Alzheimer's disease (AD) is a common
neurodegenerative disorder that is the main cause of dementia. The
clinical presentation of AD is characterized by progressive memory
disorder and cognitive dysfunction (1). The worldwide prevalence of AD was 26.6
million in 2006, and this number is predicted to quadruple by 2050.
The rapidly increased AD incidence is likely lead to a significant
burden on family and society (2).
AD is a complex disease involving the interaction of
genetic and environmental factors. It has been shown that AD
development is contributed to by several elements, such as senile
plaques, neurofibrillary tangles (NFTs), abnormally aggregated
‘reactive’ proteins like β-amyloid (Aβ) and tau, exposure to
aluminum and brain inflammation (3).
A genome-wide association study revealed that multiple mutations in
candidate genes greatly increase the chance of developing AD
(4). Genes such as brain-derived
neurotrophic factor (BDNF) and butyrylcholinesterase
(BCHE) are believed to play a significant role in AD
progression (5–7).
BDNF is a member of the neurotrophic factor family
and is encoded by a gene located on chromosome 11p13 (8). A previous study demonstrated that the
levels of BDNF and its receptor, tyrosine receptor kinase B, were
decreased in the frontal cortex and hippocampus of patients with AD
(9). BDNF is known to protect
against the neurotoxicity of the Aβ peptide and neural cell death
by the aggregation of Aβ and tau proteins (10,11).
Several single nucleotide polymorphisms (SNPs), such as Val66Met
and C270T (rs2030324), in BDNF have been reported to be
associated with AD (12–19).
Inconsistent results have been reported in the
previous studies on the association of BDNF and BCHE
polymorphisms with AD. For BDNF Val66Met, there have been
five positive results in Europeans (12–15) and
Asians (16), and 21 negative
results (among 18 studies) in Europeans (26–37),
Asians (19,38–42) and
Africans (29). For BDNF
C270T, there have been four positive results in Europeans (13,17) and
Asians (18,19) and 14 negative results in Europeans
(28–31,34–37,43–45),
Asians (40,41) and Africans (29). For BCHE-K variants, there have
been six positive results in Europeans (46–51) and
22 negative results in Europeans (5,52–67) and
Asians (68–72). Discrepancies among the previous
association studies may have been the result of limited power,
different ethnic backgrounds or the different processes and status
in patients with AD. Meta-analysis can strengthen the power by
combining data from different studies and can draw a more
comprehensive conclusion by analyzing studies in different
ethnicities (73–75). The aim of the current meta-analysis
was to assess the association between the three polymorphisms and
AD.
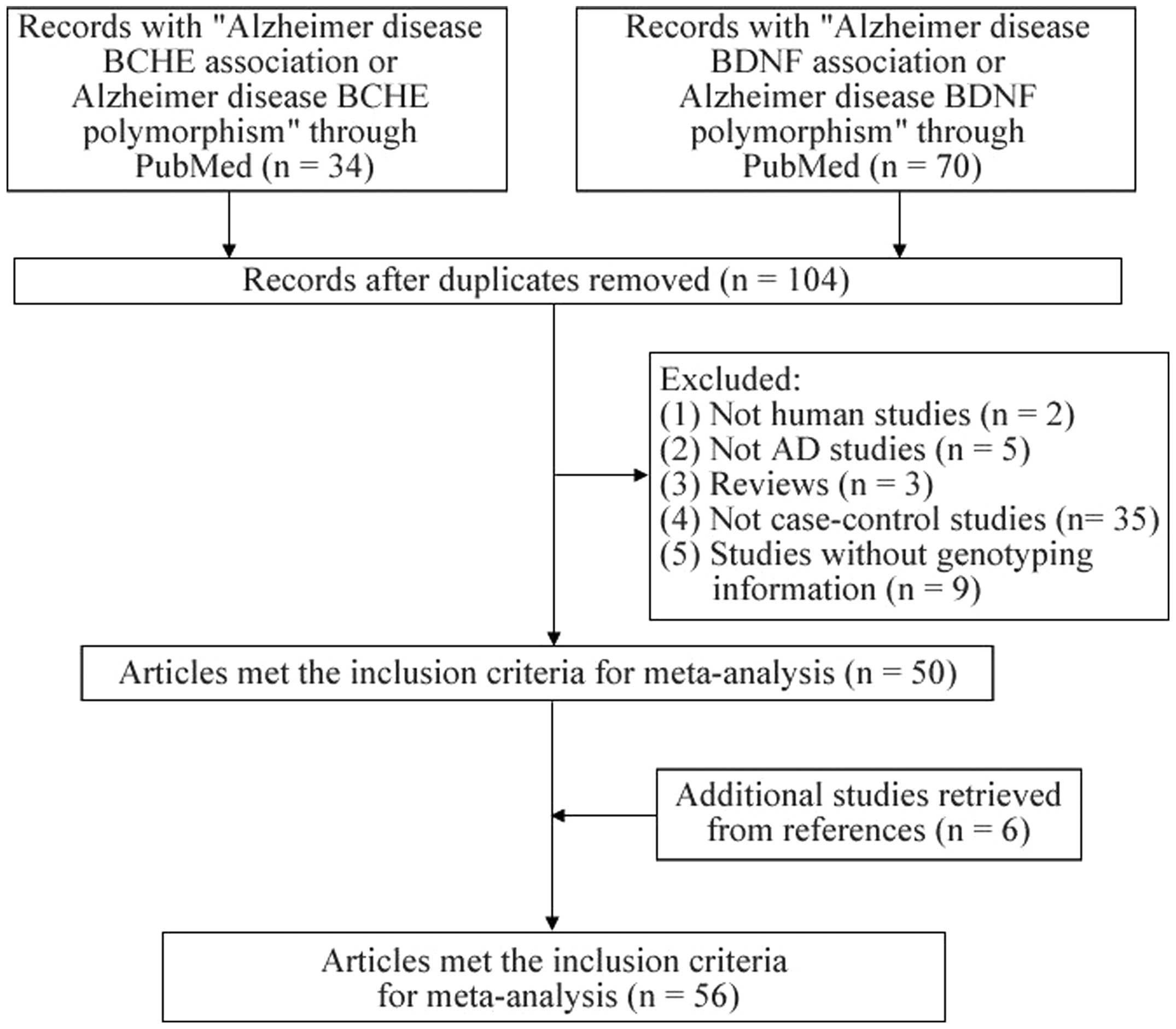
Articles were retrieved in January 2014 by searching
PubMed without time or language restrictions. The following
keywords were used: ‘Alzheimer disease BCHE association or
Alzheimer disease BCHE polymorphism’ and ‘Alzheimer disease BDNF
association or Alzheimer disease BDNF polymorphism’. The current
meta-analysis included studies that met the following criteria: i)
An original case-control study assessing the association of
BDNF and BDHE with AD in humans; ii) a study
containing sufficient information for the odds ratios (ORs) and 95%
confidence intervals (CIs) to be obtained; iii) a study in which
the genotype distribution of each polymorphism in controls met the
Hardy-Weinberg equilibrium (HWE); iv) a study in the cumulative
number of stages for one genetic locus was at least three. From
each study, the following data were extracted or calculated: First
author, publication year, country, ethnicity, number of cases and
controls, HWE for controls, reported association results and the
power of each study.
A total of 23 articles with 26 stages involving
6,504 patients with AD and 6,636 controls were included for the
meta-analysis of BDNF Val66Met. Significant statistical
heterogeneity was found at the allelic level (I2=58%),
under the dominant model (I2=56%) and the additive model
(I2=53%). The major allele frequency of BDNF
Val66Met was 0.805 in Europeans [International Haplotype Mapping
Project panel derived from Utah residents with Northern and Western
European ancestry (HapMap-CEU)], higher than the frequency in
Asians [HapMap-Han Chinese in Beijing, China (HCB), 0.733;
HapMap-Japanese in Tokyo Japan (JPT), 0.682] and Africans
[HapMap-Yoruba in Ibadan, Nigeria (YRI), 0.004]. The ethnic
differences for BDNF Val66Met were low between different
populations [Fixation index (Fst)=0.1006]; therefore,
the meta-analysis was also performed by ethnicity. No significant
association was found in the meta-analysis on allelic analysis
(P=0.99; OR, 1.00; 95% CI, 0.91–1.10; Table II) or under the other models for
combined and stratified populations (P>0.05, Table II).
A total of 17 articles with 18 stages involving
4,216 patients with AD and 4,370 controls were included for the
meta-analysis of the BDNF C270T polymorphism. Significant
heterogeneity was observed in the meta-analysis at the allelic
level (I2=63%) and under the dominant model
(I2=62%). The frequency of the BDNF C270T allele
was 0.667 in Chinese subjects (HapMap-HCB), higher than that in the
Japanese group (HapMap-JPT, 0.455), Europeans (HapMap-CEU, 0.570)
and Africans (HapMap-YRI, 0.550). Further analysis showed a low
ethnic difference for BDNF C270T (Fst=0.0230). No
significant association between BDNF C270T and AD was
observed at the allelic level (P=0.30; OR, 1.12; 95% CI, 0.91–1.37;
Table II). A further subgroup
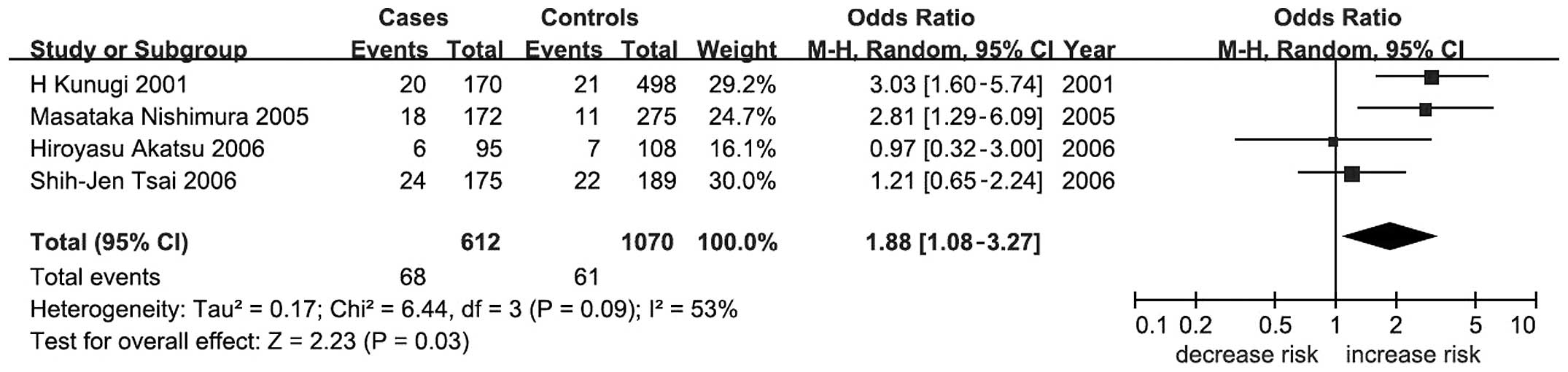
meta-analysis by ethnicity showed a significant association between
BDNF C270T and AD in Asians under a dominant model (P=0.03;
OR, 1.88; 95% CI, 1.08–3.27; Table
II and Fig. 2).
A total of 28 articles with 4,894 patients with AD
and 4,367 controls were included for the meta-analysis of the
BCHE-K variant. Although minimal ethnic differences were
found in Europeans and Asians (Fst=0.0009), significant
heterogeneity was found in the meta-analysis on the allelic level
(I2=56%) and under the dominant model
(I2=65%); however, no significant association was found
at the allelic level (P=0.31; OR, 1.07; 95% CI, 0.94–1.21; Table II). Subgroup meta-analysis also did
not yield any significant results (P>0.05, Table II).
The power analyses in this meta-analysis were
calculated under a moderate risk of AD (OR, 1.2; Tables I and II). The power was 1.000 for BDNF
Val66Met, 0.986 for BDNF C270T and 1.000 for the
BCHE-K variant (Table II),
which was considerably higher than that in each of the individual
studies (Table I). No publication
bias was found in the meta-analyses of the three SNPs (Fig. 3).
In the present study, 56 studies (72 stages) among
12,563 cases and 12,622 controls were analyzed to assess the
association of the BDNF Val66Met, BDNF C270T and
BCHE-K variants with AD. The results showed a moderate
association between BDNF C270T and AD in Asians (P=0.03; OR,
1.88; 95% CI, 1.08–3.27; Table II
and Fig. 2) but no significant
associations were observed in the other meta-analyses.
There were certain limitations in the meta-analysis.
Firstly, publication bias may exist, as the negative-result studies
are less likely to be published and may be missed, which may
influence the results. Secondly, the majority of studies
investigating the association between the three polymorphisms and
AD were carried out in the European and Asian populations. The
number of studies in other populations, such as Africans, was
limited. Future studies in other ethnic populations are warranted.
Thirdly, AD is a complex disease. Different statuses in AD may
affect the results of the study; however, no detailed information
of the AD diagnostic criteria was available from previous studies.
Future case-control studies with more comprehensive information are
required. Fourthly, there are 5,724 polymorphisms in BDNF
and 5,059 polymorphisms in BCHE. The current study only
focused on two polymorphisms of BDNF and one polymorphism of
BCHE, which may not fully show the function of those two
genes. Studies investigating a wider range of polymorphisms are
required to improve the representation of the two genes. Fifthly,
APOE is a known pivotal gene in the AD pathogenesis but no
APOE genotype was included in any of the studies. Thus, any
hidden interaction of the APOE genotype with the tested
three polymorphisms may have been missed in the current
meta-analysis. Sixthly, although a moderate association of the
BDNF C270T polymorphism with the risk of AD was observed in
Asians under a dominant model (P=0.03; OR, 1.88; 95% CI,
1.08–3.27), the significance was not retained following correction
by multiple testing. This result should therefore be taken with
caution. Finally, there was high heterogeneity in the BDNF
C270T variant under the dominant model in Asians (P=0.03,
I2=53%, Table II). We
speculated that the number of participants was the source of the
heterogeneity, as the studies with limited samples tended to
produce negative results (n<200, P>0.05) in contrast to
significant results produced in the studies with sufficient samples
(n>200, P<0.05).
In conclusion, the present comprehensive
meta-analysis suggested a moderate association between the
BDNF C270T polymorphism and AD in Asians under the dominant
model. Further studies focusing on a wider range of ethnic
populations are required to confirm the results of the study.
The study was supported by grants from the National
Natural Science Foundation of China (nos. 31100919 and 81371469),
the 973 program from the Ministry of Science and Technology of
China (no. 2013CB835100), the Natural Science Foundation of
Zhejiang Province (no. LR13H020003), the Disciplinary Project of
Ningbo University (no. B01350104900), the KC Wong Magna Fund in
Ningbo University, the Program for Professor of Special Appointment
(Eastern Scholars) at Shanghai Institutions of Higher Learning and
the Key Basic Research Foundation of Science and Technology
Commission of Shanghai Municipality (no. 13JC1403700).
|
1
|
Mucke L: Neuroscience: Alzheimer's
disease. Nature. 461:895–897. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brookmeyer R, Johnson E, Ziegler-Graham K
and Arrighi HM: Forecasting the global burden of Alzheimer's
disease. Alzheimers Dement. 3:186–191. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Armstrong RA: What causes Alzheimer's
disease. Folia Neuropathol. 51:169–188. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bertram L and Tanzi RE: Genome-wide
association studies in Alzheimer's disease. Hum Mol Genet.
18:R137–R145. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Scacchi R, Gambina G, Moretto G and Corbo
RM: Variability of AChE, BChE, and ChAT genes in the late-onset
form of Alzheimer's disease and relationships with response to
treatment with Donepezil and Rivastigmine. Am J Med Genet B
Neuropsychiatr Genet. 150B:502–507. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Borroni B, Costanzi C and Padovani A:
Genetic susceptibility to behavioural and psychological symptoms in
Alzheimer disease. Curr Alzheimer Res. 7:158–164. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Serretti A, Olgiati P and De Ronchi D:
Genetics of Alzheimer's disease. A rapidly evolving field. J
Alzheimers Dis. 12:73–92. 2007.PubMed/NCBI
|
|
8
|
Maisonpierre PC, Le Beau MM, Espinosa R
III, Ip NY, Belluscio L, de la Monte SM, Squinto S, Furth ME and
Yancopoulos GD: Human and rat brain-derived neurotrophic factor and
neurotrophin-3: Gene structures, distributions, and chromosomal
localizations. Genomics. 10:558–568. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ferrer I, Marín C, Rey MJ, Ribalta T,
Goutan E, Blanco R, Tolosa E and Martí E: BDNF and full-length and
truncated TrkB expression in Alzheimer disease. Implications in
therapeutic strategies. J Neuropathol Exp Neurol. 58:729–739. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Aicardi G, Argilli E, Cappello S, Santi S,
Riccio M, Thoenen H and Canossa M: Induction of long-term
potentiation and depression is reflected by corresponding changes
in secretion of endogenous brain-derived neurotrophic factor. Proc
Natl Acad Sci USA. 101:15788–15792. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Poo MM: Neurotrophins as synaptic
modulators. Nat Rev Neurosci. 2:24–32. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ventriglia M, Bocchio Chiavetto L, Benussi
L, Binetti G, Zanetti O, Riva MA and Gennarelli M: Association
between the BDNF 196 A/G polymorphism and sporadic Alzheimer's
disease. Mol Psychiatry. 7:136–137. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Saarela MS, Lehtimaki T, Rinne JO, Huhtala
H, Rontu R, Hervonen A, Roytta M, Ahonen JP and Mattila KM: No
association between the brain-derived neurotrophic factor 196
G>;A or 270 C>T polymorphisms and Alzheimer's or Parkinson's
disease. Folia Neuropathol. 44:12–16. 2006.PubMed/NCBI
|
|
14
|
Fehér A, Juhász A, Rimanóczy A, Kálmán J
and Janka Z: Association between BDNF Val66Met polymorphism and
Alzheimer disease, dementia with Lewy bodies, and Pick disease.
Alzheimer Dis Assoc Disord. 23:224–228. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pivac N, Nikolac M, Nedic G, et al: Brain
derived neurotrophic factor Val66Met polymorphism and psychotic
symptoms in Alzheimer's disease. Prog Neuropsychopharmacol Biol
Psychiatry. 35:356–362. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Matsushita S, Arai H, Matsui T, Yuzuriha
T, Urakami K, Masaki T and Higuchi S: Brain-derived neurotrophic
factor gene polymorphisms and Alzheimer's disease. J Neural Transm.
112:703–711. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Olin D, MacMurray J and Comings DE: Risk
of late-onset Alzheimer's disease associated with BDNF C270T
polymorphism. Neurosci Lett. 381:275–278. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kunugi H, Ueki A, Otsuka M, Isse K,
Hirasawa H, Kato N, Nabika T, Kobayashi S and Nanko S: A novel
polymorphism of the brain-derived neurotrophic factor (BDNF) gene
associated with late-onset Alzheimer's disease. Mol Psychiatry.
6:83–86. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nishimura M, Kuno S, Kaji R and Kawakami
H: Brain-derived neurotrophic factor gene polymorphisms in Japanese
patients with sporadic Alzheimer's disease, Parkinson's disease,
and multiple system atrophy. Mov Disord. 20:1031–1033. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Allderdice PW, Gardner HA, Galutira D,
Lockridge O, LaDu BN and McAlpine PJ: The cloned
butyrylcholinesterase (BCHE) gene maps to a single chromosome site,
3q26. Genomics. 11:452–454. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Arpagaus M, Kott M, Vatsis KP, Bartels CF,
La Du BN and Lockridge O: Structure of the gene for human
butyrylcholinesterase. Evidence for a single copy. Biochemistry.
29:124–131. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Perry E, McKeith I and Ballard C:
Butyrylcholinesterase and progression of cognitive deficits in
dementia with Lewy bodies. Neurology. 60:1852–1853. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Arendt T, Brückner MK, Lange M and Bigl V:
Changes in acetylcholinesterase and butyrylcholinesterase in
Alzheimer's disease resemble embryonic development - a study of
molecular forms. Neurochem Int. 21:381–396. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Carson KA, Geula C and Mesulam MM:
Electron microscopic localization of cholinesterase activity in
Alzheimer brain tissue. Brain Res. 540:204–208. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Darvesh S, Cash MK, Reid GA, Martin E,
Mitnitski A and Geula C: Butyrylcholinesterase is associated with
β-amyloid plaques in the transgenic APPSWE/PSEN1dE9 mouse model of
Alzheimer disease. J Neuropathol Exp Neurol. 71:2–14. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nacmias B, Piccini C, Bagnoli S, Tedde A,
Cellini E, Bracco L and Sorbi S: Brain-derived neurotrophic factor,
apolipoprotein E genetic variants and cognitive performance in
Alzheimer's disease. Neurosci Lett. 367:379–383. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Combarros O, Infante J, Llorca J and
Berciano J: Polymorphism at codon 66 of the brain-derived
neurotrophic factor gene is not associated with sporadic
Alzheimer's disease. Dement Geriatr Cogn Disord. 18:55–58. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lee J, Fukumoto H, Orne J, Klucken J, Raju
S, Vanderburg CR, Irizarry MC, Hyman BT and Ingelsson M: Decreased
levels of BDNF protein in Alzheimer temporal cortex are independent
of BDNF polymorphisms. Exp Neurol. 194:91–96. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Desai P, Nebes R, DeKosky ST and Kamboh
MI: Investigation of the effect of brain-derived neurotrophic
factor (BDNF) polymorphisms on the risk of late-onset Alzheimer's
disease (AD) and quantitative measures of AD progression. Neurosci
Lett. 379:229–234. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Vepsӓlӓinen S, Castren E, Helisalmi S,
Iivonen S, Mannermaa A, Lehtovirta M, Hӓnninen T, Soininen H and
Hiltunen M: Genetic analysis of BDNF and TrkB gene polymorphisms in
Alzheimer's disease. J Neurol. 252:423–428. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bodner SM, Berrettini W, van Deerlin V,
Bennett DA, Wilson RS, Trojanowski JQ and Arnold SE: Genetic
variation in the brain derived neurotrophic factor gene in
Alzheimer's disease. Am J Med Genet B Neuropsychiatr Genet.
134B:1–5. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li Y, Rowland C, Tacey K, et al: The BDNF
Val66Met polymorphism is not associated with late onset Alzheimer's
disease in three case-control samples. Mol Psychiatry. 10:809–810.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Forero DA, Benítez B, Arboleda G, Yunis
JJ, Pardo R and Arboleda H: Analysis of functional polymorphisms in
three synaptic plasticity-related genes (BDNF, COMT AND UCHL1) in
Alzheimer's disease in Colombia. Neurosci Res. 55:334–341. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang H, Ozbay F, Lappalainen J, et al:
Brain derived neurotrophic factor (BDNF) gene variants and
Alzheimer's disease, affective disorders, posttraumatic stress
disorder, schizophrenia, and substance dependence. Am J Med Genet B
Neuropsychiatr Genet. 141B:387–393. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Huang R, Huang J, Cathcart H, Smith S and
Poduslo SE: Genetic variants in brain-derived neurotrophic factor
associated with Alzheimer's disease. J Med Genet. 44:e662007.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Boiocchi C, Maggioli E, Zorzetto M,
Sinforiani E, Cereda C, Ricevuti G and Cuccia M: Brain-derived
neurotrophic factor gene variants and Alzheimer disease: An
association study in an Alzheimer disease Italian population.
Rejuvenation Res. 16:57–66. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cozza A, Melissari E, Iacopetti P,
Mariotti V, Tedde A, Nacmias B, Conte A, Sorbi S and Pellegrini S:
SNPs in neurotrophin system genes and Alzheimer's disease in an
Italian population. J Alzheimers Dis. 15:61–70. 2008.PubMed/NCBI
|
|
38
|
Tsai SJ, Hong CJ, Liu HC, Liu TY, Hsu LE
and Lin CH: Association analysis of brain-derived neurotrophic
factor Val66Met polymorphisms with Alzheimer's disease and age of
onset. Neuropsychobiology. 49:10–12. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bian JT, Zhang JW, Zhang ZX and Zhao HL:
Association analysis of brain-derived neurotrophic factor (BDNF)
gene 196 A/G polymorphism with Alzheimer's disease (AD) in mainland
China. Neurosci Lett. 387:11–16. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Akatsu H, Yamagata HD, Kawamata J, Kamino
K, Takeda M, Yamamoto T, Miki T, Tooyama I, Shimohama S and Kosaka
K: Variations in the BDNF gene in autopsy-confirmed Alzheimer's
disease and dementia with Lewy bodies in Japan. Dement Geriatr Cogn
Disord. 22:216–222. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tsai SJ, Hong CJ, Liu HC, Liu TY and Liou
YJ: The brain-derived neurotrophic factor gene as a possible
susceptibility candidate for Alzheimer's disease in a Chinese
population. Dement Geriatr Cogn Disord. 21:139–143. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
He XM, Zhang ZX, Zhang JW, Zhou YT, Tang
MN, Wu CB and Hong Z: Lack of association between the BDNF gene
Val66Met polymorphism and Alzheimer disease in a Chinese Han
population. Neuropsychobiology. 55:151–155. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Riemenschneider M, Schwarz S, Wagenpfeil
S, Diehl J, Müller U, Förstl H and Kurz A: A polymorphism of the
brain-derived neurotrophic factor (BDNF) is associated with
Alzheimer's disease in patients lacking the Apolipoprotein E
epsilon4 allele. Mol Psychiatry. 7:782–785. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Nishimura AL, Oliveira JR, Mitne-Neto M,
Guindalini C, Nitrini R, Bahia VS, de Brito-Marques PR, Otto PA and
Zatz M: Lack of association between the brain-derived neurotrophin
factor (C-270T) polymorphism and late-onset Alzheimer's disease
(LOAD) in Brazilian patients. J Mol Neurosci. 22:257–260. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cousin E, Macé S, Rocher C, et al: No
replication of genetic association between candidate polymorphisms
and Alzheimer's disease. Neurobiol Aging. 32:1443–1451. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Combarros O, Riancho JA, Infante J, Sañudo
C, Llorca J, Zarrabeitia MT and Berciano J: Interaction between
CYP19 aromatase and butyrylcholinesterase genes increases
Alzheimer's disease risk. Dement Geriatr Cogn Disord. 20:153–157.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Raygani AV, Zahrai M, Soltanzadeh A,
Doosti M, Javadi E and Pourmotabbed T: Analysis of association
between butyrylcholinesterase K variant and apolipoprotein E
genotypes in Alzheimer's disease. Neurosci Lett. 371:142–146. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Alvarez-Arcaya A, Combarros O, Llorca J,
Sánchez-Guerra M, Berciano J, Fernández-Viadero C and Peña N: The
butyrylcholinesterase K variant is a protective factor for sporadic
Alzheimer's disease in women. Acta Neurol Scand. 102:350–353. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
McIlroy SP, Crawford VL, Dynan KB,
McGleenon BM, Vahidassr MD, Lawson JT and Passmore AP:
Butyrylcholinesterase K variant is genetically associated with late
onset Alzheimer's disease in Northern Ireland. J Med Genet.
37:182–185. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wiebusch H, Poirier J, Sévigny P and
Schappert K: Further evidence for a synergistic association between
APOE epsilon4 and BCHE-K in confirmed Alzheimer's disease. Hum
Genet. 104:158–163. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lehmann DJ, Johnston C and Smith AD:
Synergy between the genes for butyrylcholinesterase K variant and
apolipoprotein E4 in late-onset confirmed Alzheimer's disease. Hum
Mol Genet. 6:1933–1936. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Simão-Silva DP, Bertolucci PH, de Labio
RW, Payão SL, Furtado-Alle L and Souza RL: Association analysis
between K and −116A variants of butyrylcholinesterase and
Alzheimer's disease in a Brazilian population. Chem Biol Interact.
203:358–360. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Mateo I, Llorca J, Infante J,
Rodríguez-Rodríguez E, Berciano J and Combarros O: Gene-gene
interaction between 14-3-3 zeta and butyrylcholinesterase modulates
Alzheimer's disease risk. Eur J Neurol. 15:219–222. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Déniz-Naranjo MC, Muñoz-Fernández C,
Alemany-Rodríguez MJ, del Carmen Pérez-Vieitez M, Aladro-Benito Y,
Irurita-Latasa J and Sánchez-García F: Butyrylcholinesterase, ApoE
and Alzheimer's disease in a population from the Canary Islands
(Spain). Neurosci Lett. 427:34–38. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Piccardi M, Congiu D, Squassina A, Manconi
F, Putzu PF, Mereu RM, Chillotti C and Del Zompo M: Alzheimer's
disease: Case-control association study of polymorphisms in ACHE,
CHAT, and BCHE genes in a Sardinian sample. Am J Med Genet B
Neuropsychiatr Genet. 144B:895–899. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Prince JA, Feuk L, Sawyer SL, Gottfries J,
Ricksten A, Nӓgga K, Bogdanovic N, Blennow K and Brookes AJ: Lack
of replication of association findings in complex disease: An
analysis of 15 polymorphisms in prior candidate genes for sporadic
Alzheimer's disease. Eur J Hum Genet. 9:437–444. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Mattila KM, Rinne JO, Röyttӓ M, Laippala
P, Pietilӓ T, Kalimo H, Koivula T, Frey H and Lehtimӓki T:
Dipeptidyl carboxypeptidase 1 (DCP1) and butyrylcholinesterase
(BCHE) gene interactions with the apolipoprotein E epsilon4 allele
as risk factors in Alzheimer's disease and in Parkinson's disease
with coexisting Alzheimer pathology. J Med Genet. 37:766–770. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Tilley L, Morgan K, Grainger J, Marsters
P, Morgan L, Lowe J, Xuereb J, Wischik C, Harrington C and
Kalsheker N: Evaluation of polymorphisms in the presenilin-1 gene
and the butyrylcholinesterase gene as risk factors in sporadic
Alzheimer's disease. Eur J Hum Genet. 7:659–663. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Grubber JM, Saunders AM, Crane-Gatherum
AR, et al: Analysis of association between Alzheimer disease and
the K variant of butyrylcholinesterase (BCHE-K). Neurosci Lett.
269:115–119. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Kehoe PG, Williams H, Holmans P, Wilcock
G, Cairns NJ, Neal J and Owen MJ: The butyrylcholinesterase K
variant and susceptibility to Alzheimer's disease. J Med Genet.
35:1034–1035. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Hiltunen M, Mannermaa A, Helisalmi S,
Koivisto A, Lehtovirta M, Ryynӓnen M, Riekkinen P and Soininen H:
Butyrylcholinesterase K variant and apolipoprotein E4 genes do not
act in synergy in Finnish late-onset Alzheimer's disease patients.
Neurosci Lett. 250:69–71. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Crawford F, Fallin D, Suo Z, Abdullah L,
Gold M, Gauntlett A, Duara R and Mullan M: The
butyrylcholinesterase gene is neither independently nor
synergistically associated with late-onset AD in clinic- and
community-based populations. Neurosci Lett. 249:115–118. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Singleton AB, Smith G, Gibson AM, Woodward
R, Perry RH, Ince PG, Edwardson JA and Morris CM: No association
between the K variant of the butyrylcholinesterase gene and
pathologically confirmed Alzheimer's disease. Hum Mol Genet.
7:937–939. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Brindle N, Song Y, Rogaeva E, et al:
Analysis of the butyrylcholinesterase gene and nearby chromosome 3
markers in Alzheimer disease. Hum Mol Genet. 7:933–935. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Russ C, Powell J, Lovestone S and Holmes
C: K variant of butyrycholinesterase and late-onset Alzheimer's
disease. Lancet. 351:8811998. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Helbecque N: The butyrylcholinesterase K
variant is not associated with Alzheimer's. Alzheimers Rep.
1:309–313. 1998.
|
|
67
|
Laws SM: Evidence that the
butyrylcholinesterase K variant can protect against late-onset
Alzheimer's disease. Alzheimers Rep. 2:219–223. 1999.
|
|
68
|
Bi S, Zhang Y, Wu J, Wang D and Zhao Q:
Association between low-density lipoprotein receptor-related
protein gene, butyrylcholinesterase gene and Alzheimer's disease in
Chinese. Chin Med Sci J. 16:71–75. 2001.PubMed/NCBI
|
|
69
|
Lee DW, Liu HC, Liu TY, Chi CW and Hong
CJ: No association between butyrylcholinesterase K-variant and
Alzheimer disease in Chinese. Am J Med Genet. 96:167–169. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Yamamoto Y, Yasuda M, Mori E and Maeda K:
Failure to confirm a synergistic effect between the K-variant of
the butyrylcholinesterase gene and the epsilon4 allele of the
apolipoprotein gene in Japanese patients with Alzheimer's disease.
J Neurol Neurosurg Psychiatry. 67:94–96. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Ki CS, Na DL, Kim JW, Kim HJ, Kim DK and
Yoon BK: No association between the genes for butyrylcholinesterase
K variant and apolipoprotein E4 in late-onset Alzheimer's disease.
Am J Med Genet. 88:113–115. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Yamada M, Sodeyama N, Itoh Y, Suematsu N,
Otomo E, Matsushita M and Mizusawa H: Butyrylcholinesterase K
variant and cerebral amyloid angiopathy. Stroke. 29:2488–2490.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Xu X, Wang Y, Wang L, et al: Meta-analyses
of 8 polymorphisms associated with the risk of the Alzheimer's
disease. PLoS One. 8:e731292013. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Dai D, Wang Y, Wang L, et al:
Polymorphisms of DRD2 and DRD3 genes and Parkinson's disease: A
meta-analysis. Biomed Rep. 2:275–281. 2014.PubMed/NCBI
|
|
75
|
Zhou J, Huang Y, Huang RS, et al: A
case-control study provides evidence of association for a common
SNP rs974819 in PDGFD to coronary heart disease and suggests a
sex-dependent effect. Thromb Res. 130:602–606. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Excoffier L, Laval G and Schneider S:
Arlequin (version 3.0): An integrated software package for
population genetics data analysis. Evol Bioinform Online. 1:47–50.
2005.
|
|
77
|
Gibson E, Fenster A and Ward AD: The
impact of registration accuracy on imaging validation study design:
A novel statistical power calculation. Med Image Anal. 17:805–815.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Coory MD: Comment on: Heterogeneity in
meta-analysis should be expected and appropriately quantified. Int
J Epidemiol. 39:932–933. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Kawalec P, Mikrut A, Wiśniewska N and Pilc
A: The effectiveness of tofacitinib, a novel Janus kinase
inhibitor, in the treatment of rheumatoid arthritis: a systematic
review and meta-analysis. Clin Rheumatol. 32:1415–1424. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
McHughen SA, Rodriguez PF, Kleim JA, Kleim
ED, Marchal Crespo L, Procaccio V and Cramer SC: BDNF val66met
polymorphism influences motor system function in the human brain.
Cereb Cortex. 20:1254–1262. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Pezawas L, Verchinski BA, Mattay VS,
Callicott JH, Kolachana BS, Straub RE, Egan MF, Meyer-Lindenberg A
and Weinberger DR: The brain-derived neurotrophic factor val66met
polymorphism and variation in human cortical morphology. J
Neurosci. 24:10099–10102. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Fukumoto N, Fujii T, Combarros O, et al:
Sexually dimorphic effect of the Val66Met polymorphism of BDNF on
susceptibility to Alzheimer's disease: New data and meta-analysis.
Am J Med Genet B Neuropsychiatr Genet. 153B:235–242.
2010.PubMed/NCBI
|
|
83
|
Nagata T, Shinagawa S, Nukariya K, Ochiai
Y, Kawamura S, Agawa-Ohta M, Kasahara H, Nakayama K and Yamada H:
Association between brain-derived neurotrophic factor (BDNF) gene
polymorphisms and executive function in Japanese patients with
Alzheimer's disease. Psychogeriatrics. 11:141–149. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Guillozet AL, Smiley JF, Mash DC and
Mesulam MM: Butyrylcholinesterase in the life cycle of amyloid
plaques. Ann Neurol. 42:909–918. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Lehmann DJ, Nagy Z, Litchfield S, Borja MC
and Smith AD: Association of butyrylcholinesterase K variant with
cholinesterase-positive neuritic plaques in the temporal cortex in
late-onset Alzheimer's disease. Hum Genet. 106:447–452. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Lehmann DJ, Williams J, McBroom J and
Smith AD: Using meta-analysis to explain the diversity of results
in genetic studies of late-onset Alzheimer's disease and to
identify high-risk subgroups. Neuroscience. 108:541–554. 2001.
View Article : Google Scholar : PubMed/NCBI
|