also been found to
suppress the epithelial-mesenchymal transition and metastasis of
hepatocellular carcinoma cells by targeting Met/Snail signaling
(13,14). As one miRNA can directly bind to
numerous target mRNAs, whether other target genes exist in
hepatocellular carcinoma remains unclear.
The present study aimed to explore the role of
miR-148a in the regulation of hepatocellular carcinoma cell
invasion. Furthermore, it was determined whether
sphingosine-1-phosphate receptor 1 (S1PR1) acted as a downstream
effector of miR-148a in hepatocellular carcinoma cells.
Materials and methods
Tissue specimen collection
This study was approved by the Ethics Committee of
Shandong University (Jinan, China). Informed consent was obtained
from the patients. Twenty hepatocellular carcinoma tissues and
their matched adjacent normal tissues were collected in the
Department of General Surgery, Qilu Hospital of Shandong University
(Jinan, China). Tissue samples were immediately frozen in liquid
nitrogen following surgical removal.
Cell culture
Human hepatocellular carcinoma HepG2 cells were
obtained from the Cell Bank of Shandong University and cultured in
Dulbecco’s modified Eagle’s medium (Life Technologies, Carlsbad,
CA, USA) with 10% fetal bovine serum (FBS; Pierce Chemical,
Rockford, IL, USA) at 37°C in a humidified incubator containing 5%
CO2.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) assay
Total RNA was extracted using TRIzol®
reagent (Invitrogen Life Technologies, Carlsbad, CA, USA). An miRNA
Reverse Transcription kit (Invitrogen Life Technologies) was used
to convert RNA into cDNA, according to the manufacturer’s
instructions. RT-qPCR was then performed using an miRNA qPCR
Detection kit (GeneCopoeia, Rockville, MD, USA) on an ABI 7500
thermocycler (Applied Biosystems, Carlsbad, CA, USA). The PCR
conditions were set as follows: 94°C for 3 min; followed by 40
cycles of 94°C for 30 sec, 60°C for 15 sec and 72°C for 30 sec; and
a final step of 82°C for 5 sec. U6 gene was used as an internal
reference. The relative expression was analyzed by the
2−ΔΔCt method.
Western blotting
Tissues or cells were solubilized in cold
radioimmunoprecipitation assay lysis buffer. Proteins were
separated with 12% SDS-PAGE and transferred onto a polyvinylidene
difluoride (PVDF) membrane, which was then incubated with
Tris-buffered saline and Tween 20 (Sigma-Aldrich, St. Louis, MO,
USA) containing 5% milk at room temperature for 3 h. The PVDF
membrane was subsequently incubated with rabbit monoclonal
anti-S1PR1 (ab125074; 1:200 dilution) and -GAPDH (ab181602; 1:200
dilution) primary antibodies (Abcam, Cambridge, UK) at room
temperature for 3 h. Following washing three times with
phosphate-buffered saline-Tween 20, the membrane was incubated with
the goat anti-rabbit secondary antibodies (Abcam) at room
temperature for 40 min. Chemiluminescent detection was performed
using an enhanced chemiluminescence kit (Pierce Chemical). The
relative protein expression was analyzed by Image-Pro®
Plus software 6.0 (Media Cybernetics, Inc., Silver Spring, MD,
USA), and presented as the density ratio versus GAPDH.
Transfection
Transfection was performed using
Lipofectamine® 2000 (Invitrogen Life Technologies), in
accordance with the manufacturer’s instructions. For miR-148a
functional analysis, the HepG2 cells were transfected with the
scrambled miRNA as a negative control, miR-148a mimics or miR-148a
inhibitor (Invitrogen Life Technologies). For S1PR1 functional
analysis, the HepG2 cells were transfected with S1PR1-specific
small interfering (si)RNA or pcDNA3.1-S1PR1 plasmid (Nlunbio,
Changsha, China).
Dual-luciferase reporter assay
A QuikChange® Site-Directed Mutagenesis
kit (Stratagene, La Jolla, CA, USA) was used to generate a
mutant-type 3-UTR of S1PR1, according to the manufacturer’s
instructions. The wild- or mutant-type 3-UTR of S1PR1 was inserted
into the psiCHECK™-2 vector (Promega Corp., Madison, WI, USA). Once
HepG2 cells were cultured to ~70% confluence, they were transfected
with psiCHECK™-2-S1PR1-3-UTR or psiCHECK™-2-mutant S1PR1-3-UTR
vector, with or without 100 nM miR-148a mimics. Following
transfection for 48 h, the luciferase activities were determined on
an LD400 luminometer (Beckman Coulter, Fullerton, CA, USA). Renilla
luciferase activity was normalized to firefly luciferase
activity.
Invasion assay
A cell suspension containing 5×105
cells/ml was prepared in serum-free media; 300 μl was added into
the upper chamber and 500 μl RPMI-1640 containing 10% FBS was added
into the lower chamber. Following incubation for 24 h, non-invading
cells as well as the matrix gel (BD Biosciences, Franklin Lakes,
NJ, USA) on the interior of the inserts was removed using a
cotton-tipped swab. Invasive cells on the lower surface of the
membrane were stained with crystal violet (Sigma-Aldrich) for 20
min, rinsed with water and dried in air. Five fields were randomly
selected and the cell number was counted under an inverted
microscope (IX83; Olympus Corporation, Tokyo, Japan)
(magnification, ×100).
Statistical analysis
The results are expressed as the mean ± standard
deviation of three independent experiments. Statistical analysis of
differences was performed by one-way analysis of variance using
SPSS software (version 17; SPSS Inc., Chicago, IL, USA). P<0.05
was considered to indicate a statistically significant
difference.
Results
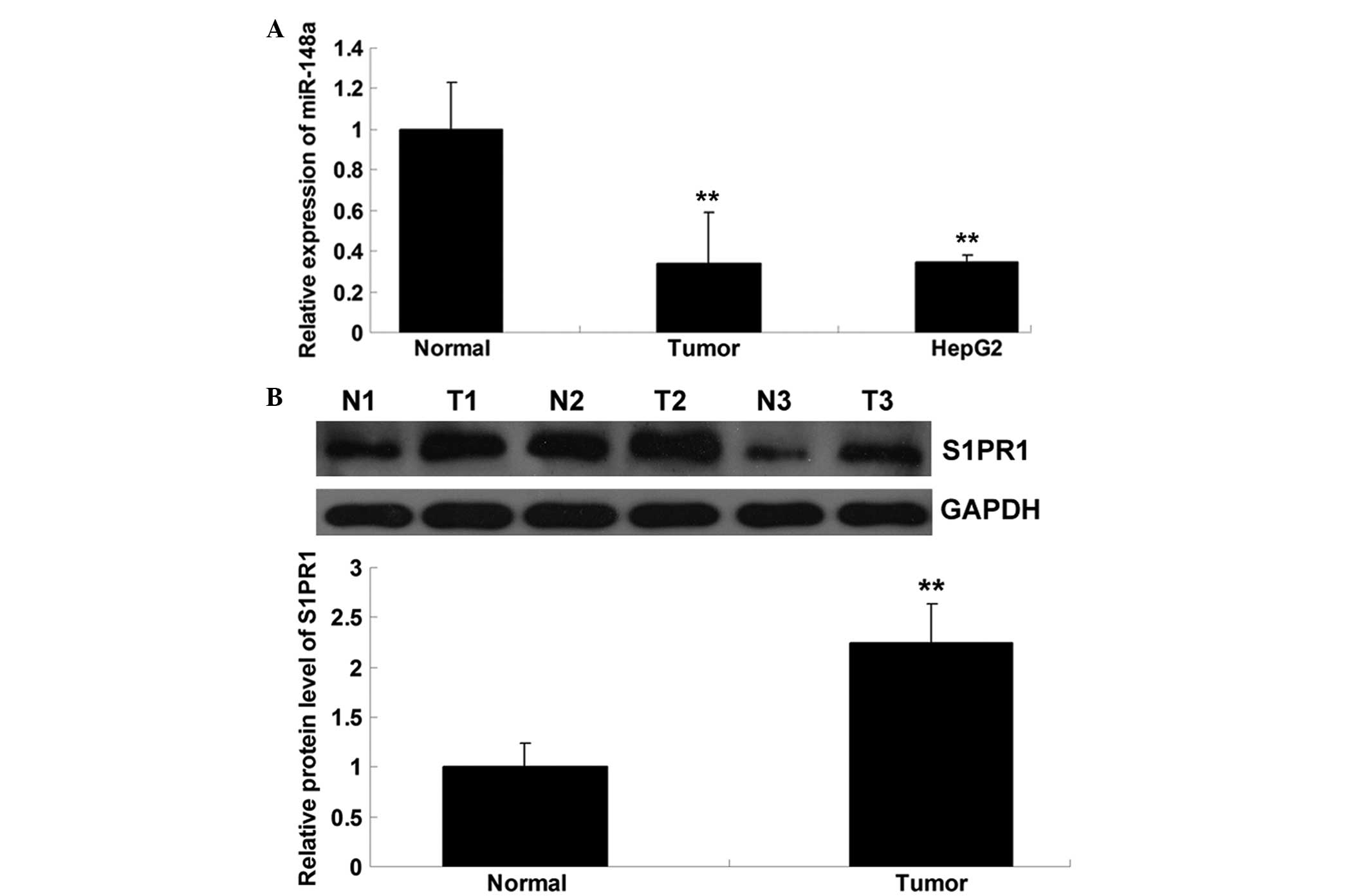
miR-148a is downregulated and S1PR1 is
upregulated in hepatocellular carcinoma tissues and cells
Firstly, the expression level of miR-148a in
hepatocellular carcinoma tissues, their matched adjacent normal
tissues and hepatocellular carcinoma HepG2 cells was examined. As
shown in Fig. 1A, the expression
level of miR-148a in the hepatocellular carcinoma tissues was
significantly reduced compared with that in the normal tissues.
Consistently, the expression of miR-148a was downregulated in the
hepatocellular carcinoma HepG2 cells. The protein expression of
S1PR1 was determined by performing western blotting, which showed
that the protein level of S1PR1 was upregulated in the
hepatocellular carcinoma tissues compared with that in their
matched normal adjacent tissues (Fig.
1B). Accordingly, the present data suggest that miR-148a is
downregulated whereas S1PR1 is upregulated in hepatocellular
carcinoma.
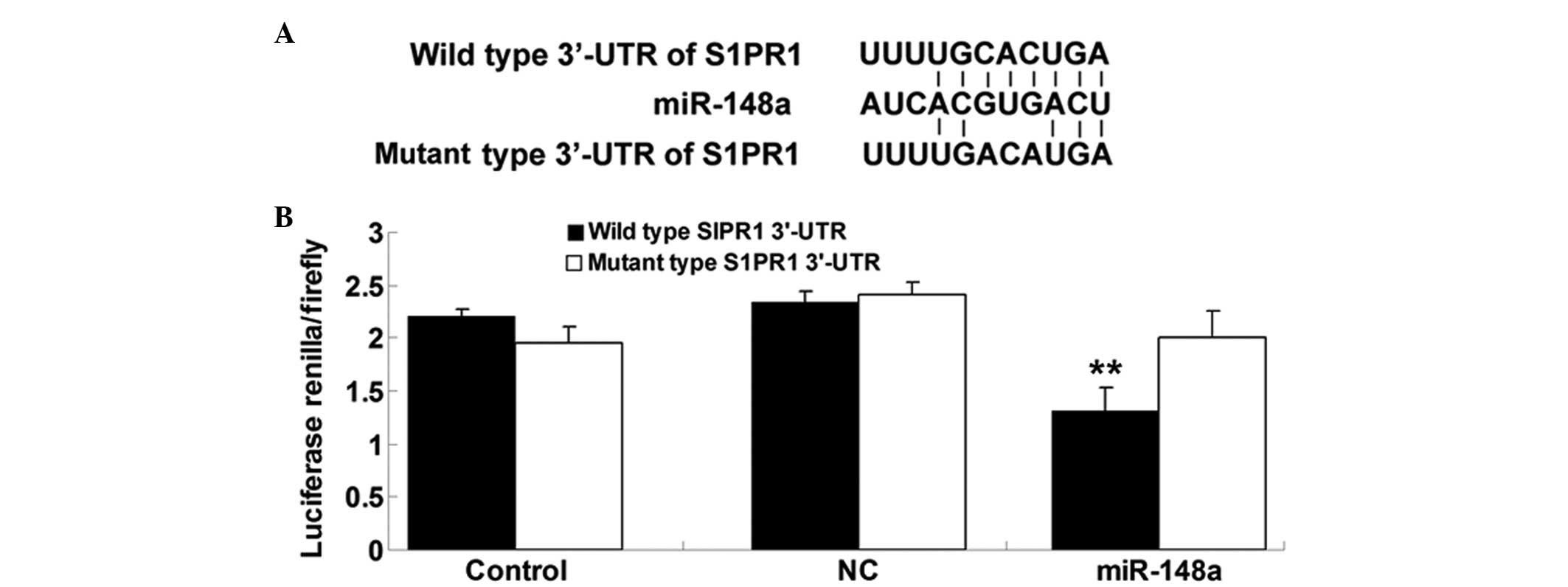
S1PR1 is a direct target of miR-148a
Bioinformatical predication was performed using
TargetScan online software (http://www.targetscan.org/) and the findings showed
that the putative seed sequences for miR-148a at the 3′-UTR of
S1PR1 were highly conserved (Fig.
2A). To verify whether S1PR1 was a direct target of miR-148a,
the wild and mutant types of S1PR1 3′-UTR were generated. The
dual-luciferase reporter assay was subsequently performed in
hepatocellular carcinoma HepG2 cells. As shown in Fig. 2B, the luciferase activity was
significantly reduced in HepG2 cells co-transfected with the
wild-type 3′-UTR of S1PR1 and miR-148a mimics, but unchanged in
HepG2 cells co-transfected with the mutant S1PR1 3 UTR and miR-148a
mimics, indicating that miR-148a directly binds to the 3′-UTR of
S1PR1 in HepG2 cells.
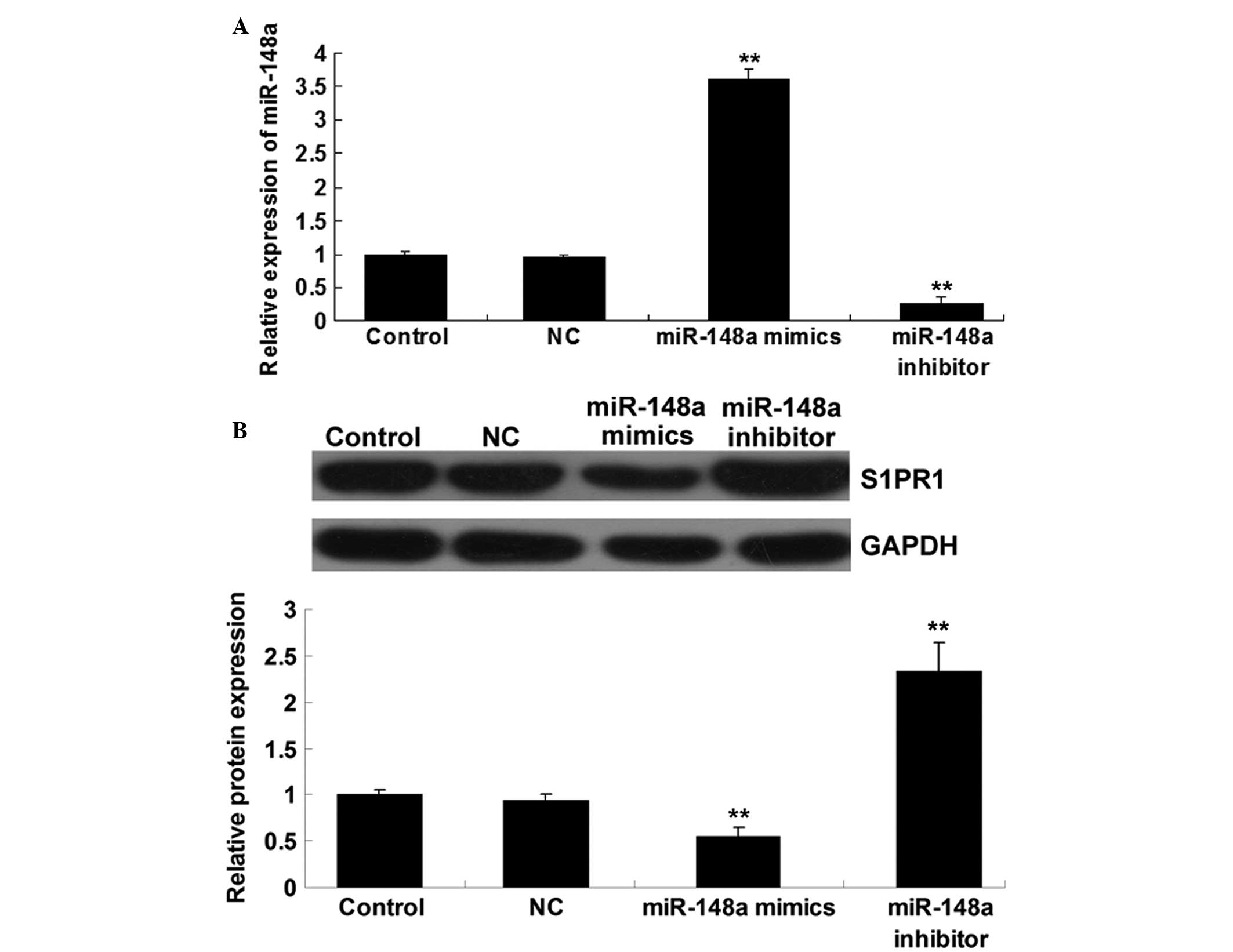
S1PR1 expression is negatively regulated
by miR-148a at a transcriptional level in hepatocellular carcinoma
cells
To further investigate the role of miR-148a in the
regulation of S1PR1 expression in hepatocellular carcinoma cells,
HepG2 cells were transfected with scrambled miRNA, miR-148a mimics
and miR-148a inhibitor, respectively. The transfection efficiency
was satisfactory (Fig. 3A). The
findings showed that the protein level of S1PR1 was significantly
reduced following the upregulation of miR-148a but was increased in
HepG2 cells transfected with miR-148a inhibitor (Fig. 3B). These findings indicate that
miR-148a plays a negative role in the regulation of S1PR1
expression at a post-transcriptional level in hepatocellular
carcinoma cells.
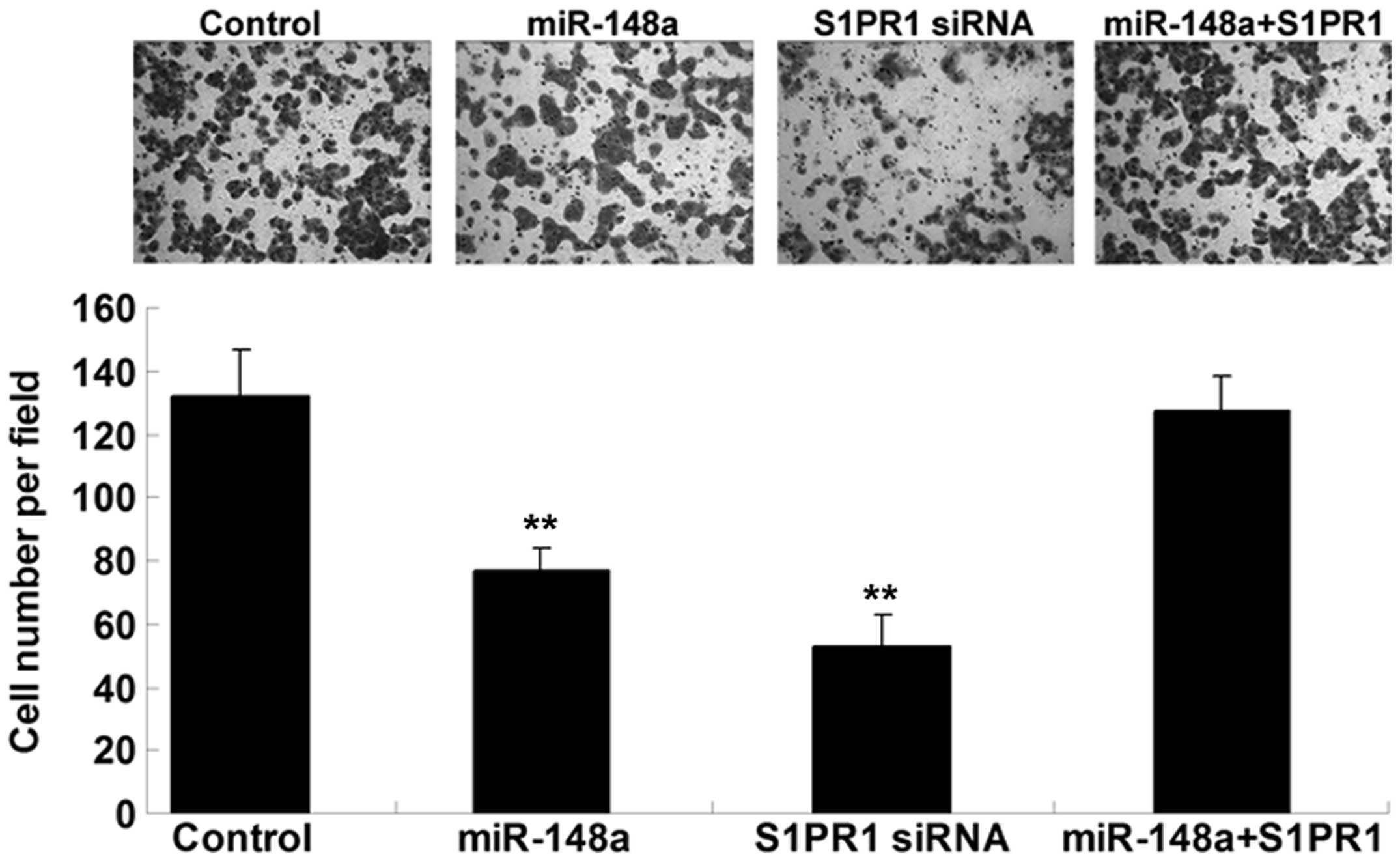
S1PR1 acts as a downstream effector in
the miR-148a-induced inhibition of hepatocellular carcinoma cell
invasion
The inhibition of S1PR1 expression in HepG2 cells
was used to determine whether miR-148a played a suppressive role in
hepatocellular carcinoma cell invasion. HepG2 cells were
transfected with miR-148a mimics or S1PR1-specific siRNA or
co-transfected with miR-148a mimics and S1PR1 plasmid. A cell
invasion assay was then performed. As shown in Fig. 4, the upregulation of miR-148a and
inhibition of S1PR1 notably inhibited HepG2 cell invasion. In
addition, the suppressive effect of miR-148a upregulation on cell
invasion was reversed by S1PR1 overexpression. Based on these
findings we suggest that S1PR1 acts as a downstream effector in the
miR-148a-induced inhibition of hepatocellular carcinoma cell
invasion.
Discussion
In the present study, the expression level of
miR-148a was shown to be notably reduced in hepatocellular
carcinoma tissues and cells compared with that in normal tissues;
however, the protein expression of S1PR1 was markedly upregulated.
Further investigation identified S1PR1 as a direct target of
miR-148a, and the protein expression of S1PR1 was negatively
regulated by miR-148a in hepatocellular carcinoma cells. In
addition, the current study suggested that the suppressive effect
of miR-148a on hepatocellular carcinoma cell invasion is, at least
partly, via the inhibition of S1PR1.
It has been well established that miRNAs can
regulate various biological processes by modulating the expression
of their targets at a post-transcriptional level
(15). In addition, deregulation of miRNAs has been
demonstrated to be associated with the development and progression
of various human malignancies (16). In the present
study, the expression level of miR-148a was shown to be notably
reduced in hepatocellular carcinoma tissues and cells compared with
that in normal tissues. These findings were consistent with others
(17). Magrelli et al
(17) performed a microarray and
RT-qPCR to examine the miRNA expression in hepatoblastoma tissues.
It was found that miR-148a was the only downregulated miRNA in
hepatoblastoma tissues (17).
Following the study by Magrelli et al (17), Yuan et al (18) suggested that miR-148a may be
associated with hepatitis-B-virus-associated hepatocellular
carcinoma. According to these and the current findings, we suggest
that deregulation of miR-148a is involved in the development and
progression of hepatocellular carcinoma; however, the detailed role
of miR-148a in hepatocellular carcinoma, particularly the molecular
regulatory mechanism, remains to be fully elucidated.
Recently, Gailhouste et al (14) showed that miR-148a could promote
the hepatospecific phenotype, and acted as a tumor suppressor by
targeting the c-Met oncogene. It was found that overexpression of
miR-148a led to a notable inhibition of the invasive properties of
hepatocellular carcinoma cells, whereas silencing of miR-148a
promoted hepatocellular carcinoma cell invasion (14). In the present study, it was also
found that miR-148a upregulation had an inhibitory effect on
hepatocellular carcinoma HepG2 cell invasion. Furthermore, it was
shown that S1PR1 was an important downstream effector of the
miR-148a-induced inhibition of cell invasion in hepatocellular
carcinoma HepG2 cells. Gailhouste et al (14) demonstrated that miR-148a exerted
its tumor-suppressive effect by directly targeting the c-Met
oncogene.
Bioactive sphingolipids, including sphingosine
kinases (SKs) and their product S1P, have been demonstrated to be
involved in the regulation of cancer growth, metastasis and drug
resistance (19). It has been well
established that S1P exerts its intracellular and extracellular
pro-survival and drug resistance functions through S1PR1 (20,21);
therefore, SK/S1P/S1PR1 signaling appears to be a promising
therapeutic target for cancer. In the present study, it was shown
that S1PR1 was significantly upregulated in hepatocellular
carcinoma tissues, consistent with a previous study (9). The role of S1PR1 in the regulation of
cancer cell invasion has also been previously demonstrated. For
instance, S1P/S1PR1 signaling has been shown to promote cell
migration and invasion via the activation of stat3 in prostate
cancer cells (22). In addition,
S1P/S1P1 signaling has been found to mediate Wilms tumor cell
migration and invasion (23). In
the present study, S1PR1 was found to be involved in the
miR-148a-mediated inhibition of hepatocellular carcinoma invasion.
Based on previous and the current findings, we suggest that S1PR1
could become an effective target for the prevention of
hepatocellular carcinoma metastasis.
In conclusion, the present study identified S1PR1 as
a direct target of miR-148a in hepatocellular carcinoma cells, and
suggested that miR-148a plays a suppressive role in the regulation
of hepatocellular carcinoma cell invasion, at least partially
through the direct downregulation of S1PR1 expression. miR-148a
may, therefore, serve as a potential therapeutic agent for
hepatocellular carcinoma.
References
|
1
|
Hung CH, Chiu YC, Chen CH and Hu TH:
MicroRNAs in hepatocellular carcinoma: carcinogenesis, progression,
and therapeutic target. Biomed Res Int. 2014:4864072014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Callegari E, Elamin BK, Sabbioni S,
Gramantieri L and Negrini M: Role of microRNAs in hepatocellular
carcinoma: a clinical perspective. Onco Targets Ther. 6:1167–1178.
2013.PubMed/NCBI
|
|
3
|
Khare S, Zhang Q and Ibdah JA: Epigenetics
of hepatocellular carcinoma: role of microRNA. World J
Gastroenterol. 19:5439–5445. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huang X, Liang M, Dittmar R and Wang L:
Extracellular microRNAs in urologic malignancies: chances and
challenges. Int J Mol Sci. 14:14785–14799. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cui ZH, Shen SQ, Chen ZB and Hu C: Growth
inhibition of hepatocellular carcinoma tumor endothelial cells by
miR-204-3p and underlying mechanism. World J Gastroenterol.
20:5493–5504. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chang RM, Yang H, Fang F, Xu JF and Yang
LY: MicroRNA-331-3p promotes proliferation and metastasis of
hepatocellular carcinoma by targeting PH domain and leucine-rich
repeat protein phosphatase. Hepatology. 60:1251–1263. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tsang FH, Au V, Lu WJ, et al: Prognostic
marker microRNA-125b inhibits tumorigenic properties of
hepatocellular carcinoma cells via suppressing tumorigenic molecule
eIF5A2. Dig Dis Sci. 59:2477–2487. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang Z, Zheng W and Hai J: MicroRNA-148b
expression is decreased in hepatocellular carcinoma and associated
with prognosis. Med Oncol. 31:9842014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Heo MJ, Kim YM, Koo JH, et al:
microRNA-148a dysregulation discriminates poor prognosis of
hepatocellular carcinoma in association with USP4 overexpression.
Oncotarget. 5:2792–2806. 2014.PubMed/NCBI
|
|
10
|
Xia J, Guo X, Yan J and Deng K: The role
of miR-148a in gastric cancer. J Cancer Res Clin Oncol.
140:1451–1456. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li J, Song Y, Wang Y, Luo J and Yu W:
MicroRNA-148a suppresses epithelial-to-mesenchymal transition by
targeting ROCK1 in non-small cell lung cancer cells. Mol Cell
Biochem. 380:277–282. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Takahashi M, Cuatrecasas M, Balaguer F, et
al: The clinical significance of MiR-148a as a predictive biomarker
in patients with advanced colorectal cancer. PLoS One.
7:e466842012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang JP, Zeng C, Xu L, Gong J, Fang JH
and Zhuang SM: MicroRNA-148a suppresses the epithelial-mesenchymal
transition and metastasis of hepatoma cells by targeting Met/Snail
signaling. Oncogene. 33:4069–4076. 2014. View Article : Google Scholar
|
|
14
|
Gailhouste L, Gomez-Santos L, Hagiwara K,
et al: miR-148a plays a pivotal role in the liver by promoting the
hepatospecific phenotype and suppressing the invasiveness of
transformed cells. Hepatology. 58:1153–1165. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Magrelli A, Azzalin G, Salvatore M, et al:
Altered microRNA expression patterns in hepatoblastoma patients.
Transl Oncol. 2:157–163. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yuan K, Lian Z, Sun B, Clayton MM, Ng IO
and Feitelson MA: Role of miR-148a in hepatitis B associated
hepatocellular carcinoma. PLoS One. 7:e353312012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Selvam SP and Ogretmen B: Sphingosine
kinase/sphingosine 1-phosphate signaling in cancer therapeutics and
drug resistance. Handb Exp Pharmacol. 3–27. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cuvillier O: Sphingosine 1-phosphate
receptors: from biology to physiopathology. Med Sci (Paris).
28:951–957. 2012.(In French). View Article : Google Scholar
|
|
21
|
Pyne NJ, Tonelli F, Lim KG, Long JS,
Edwards J and Pyne S: Sphingosine 1-phosphate signalling in cancer.
Biochem Soc Trans. 40:94–100. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sekine Y, Suzuki K and Remaley AT: HDL and
sphingosine-1-phosphate activate stat3 in prostate cancer DU145
cells via ERK1/2 and S1P receptors, and promote cell migration and
invasion. Prostate. 71:690–699. 2011. View Article : Google Scholar
|
|
23
|
Li MH, Sanchez T, Yamase H, et al:
S1P/S1P1 signaling stimulates cell migration and invasion in Wilms
tumor. Cancer Lett. 276:171–179. 2009. View Article : Google Scholar : PubMed/NCBI
|