Introduction
Distal-less genes (DLX) are divergent homeobox genes
of the Hox gene family, including DLX1-6, which play important
roles in regulating organism development (1,2). DLX2
overexpression occurs in the middle area of the first branchial
arch, namely the maxillary process, between days 9.5 and 10.5 of
mouse embryo development. This overexpression is crucial to the
differentiation and development of the primordium, as demonstrated
by experimental embryology, since it determines the subsequent
development and phenotype of the maxillofacial skeletal patterns
(3). Thereafter, DLX2 is the primary
candidate gene that regulates the development of the first
branchial arch (4). Craniofacial
bone defects, cranial parietal bone ossification delay and cortical
bone dysplasia in long bones have been observed in DLX2 knockout
mouse models (5,6), indicating that the main function of
DLX2 is to regulate the proper migration of ectomesenchymal cells
and the normal osteogenic differentiation of preosteoblast cells.
However, the function of DLX2 in regulating osteoblast
differentiation and the underlying molecular mechanism remain
controversial (7–9).
Therefore, the aim of the present study was to
investigate the effect of DLX2 on osteogenic differentiation, and
the possible underlying molecular mechanism, through in
vitro transfection of a murine stem cell virus containing DLX2
(pMSCV-DLX2) into the preosteoblast cell line, MC3T3-E1.
Materials and methods
Materials
An MC3T3-E1 cell line was purchased from the Cell
Bank of the Chinese Academy of Sciences (Shanghai, China). Fetal
calf serum (FCS) was obtained from the Beyotime Institute of
Biotechnology (Haimen, China) and α-Mimimal Essential Medium (MEM)
was purchased from Gibco Life Technologies (Carlsbad, CA, USA).
α-MEM osteogenesis induction culture medium (α-MEM plus
1.0×10−7mol/l dexamethasone, 2.5×10−4 mol/l
vitamin C and 1.0×10−2mol/l β-sodium glycerophosphate)
and ρ-nitrophenylphosphate (PNPP) were purchased from Sigma-Aldrich
(St. Louis, MO, USA). A 3180P INC Heraeus Hera Cell CO2
incubator was obtained from Heraeus Holding GmbH (Hanau, Germany)
and a Leica DM IRB inverted phase contrast microscope was purchased
from Leica Microsystems GmbH (Wetzlar, Germany). The Z2 Cell
Counter was purchased from Beckman Coulter (Brea, CA, USA). A Prime
Script™ RT Reagent Kit and SYBR Premix Ex Taq™ were purchased from
Takara Biotechnology Co., Ltd. (Dalian, China). CAGGS/DLX2 was
obtained from Dr Rubenstein (University of California, San
Francisco, CA, USA).
Establishment of pMSCV-puro-DLX2
Restriction enzyme identification with XhoI
and EcoRI (Takara Biotechnology Co., Ltd.) was applied to
pCAGGS/DLX2 and pMSCV-puro (GeneCopoeia, Rockville, MD, USA),
respectively. The target fragments were collected through gel
extraction according to the manufacturer's instructions (QIAquick
Gel extraction kit, Qiagen, Hilden, Germany) and stored at 4°C
overnight. pMSCV-puro and DLX2 were subsequently combined by T4
ligase (Takara Biotechnology Co. Ltd.) at 16°C overnight and
sequenced.
293FT viral packaging
Transfection was performed when the cell density of
the 293FT cell line reached 80–90%. The 293FT cell line was used to
construct the retrovirus. Vesicular stomatitis virus glycoprotein
(10 µg), GAG-pol (15 µg) and pMSCV (15 µg) were prepared for the
retroviral packaging system. At 4–6 h after transfection, the
culture medium in the T75 flask was replaced with 10 ml α-MEM
containing 10% FCS, after which the virus was collected. This was
done by collecting the pseudovirus-containing medium from the 293FT
cell culture into sterile capped tubes for 48 h post-transfection.
The tubes were then centrifuged at 550 × g for 10 min to remove
cell debris and the supernatant was filtered through 45 µm
polyethersulfone low protein-binding filters. The virus pellet was
resuspended in phosphate-buffered saline (PBS) and stored at
4°C.
MC3T3-E1 transfection
The virus used for transfection was determined
through dosage titration at 1:1, 10:1, 100:1 and 1,000:1. Polybrene
(1/1,000; Sigma-Aldrich) was added to the medium, after which the
plate was centrifuged for 3 min at 300 × g at 37°C. The culture
medium was replaced 24 h after transfection. Following incubation
in the culture medium for 48 h, 3 µg/ml puromycin was applied for
growth selection. The cells that survived the selection were
collected and passaged. Controls included pMSCV (dosage titration
at 1:1, 10:1 and 100:1).
DLX2 expression detection with
quantitative polymerase chain reaction (PCR) and western blot
analysis
At 72 h after puromycin selection, the total RNA was
extracted for quantitative PCR. This was performed using the SYBR
Green system (Takara Bio, Inc, Shiga, Japan) with a total volume of
20 µl in a 96-well microwell plate. The reaction consisted of 10 µl
SYBR Premix Ex Taq (Takara Biotechnology Co. Ltd.), 0.8 µl mixed
primers, 1 µl cDNA template and 8.2 µl RNase-free distilled water.
The PCR program was set at 95°C for 4 min followed by 40 cycles at
95°C for 20 sec, 60 °C for 15 sec and 72 °C for 20 sec. The
threshold cycle values were calculated using the iQ5 software
(Bio-Rad, Hercules, CA, USA). All the primers used in the
experiments are shown in Table I.
The analysis of the data was based on calculating the relative
expression levels of these genes compared to the expression of the
control (glyceraldehyde 3-phosphate dehydrogenase, a housekeeping
gene).
 | Table I.Primers for used for quantitative
polymerase chain reaction. |
Table I.
Primers for used for quantitative
polymerase chain reaction.
| Gene | Forward sequence | Reverse sequence |
|---|
| DLX2 |
5′-CATGGGCTCCTACCAGTACCAC-3′ |
5′-TCGGATTTCAGGCTCAAGGTC-3′ |
| GAPDH |
5′-GGTGAAGGTCGGTGTGAACG-3′ |
5′-CTCGCTCCTGGAAGATGGTG-3′ |
| ALP |
5′-TGGGCATTGTGACTACCACTCGG-3′ |
5′-CCTCTGGTGGCATCTCGTTATCC-3′ |
| OCN |
5′-GGACCATCTTTCTGCTCACTCTG-3′ |
5′-GTTCACTACCTTATTGCCCTCCTG-3′ |
| RUNX2 |
5′-AACTTCCTGTGCTCCGTGCTG-3′ |
5′-TCGTTGAACCTGGCTACTTGG-3′ |
| MSX2 |
5′-GGAGCACCGTGGATACAGGA-3′ |
5′-AGGCTAGAAGCTGGGATGTGG-3′ |
Cells were collected, washed 3 times with pre
ice-cold PBS and lysed in SDS-lysis buffer containing protease
inhibitors (Roche Diagnostics GmbH, Mannheim, Germany) for 20 min
on ice. The lysates were centrifuged at 12,000 × g at 4°C for 30
min and the supernatants were boiled in sodium dodecyl sulfate
sample buffer containing 0.5 M of mercaptoethanol. The samples were
then separated on a 10% sodium dodecyl sulfate polyacrylamide gel
and transferred to a polyvinylidene difluoride membrane using a
semi-dry transfer apparatus (Bio-Rad, Hercules, CA, USA). The
membrane was blocked with 5% milk for 1 h and then incubated with
primary antibodies in modified D-PBS tween-20 buffer overnight. The
primary antibodies were anti-DLX2 (1:800; Abcam, Cambridge, UK) and
β-actin (1:1,000; Sigma-Aldrich). Following the primary antibody
incubation, they were incubated with horse-radish
peroxidase-conjugated secondary antibodies, including goat
anti-rabbit and goat anti-mouse IgG (1:5,000; Beyotime Institute of
Biotechnology, Haimen, China). The protein bands were visualised
using an enhanced chemilluminescent western blot analysis kit
(Pierce Biotechnology, Inc., Rockford, IL, USA).
Detection of DLX2 expression in the
stable cell line following transfection
Following transfection and selection, the stable
cell line was seeded into a six-well plate at a cell density of
1.5×104/cm2. α-MEM osteogenesis induction
culture medium was added to the wells and the cells were cultured
in conditions of 5% CO2 at 37°C. Cells were collected
and protein samples were obtained on days 1, 4, 7 and 14 for
western blot analysis.
Analysis of effect of DLX2
overexpression on osteogenesis-associated genes via quantitative
PCR
Stable cell lines, following transfection and
selection, were seeded into a six-well plate at a cell density of
1.5×104/cm2. The wells contained α-MEM
osteogenesis induction culture medium and the cells were cultured
in 5% CO2 at 37°C. Cells were collected and the total
RNA was extracted on days 1, 4, 7 and 14 for quantitative PCR. The
primers used for the PCR assays of ALP, osteocalcin (OCN),
runt-related transcription factor (RUNX)2 and MSX2 are included in
Table I.
ALP activity detection
Stable cell lines, following transfection and
selection, were seeded into a 24-well plate at a cell density of
1.5×104/cm2. α-MEM osteogenesis induction
culture medium was added to the wells and the cells were cultured
in 5% CO2 at 37°C. ALP activity detection was performed
on days 4, 7 and 14 using PNPP. The cells were washed with PBS and
lysed with ALP buffer containing 0.2% Triton X-100, 42.74 mg
MgCl2·6H2O, 20 µl HCl (10 mol/l), 19.4 ml
diethylamine and 40 mg sodium azide dissolved in 200 ml distilled
water. After 4 h in the incubator at 37°C, 190 µl supernatant was
collected and added to a 96-well plate containing 190 µl substrate
buffer per well. The optical density (OD) value was measured at 405
nm following incubation for 30 min at 37°C. A 10-µl sample of the
supernatant was added to 200 µl protein analysis buffer and the OD
value at 630 nm was detected after 5 min. The intracellular protein
content was determined using a Micro BCA protein assay kit (Thermo
Fisher Scientific, Waltham, MA, USA) and applied for the standard
curve. The alkaline phosphatase activity was determined by
measuring the optical density values for absorbance at 405 nm after
incubation with p-nitrophenyl phosphate for 30 min at 37°C
(Sigma-Aldrich).
Alizarin red staining
Alizarin red staining was used in a biochemical
assay to determine, quantitatively by colorimetry, the presence of
calcific deposition by cells of an osteogenetic lineage. Following
transfection and selection, the stable cell lines were seeded into
a six-well plate at a cell density of
1.5×104/cm2 with α-MEM osteogenesis induction
culture medium. The cells were cultured in conditions of 5%
CO2 at 37°C. Alizarin red staining (Sigma-Aldrich) was
performed on days 3, 21 and 27, however, the stain was not that
notable amongst the blank, pMSCV and pMSCV-Dl×2 groups. Each group
was scanned for detection by a Canon LiDE 110 scanner (Canon,
Beijing, China) following the staining.
Statistical analysis
Data were analyzed by analysis of variance using the
Student-Newman-Keuls method. Statistical analysis was performed
using SAS 6.04 software package (SAS Institute, Cary, NC, USA),
where P<0.05 was considered to indicate a statistically
significant difference.
Results
DLX2 expression in the stably
transfected cell line
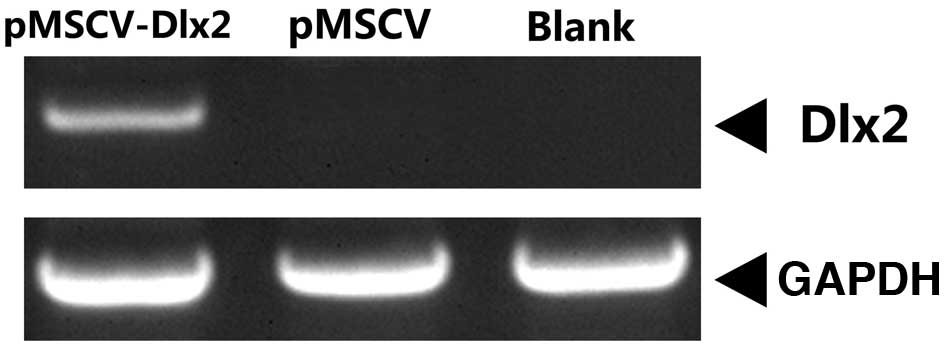
At 72 h after puromycin selection, mRNA expression
of DLX2 was observed in the pMSCV-DLX2 group, but not in the pMSCV
or non-transfection groups (Fig. 1).
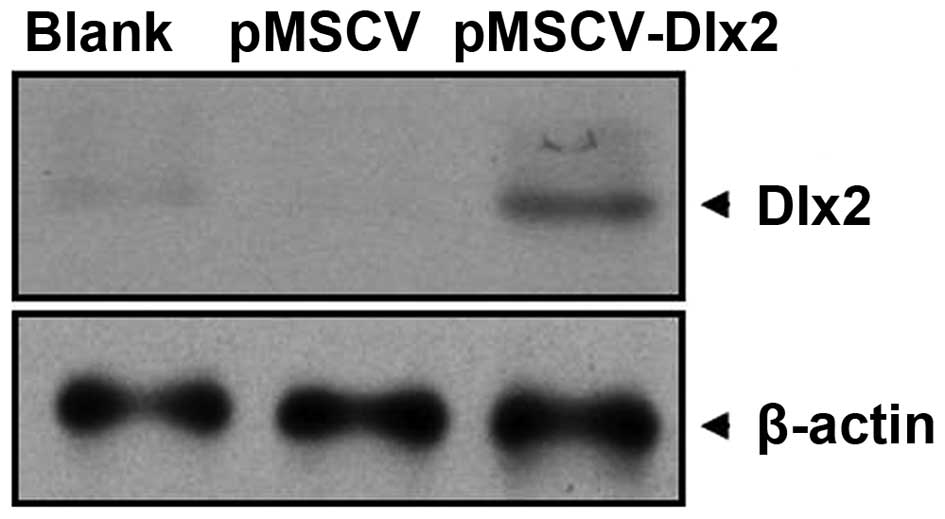
As shown in the western blot analysis, a 33-kDa band presented in
the pMSCV-DLX2 group; however, this band was not observed in the
pMSCV or non-transfection groups (Fig.
2). These observations indicated that the transfection was
successful and expression of DLX2 had been induced.
DLX2 expression in the stable clones
selected with osteogenesis induction
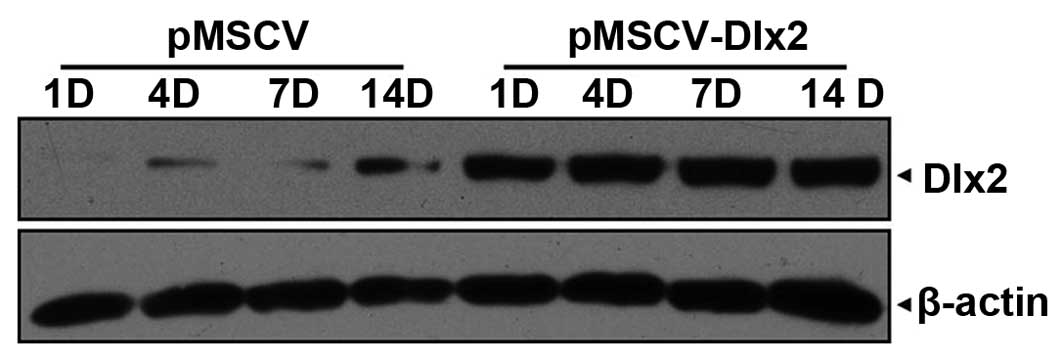
On day 1 following osteogenesis induction, DLX2
expression was detected in the pMSCV-DLX2 group, which was shown to
remain stable until day 14 post-transfection. Increasing endogenic
DLX2 expression was observed in the pMSCV group (Fig. 3).
Effect of DLX2 overexpression on the
expression levels of osteogenesis-associated genes
Expression levels of osteogenesis-associated genes
are shown in Fig. 4. The relative
expression levels were calculated, as compared with the control
group on day 1, and are outlined in Table II. Statistically significant
differences between the control and experimental cells were
observed in the expression of ALP (P<0.01), OCN (P<0.05, on
day 14) and MSX2 (P<0.01), but not in the expression of RUNX2
(P>0.05).
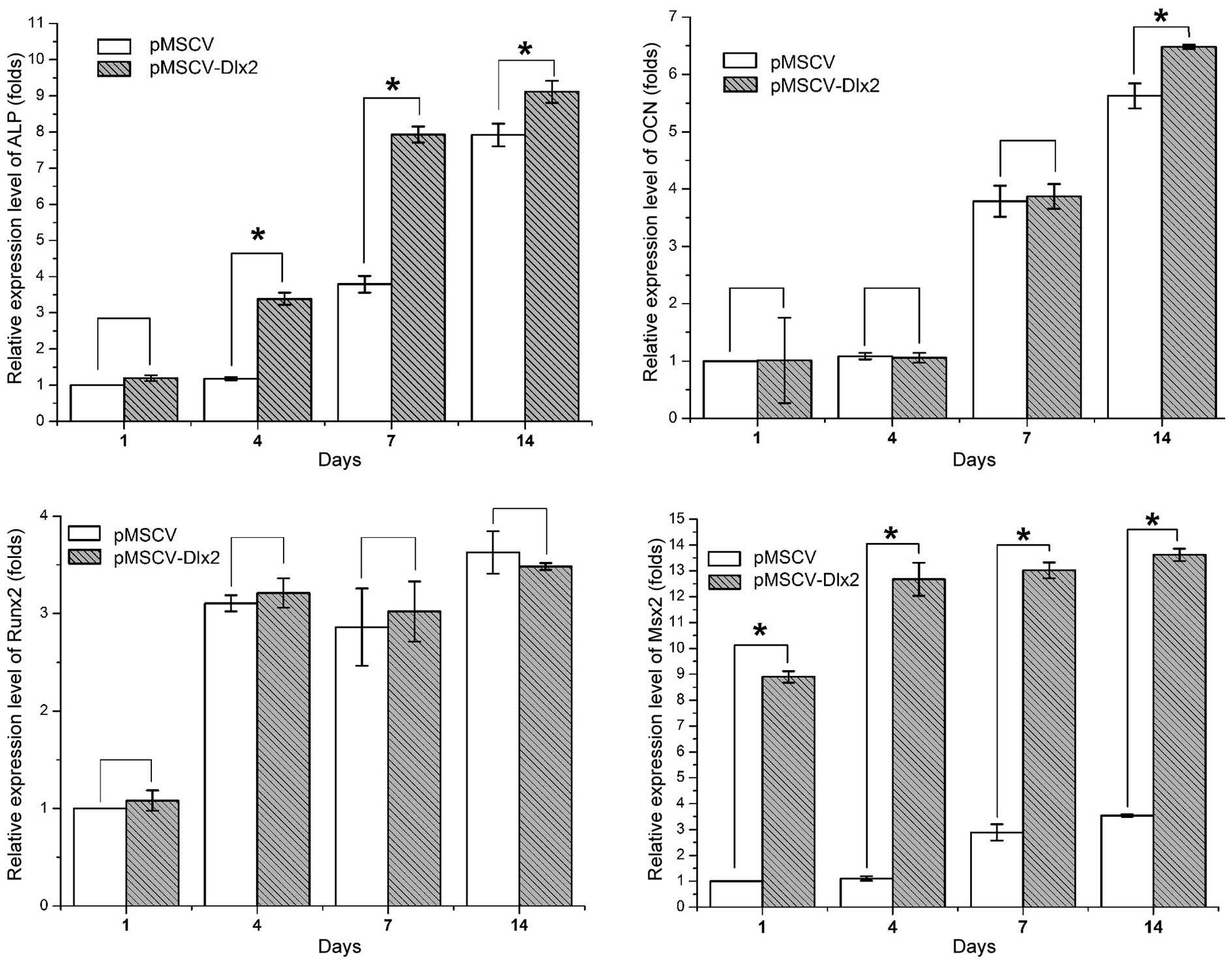 | Figure 4.Quantitative polymerase chain reaction
analysis showing the relative mRNA expression levels of the bone
formation-associated genes, ALP, OCN, RUNX2 and MSX2. Values were
normalized against the mRNA expression level of the control group
on day 1. *P<0.05, pMSCV-DLX2 group vs. pMSCV group (n=3). DLX,
distal-less gene; ALP, alkaline phosphatase; OCN, osteocalcin;
RUNX2, runt-related transcription factor; MSX, Msh homeobox; MSCV,
murine stem cell virus. |
 | Table II.Quantitative polymerase chain reaction
analysis of the relative expression levels of bone
formation-associated genes. |
Table II.
Quantitative polymerase chain reaction
analysis of the relative expression levels of bone
formation-associated genes.
| Gene | Day 1 | Day 4 | Day 7 | Day 14 |
|---|
| ALP |
|
pMSCV | 1 |
1.18±0.045 |
3.719±0.23 |
7.971±0.32 |
|
pMSCV-DLX2 |
1.193±0.076 |
3.387±0.172 |
7.93±0.223 |
9.117±0.305 |
| OCN |
|
pMSCV | 1 |
1.084±0.058 |
3.79±0.271 |
5.56±0.217 |
|
pMSCV-DLX2 |
1.013±0.075 |
1.057±0.087 |
3.873±0.215 |
6.482±0.035 |
| RUNX2 |
|
pMSCV | 1 |
3.104±0.083 |
2.86±0.395 |
3.626±0.217 |
|
pMSCV-DLX2 |
1.081±0.104 |
3.209±0.149 |
3.02±0.308 |
3.481±0.03 |
| MSX2 |
|
pMSCV | 1 |
1.104±0.083 |
2.882±0.313 |
3.54±0.046 |
|
pMSCV-DLX2 |
8.901±0.223 |
12.676±0.641 |
13.02±0.308 |
13.615±0.236 |
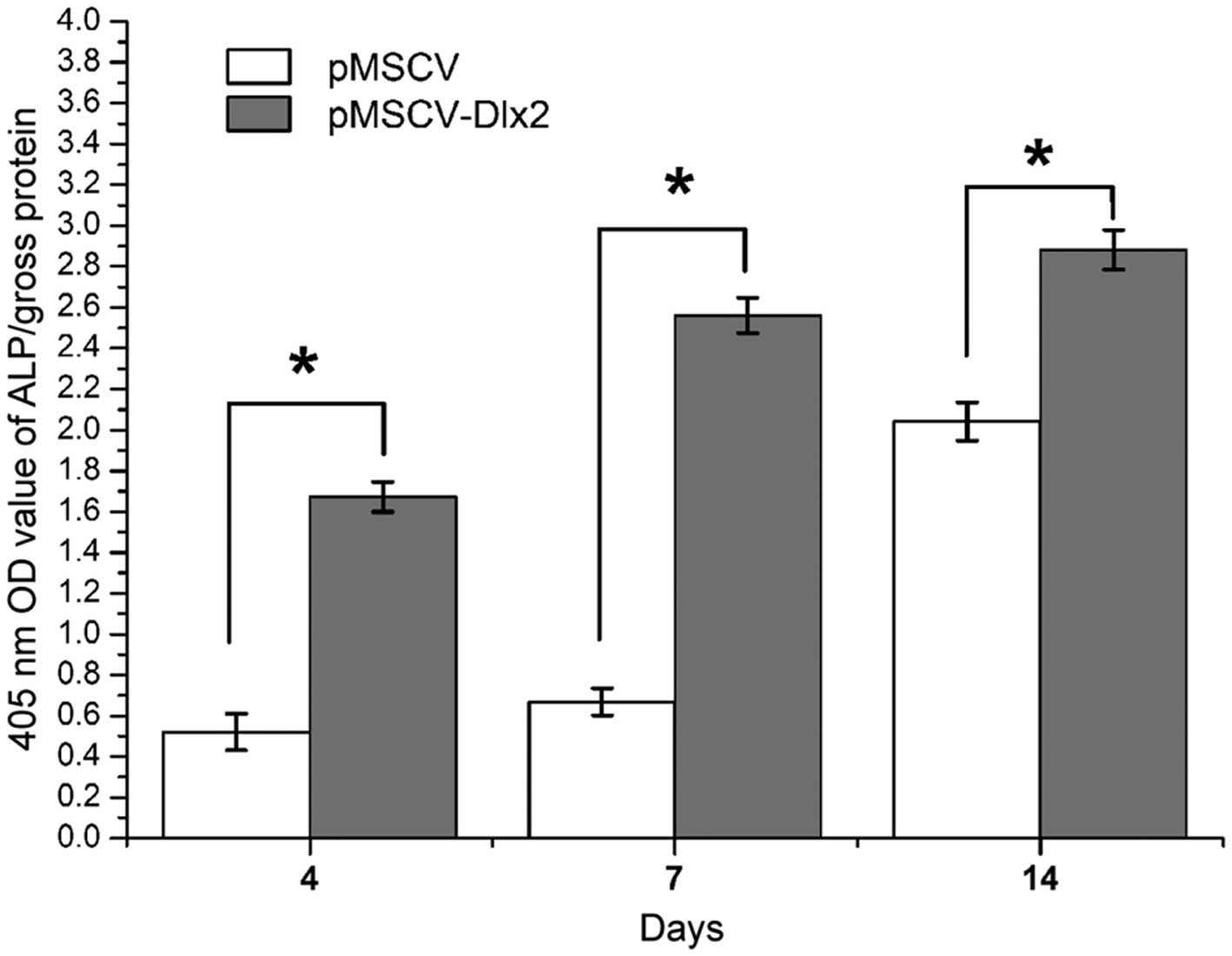
ALP activity
Higher ALP activity levels were observed on days 4,
7 and 14 after osteogenesis induction in the transfection group
(pMSCV-DLX2), and statistically significant differences were
observed when compared with the control group (P<0.01). The
highest activity was observed on day 14, as shown in Fig. 5. The ALP activity levels are listed
in Table III.
 | Table III.Semi-quantitative analysis of ALP
activity. |
Table III.
Semi-quantitative analysis of ALP
activity.
| Group | Day 4 | Day 7 | Day 14 |
|---|
| pMSCV |
0.521±0.089 |
0.668±0.066 |
2.041±0.094 |
| pMSCV-DLX2 |
1.673±0.073 |
2.561±0.087 |
2.881±0.097 |
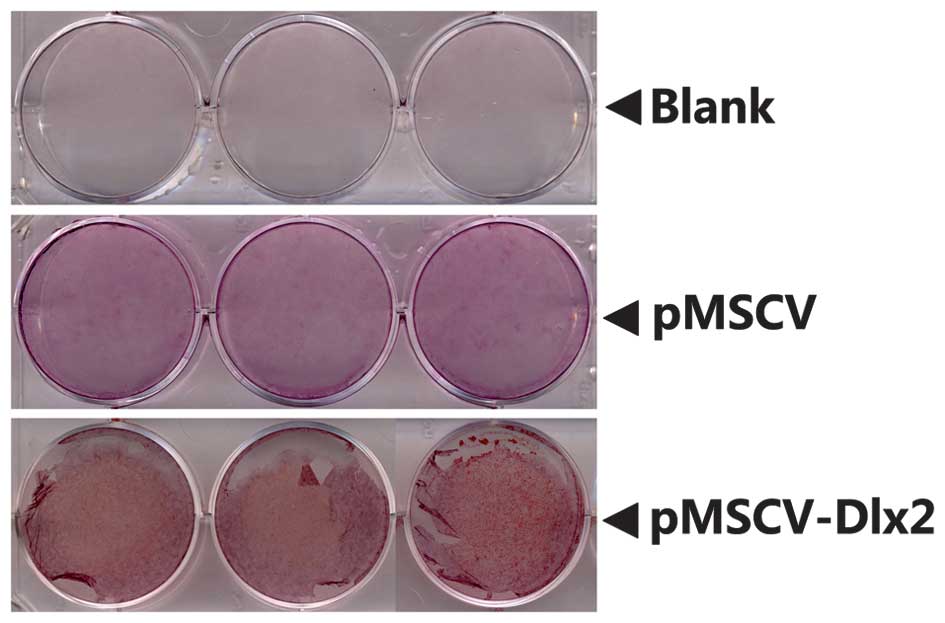
Alizarin red staining
Positive results from the Alizarin red staining were
observed on day 21 following osteogenesis induction, as shown in
Fig. 6. The results for staining at
day 27 were almost the same to those on day 21 and are therefore
not discussed. In the DLX2 transfection group, the alizarin red
staining was used to show the calcium nodule which indicated that
DLX2 promoted the osteogenic differentiation of MC3T3-E1 cells.
Discussion
DLX are divergent homeobox genes that are part of
the Hox gene family, which are involved in the regulation of
maxillofacial bone development. However, the function of DLX
remains controversial and the regulatory mechanisms are yet to be
elucidated. In two previous studies, Qiu et al (5,6) created
a DLX2−/− mouse model through gene knockout technology.
Newborn DLX2−/− mice were found to die after birth with
structural abnormalities observed in the bones originating from the
first branchial arch maxillary process, such as the basisphenoid,
greater wing of the sphenoid bone and pterygoid lamina, which was
accompanied by a cleft palate, cranial parietal bone ossification
delay and cortical bone dysplasia in long bones. These observations
indicated that DLX2 was crucial in regulating craniofacial bone
development and differentiation. In addition, gene expression
microarray analysis revealed that DLX2 is an early response gene in
the regulation of bone morphogenetic protein (BMP)2-mediated
osteogenic differentiation (10).
Therefore, the DLX knockout mouse models and DLX overexpression
experiments indicated that DLX promotes skeleton formation;
however, the effect of DLX on osteogenic differentiation remains
unknown. Lee et al (11)
found that DLX5, which is closely associated to DLX2, suppressed
the expression of OCN, in contrast to DLX2. In addition,
overexpression of DLX5 was reported to result in bone formation
disorder in immunodeficient mice and mineralized matrix deposition
of cells cultured in vitro. In vitro, overexpression
of DLX5 was shown to have no effect on chondroplast differentiation
(12). In the present study,
MC3T3-E1 cells were transfected with pMSCV-DLX2 and stable clones
were selected with puromycin. Subsequently, the mRNA and protein
expression levels of DLX2 were determined by quantitative PCR and
western blot analysis, respectively. This stable transfection cell
line established the basis for investigating the effect of DLX2
overexpression on osteogenic differentiation and the possible
underlying mechanisms.
ALP is hypothesized to be an indicator of early
osteogenesis (13), and is known to
hydrolyze numerous types of phosphates under alkaline conditions to
promote cell maturation and calcification (14). ALP has been shown to be highly
expressed during early osteogenic differentiation (day 7) through
induction by morphogenetic proteins (15,16). In
the present study, ALP activity in the MC3T3-E1 cell line was
upregulated in the early stage of osteogenesis between days 7 and
14, as induced by the osteogenesis culture medium. As indicated in
the quantitative PCR and ALP activity experiments, DLX2 upregulated
ALP activity as early as day 4. On day 14, no statistically
significant difference was observed between the groups, indicating
that DLX2 primarily functions in the early stages of osteogenic
differentiation, which is different from that of BMPs (15). OCN is the most abundant
non-procollagen protein in the bone tissue, and can promote
osteogenic differentiation by combining with minerals (17). In osteoblasts cultured in
vitro, OCN upregulation has been observed during the
mineralization stage; however, OCN expression has subsequently
decreased until the end of osteogenic differentiation (13,18,19). The
results of the present study demonstrated that low expression
levels of OCN were observed in each group during the early stage of
osteogenesis on days 1, 4 and 7, and no statistically significant
differences were observed among the groups. However, on day 14,
higher expression levels of OCN were observed in the transfection
group (P<0.05). In addition, positive Alizarin red staining
indicated that DLX2 promoted the expression of OCN and osteogenesis
at the later stage.
RUNX2 belongs to the Runt domain gene family and has
been demonstrated to be the one of the most important transcription
factors involved in osteogenic differentiation (20). However, no statistically significant
difference in RUNX2 expression was observed at any of the four time
points when comparing the transfection and control groups
(P>0.05), which indicated that the RUNX2 pathway may not be
involved in the mechanism underlying DLX2-induced promotion of
MC3T3-E1 osteogenesis. MSX2 belongs to the Hox gene family and is
involved in the regulation of osteogenesis. Downregulation of MSX2
has been shown to result in preosteoblast dysmaturity (21). DLX genes have been reported to
function in transforming growth factor-β (TGF-β)/BMP-mediated
osteogenesis regulation through combining with and antagonizing
MSX2 (22,23). In the present study, overexpression
of DLX2 was shown to upregulate MSX2 expression. Thus, it was
hypothesized that DLX2 and MSX2 regulate the transcription of
osteogenesis-associated genes in a synergistic manner by forming a
transcription factor complex. However, future investigation is
required to further support this hypothesis.
In conclusion, through DLX2 transfection,
examination of osteogenesis-associated gene expression, ALP
activity and Alizarin red staining, the present study demonstrated
that DLX2 overexpression induces the osteogenic differentiation of
MC3T3-E1 cells via upregulating bone formation-associated genes,
such as ALP and MSX2. Future studies will focus on
osseointeractions between DLX2 and MSX2. Further studies will aim
to investigate the role of the TGF-β signalling pathway on the
induction of osteogenesis by DLX2. Such a study may contribute to
potential clinical application of bone mesenchymal stem cells
transduce with Dlx2 for bone augmentation in patients suffering
from bone deficiency.
Acknowledgements
This study was supported by grants from the Shanghai
Leading Academic Discipline Project (no. S30206), the Research Fund
of Science and Technology Commission of Shanghai Municipality (no.
10JC1408700), the Research Fund of Shenkang Company (no.
SHDC12010205), the Research Fund of Shanghai Municipal Health
Bureau (no. 2009077), the Combined Engineering and Medicine Project
of Shanghai Jiao Tong University (nos. YG2010MS55 and YG2014QN02)
and the National Nature Science Foundation of China (no.
81271122).
Abbreviations:
|
DLX
|
distal-less gene
|
|
ALP
|
alkaline phosphatase
|
|
OCN
|
osteocalcin
|
|
RUNX2
|
runt-related transcription factor
2
|
|
MSX2
|
Msh homeobox 2
|
|
BMP
|
bone morphogenetic protein
|
|
MSCV
|
murine stem cell virus
|
|
PCR
|
polymerase chain reaction
|
|
FCS
|
fetal calf serum
|
|
PNPP
|
ρ-nitrophenylphosphate
|
|
MEM
|
Minimal Essential Medium
|
|
PBS
|
phosphate-buffered saline
|
|
OD
|
optical density
|
References
|
1
|
Depew MJ, Simpson CA, Morasso M and
Rubenstein JL: Reassessing the Dlx code: the genetic regulation of
branchial arch skeletal pattern and development. J Anat.
207:501–561. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jin Y: Mouse Development Biology and
Embryo Research Method. Volume 21st. People's Medical Publishing
House; Beijing: pp. 70–73. 2005
|
|
3
|
Ferguson CA, Tucker AS and Sharpe PT:
Temporospatial cell interactions regulating mandibular and
maxillary arch patterning. Development. 127:403–412.
2000.PubMed/NCBI
|
|
4
|
Depew MJ, Lufkin T and Rubenstein JL:
Specification of jaw subdivisions by Dlx genes. Science.
298:381–385. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Qiu M, Bulfone A, Ghattas I, et al: Role
of the Dlx homeobox genes in proximodistal patterning of the
branchial arches: mutations of Dlx-1, Dlx-2 and Dlx-1 and −2 alter
morphogenesis of proximal skeletal and soft tissue structures
derived from the first and second arches. Dev Biol. 185:165–184.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Qiu M, Bulfone A, Martinez S, et al: Null
mutation of Dlx-2 results in abnormal morphogenesis of proximal
first and second branchial arch derivatives and abnormal
differentiation in the forebrain. Genes Dev. 9:2523–2538. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Merlo GR, Zerega B, Paleari L, et al:
Multiple functions of Dlx genes. Int J Dev Biol. 44:619–626.
2000.PubMed/NCBI
|
|
8
|
Panganiban G and Rubenstein JL:
Developmental functions of the Distal-less/Dlx homeobox genes.
Development. 129:4371–4386. 2002.PubMed/NCBI
|
|
9
|
Li H, Marijanovic I, Kronenberg MS, et al:
Expression and function of Dlx genes in the osteoblast lineage. Dev
Biol. 316:458–470. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Harris SE, Guo D, Harris MA, et al:
Transcriptional regulation of BMP-2 activated genes in osteoblasts
using gene expression microarray analysis: role of Dlx2 and Dlx5
transcription factors. Front Biosci. 8:s1249–s1265. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee MH, Kwon TG, Park HS, et al:
BMP-2-induced Osterix expression is mediated by Dlx5 but is
independent of Runx2. Biochem Biophys Res Commun. 309:689–694.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Muraglia A, Perera M, Verardo S, et al:
DLX5 overexpression impairs osteogenic differentiation of human
bone marrow stromal cells. Eur J Cell Biol. 87:751–761. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Karsenty G and Wagner EF: Reaching a
genetic and molecular understanding of skeletal development. Dev
Cell. 2:389–406. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Beck GR Jr, Zerler B and Moran E:
Phosphate is a specific signal for induction of osteopontin gene
expression. Proc Natl Acad Sci USA. 97:8352–8357. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hu Z, Peel SA, Ho SK, et al: Role of
bovine bone morphogenetic proteins in bone matrix protein and
osteoblast-related gene expression during rat bone marrow stromal
cell differentiation. J Craniofac Surg. 16:1006–1014. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vaes BL, Dechering KJ, Feijen A, et al:
Comprehensive microarray analysis of bone morphogenetic protein
2-induced osteoblast differentiation resulting in the
identification of novel markers for bone development. J Bone Miner
Res. 17:2106–2118. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Glowacki J, Rey C, Glimcher MJ, et al: A
role for osteocalcin in osteoclast differentiation. J Cell Biochem.
45:292–302. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hu JZ, Jiang XQ, Zhang ZY, et al: Effects
of Nell-1 gene on bone formation-related gene expression during
osteoblastic differentiation of rat bone marrow stromal cells by
real-time PCR. Zhongguo Kou Qiang He Mian Wai Ke Za Zhi. 6:48–53.
2008.[(In Chinese)].
|
|
19
|
Hu JZ, Jiang XQ, Zhang ZY and Zhang XL:
Effect of Nell-1 gene on osteogenic differentiation of rat bMSCs:
An in vitro study. Zhongguo Kou Qiang He Mian Wai Ke Za Zhi.
7:38–43. 2009.[(In Chinese)].
|
|
20
|
Li P, Yu SH, Chen D, et al: Overexpression
of Runx2 induces osteogenic differentiation in C2C12 cells.
Zhongguo Sheng Wu Hua Xue Yu Fen Zi Sheng Wu Xue Bao. 26:236–242.
2010.[(In Chinese)].
|
|
21
|
Cheng SL, Shao JS, Charlton-Kachigian N,
et al: MSX2 promotes osteogenesis and suppresses adipogenic
differentiation of multipotent mesenchymal progenitors. J Biol
Chem. 278:45969–45977. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bendall AJ and Abate-Shen C: Roles for Msx
and Dlx homeoproteins in vertebrate development. Gene. 247:17–31.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ryoo HM, Lee MH and Kim YJ: Critical
molecular switches involved in BMP-2-induced osteogenic
differentiation of mesenchymal cells. Gene. 366:51–57. 2006.
View Article : Google Scholar : PubMed/NCBI
|