Introduction
Hypoxia, defined as areas with oxygen tension values
of 10 mmHg or lower, is a common feature of solid tumors that
occurs across a wide variety of malignancies (1,2).
Once cells encounter hypoxia, a large number of genes that
stimulate adaptation to oxygen deprivation are induced. These
hypoxia-inducible genes are primarily controlled by the
transcription factor hypoxia-inducible factor 1 (HIF-1), including
those involved in cell cycle arrest, apoptosis, erythropoiesis,
angiogenesis, glycolytic metabolism, and tumor invasiveness
(2,3). HIF-1 is a heterodimer consisting of
a constitutively expressed HIF-1β subunit and a HIF-1α subunit that
is regulated in an oxygen-dependent manner (3). Under hypoxia, HIF-1α can block its
own degradation and thus induce its accumulation (3). Therefore, HIF-1α is overexpressed in
many cancer types and is associated with poor prognosis in cancers
of the breast, brain, cervix, ovary, and uterus (3).
HIF-1α activation in response to hypoxia stimulates
the transcription of several of its target genes. Vascular
endothelial growth factor (VEGF), which is known to promote
angiogenesis, is strongly induced by HIF-1α (4). In addition, matrix metalloproteinase
(MMP)-1 and MMP-2 are induced to promote intravasation (5,6).
Additionally, the epithelial-mesenchymal transition-inducing genes
E-cadherin, SNAIL, TWIST, TCF3, and ZEB1 are up-regulated in a
HIF-1α-dependent manner (7–9).
Interestingly, HIF-1α also induces cell cycle arrest by
up-regulating p53, p21, and E1B 19K/Bcl-2 binding protein Nip3
expression (10–12). These findings suggest that there
are different adaptive responses to oxygen deprivation.
Cobalt chloride (CoCl2), a commonly used
hypoxia-mimetic agent, artificially induces hypoxia and can block
the degradation and thus induce the accumulation of HIF-1α protein
(13,14). Many reports have indicated that
both CoCl2 and hypoxia regulate a similar group of genes
on a global gene expression level (15–17). In addition, the reports
demonstrated that hypoxia and CoCl2 induced different
effects in many different types of cells. For example,
CoCl2 induced apoptosis in rat C6 glioma cells, human
alveolar macrophages, neuronal PC12 cells, and HeLa human cervical
cancer cells, although it inhibited apoptotic death in HepG2
hepatoma cells (18–22). Moreover, CoCl2 induced
prostate tumor cell adhesion, metastasis, and angiogenesis
(23–25) and stimulated oral squamous
carcinoma cell growth (26).
Multiple myeloma (MM) is a B-cell malignancy
selectively localized in the bone marrow (BM) (27). Unlike other organs, the normal BM
microenvironment is hypoxic. This physiological hypoxia is crucial
for normal marrow hematopoiesis (28). In several studies, oxygen levels
in BM in mice with MM were depressed (29,30). This hypoxia is an important
microenvironmental stimulus for activities critical for MM disease
progression such as MM cell growth, drug resistance, and
angiogenesis (29,30). However, how hypoxia and MM affect
each other is poorly understood. Therefore, an investigation of the
effects of hypoxia on myeloma cells is necessary.
Materials and methods
Cell culture
The human multiple myeloma cell line U266 was
purchased from the Korean Cell Line Bank (Seoul, Korea) and
cultured in RPMI-1640 medium containing 10% fetal bovine serum and
antibiotics at 37°C in a humidified chamber containing 5%
CO2. Cells were seeded into 60-mm culture dishes
(4×105 cells per dish) 1 day before CoCl2
(Sigma, St. Louis, MO, USA) treatment.
Hypoxia induction
CoCl2 was dissolved in dimethylsulfoxide
(DMSO; Sigma) and added to U266 cultures at the indicated final
concentrations. DMSO was added to culture medium alone as a vehicle
control. Cells were treated for 24 h.
RNA preparation and cDNA synthesis
Total RNA was extracted from cells by using TRIzol
reagent (Invitrogen, Carlsbad, CA, USA) according to the
manufacturer’s instructions after the 24-h CoCl2
treatment. For the microarray studies, both the quality and
concentration of the RNA samples were determined using an Agilent
2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and
an Ultrospec 3300 Pro UV/Visible Spectrophotometer (Amersham
Biosciences, UK). The recommended RNA quality parameters for
microarray analysis are as follows: a UV spectroscopy A260/A280
ratio of 1.8–2.0 and an A260/A230 ratio >1.8, an 18S/28S rRNA
ratio of 1.8–2.1, and an RNA integrity number >8.0. To
synthesize cDNA, 1 μg of RNA was incubated with oligo-dT primers at
94°C for 10 min and reverse-transcribed with reverse transcriptase
(Enzynomics, Seoul, Korea) at 37°C for 1 h.
DNA microarray analysis
DNA microarray analysis was performed using the
HumanHT-12 v4.0 Expression Beadchip kit (Illumina, San Diego, CA,
USA) according to the instruction manual. Derived data were
analyzed using Genespring GX 11 (Agilent Technologies). The raw
data were filtered using FLAG and t-tests. Significant genes were
determined using the fluorescence ratio between the control and
CoCl2-treated samples, and genes displaying a >2-fold
increase or decrease were selected for analysis.
Immunoblotting and polymerase chain
reaction (PCR)
Immunoblot analysis was performed as described
previously (31). A primary
antibody specific for HIF-1α was purchased from Cell Signaling
Technology (Danvers, MA, USA). Anti-β-actin antibody was purchased
from Sigma. The expression level of VEGF mRNA was determined by
quantitative real-time (RT)-PCR using specific primers (forward,
5′-AAGGAGGAGGGCAGAATCAT-3′; reverse, 5′-GCTGTAGGAAGCTCATCTCT-3′).
RT-PCR analysis was performed using Line-Gene K software (Bioer
Technology Co. Ltd., Hangzhou, China).
Cell viability assay and FACS
analysis
Cell proliferation was determined using the WST-1
assay (EZ-Cytox Cell Viability Assay kit, ITSBIO, Korea) according
to the manufacturer’s instructions. In brief, 4×105
cells were seeded in 60-mm dishes. The cells were cultured for 24 h
and treated with the indicated concentrations of CoCl2
for 24 h. At each time-point, the kit solution was added to
cultured cells, which were incubated at 37°C for 30 min. Cell
viability was measured using an iMark microplate reader (Bio-Rad,
Hercules, CA, USA) at 450 nm by using a 620-nm reference filter.
Cell cycle distribution was determined using FACS analysis. U266
cells were collected and fixed by resuspending them in 70% ethanol
for 1 h, centrifuged, and washed in cold PBS. The cell pellets were
resuspended in PBS containing 50 μg/ml propidium iodide (Sigma) and
100 μg/ml RNase, incubated at room temperature for 30 min, and then
analyzed using a FACScalibur (BD Biosciences, San Jose, CA,
USA).
Statistical analysis
Statistical analysis was performed using the
χ2 test or Fisher’s exact test and Spearman rank
correlation coefficient analysis. p<0.05 was considered
significant.
Results and Discussion
We first examined whether CoCl2
stimulation induces hypoxia in MM U266 cells. The cells were
treated with the indicated concentrations of CoCl2 for 4
h, and then the level of hypoxia was determined by immunoblotting
against the hypoxia marker HIF-1α. As shown in Fig. 1A, the cellular protein levels of
HIF-1α were greatly increased in a CoCl2
concentration-dependent manner. Further, to test whether the
accumulated HIF-1α is functionally active in the cells, the
expression level of a well-known HIF-1α target gene, VEGF,
was verified by RT-PCR using its specific primers (see Materials
and methods). As shown in Fig.
1B, the mRNA expression levels of VEGF were up-regulated by
CoCl2 stimulation in U266 cells. Therefore, the
hypoxia-mimetic agent CoCl2 induced hypoxia in U266
cells.
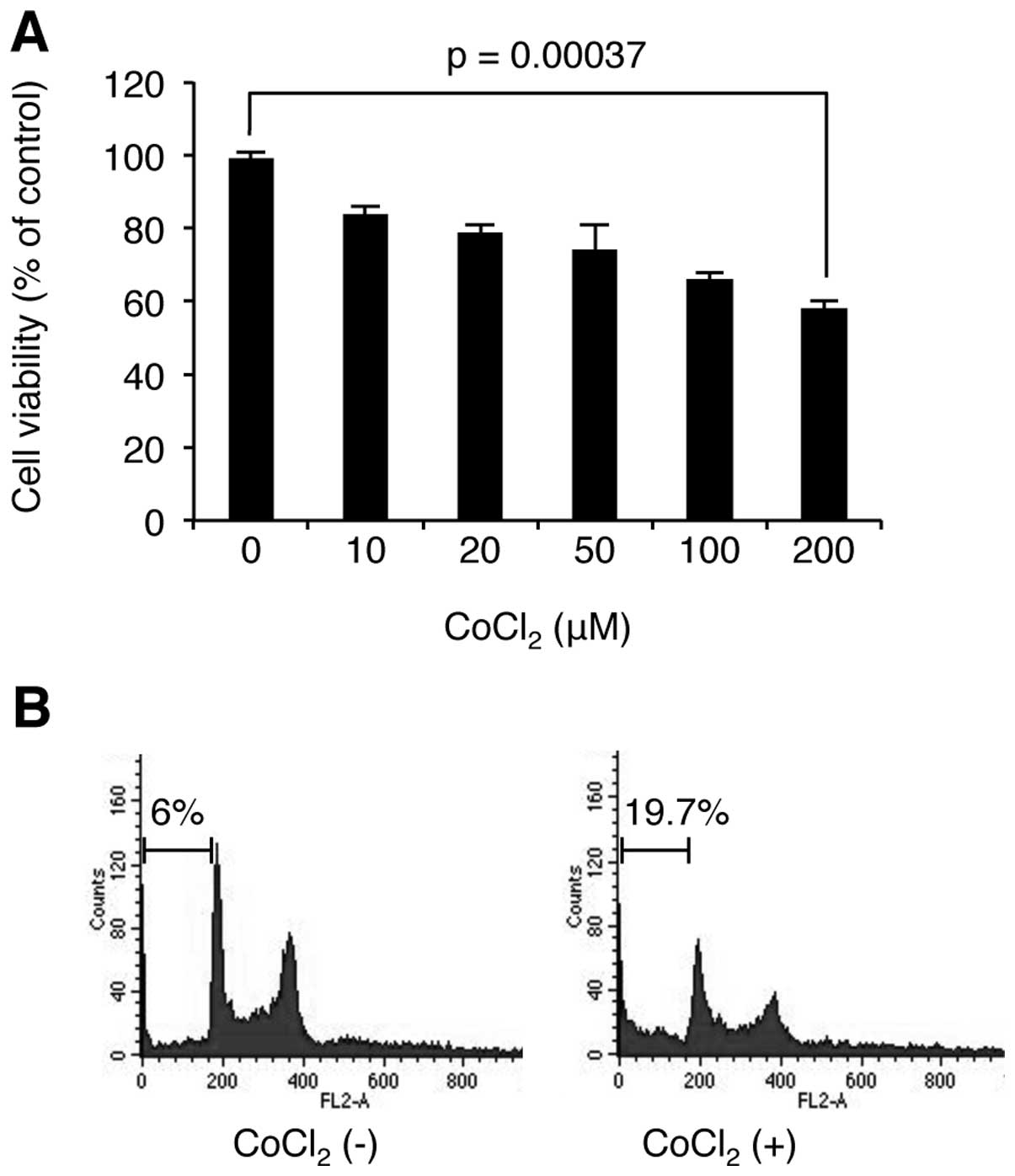
As hypoxia has dual roles in cell proliferation and
cell death, we examined the effect of hypoxia on MM cell
proliferation. U266 cells were stimulated with CoCl2 at
the indicated concentrations for 24 h, and then WST-1-based cell
viability assay was used to determine the level of cell
proliferation. As shown in Fig.
2, CoCl2 decreased cell viability in a
concentration-dependent manner. We next examined whether the loss
of cell viability is related to cell cycle arrest or apoptosis.
Interestingly, FACS analysis revealed that CoCl2
stimulation induces apoptosis, but not cell cycle arrest. These
results indicate that the hypoxia induced cell death in U266
cells.
Although hypoxia alters cell type-specific gene
expression patterns (32), its
effects have been not fully studied in MM cells. Therefore, we
analyzed mRNA profiles using Illumina Human HT-12 v4.0 Beadchip
kits in hypoxia-stimulated and control U266 cells. A total of
47,000 human mRNAs were selected to analyze gene expression
profiles. Those human genes were continually filtered with FLAG to
obtain more defined data using Agilent GeneSpring GX 11 software.
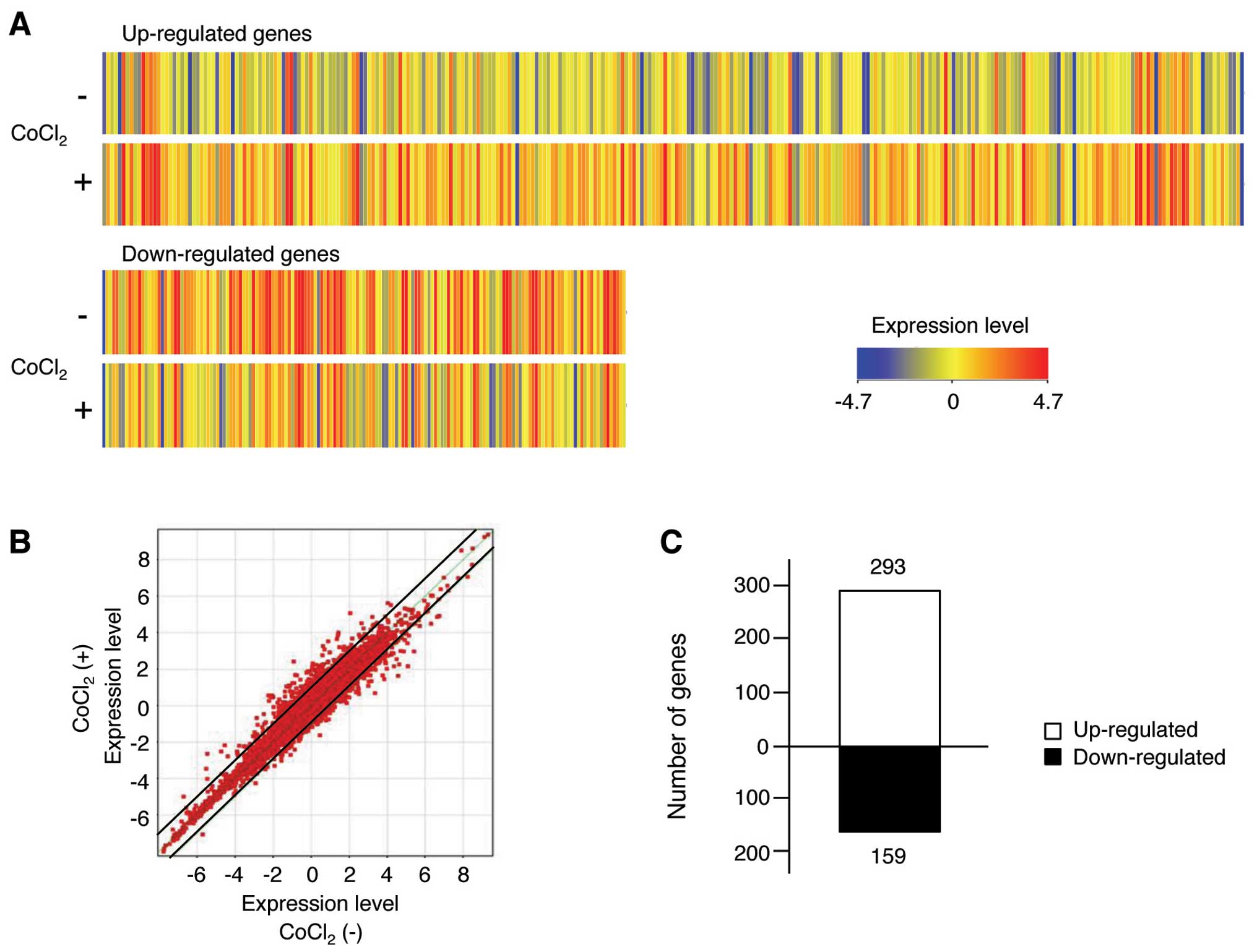
We selected total 452 human mRNAs as a result of FLAG filtration,
and the genes displaying >2-fold differences in expression
between the control and CoCl2-treated cells are shown in
Fig. 3A. In addition, the mRNA
microarray was visualized on a scatter plot (Fig. 3B). The data of the scatter blot
demonstrated that many genes displayed >2-fold differences in
expression levels between control and CoCl2-treated
cells, as indicated by their distance from the diagonal line. The
numbers of up- and downregulated mRNAs exhibiting >2-fold
changes were derived from the expression patterns in each group. We
found that 293 up-regulated and 159 down-regulated mRNA displayed
meaningful changes in their transcription profiles in response to
CoCl2 treatment (Fig.
3C). The 50 genes for which their expression was most strongly
altered by CoCl2 are listed in Table I. To further analyze the changes
in gene expression patterns, we sorted the genes displaying
>2-fold changes in expression after CoCl2 treatment
into several groups according to common features such as homology
or biochemical activity by using the Gene Set Enrichment Analysis
bioinformatics tool (www.broadiinstitute.org/gsea), and we revealed that
the sorted gene sets were categorized into transcription factors,
cell differentiation markers, cytokines and growth factors, protein
kinases, tumor suppressors, and oncogenes (Table II). Moreover, we further
identified the cellular process pattern of CoCl2-induced
gene expression changes by using the gene ontology database.
Although the analysis of cellular process pattern was firstly
performed using genes with >2-fold changes in expression, the
pattern results were not sufficient for elucidation because the
p-values of each category were >0.05; therefore, the analysis
was re-performed on genes with >1.5-fold changes in expression,
and the patterns with p-values <0.05 were selected. As shown in
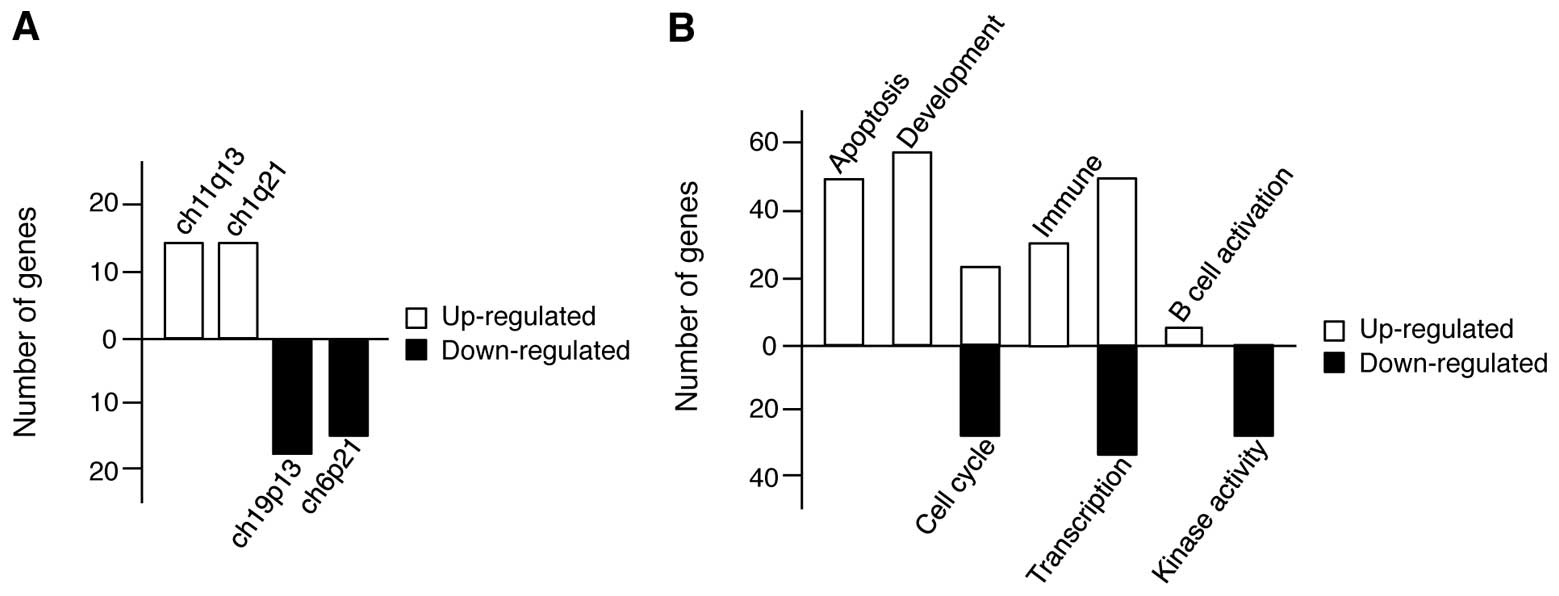
Fig. 4A, we first identified the
genomic region responsible for gene expression changes induced in
response to CoCl2 treatment in U266 cells by sorting the
genes according to their chromosomal positions and revealed that
the regions within ch11q13 and ch11q21 (up-regulated genes) and
ch19p13 and ch6p21 (down-regulated genes) were the top two
chromosome regions. Next, we grouped the genes according to
cellular process and found that the up-regulated genes were related
to apoptosis, development, cell cycle regulation, immunity,
transcription, and B cell activation; however, the downregulated
genes were related to cell cycle regulation, transcription, and
kinase activity (Fig. 4B). In
particular, there are more changes among genes associated with both
apoptosis and development, implying that in MM, hypoxia exerts both
antitumor effects, and positively regulates cell proliferation
pathways, although cell viability was decreased in hypoxic
conditions.
 | Table I.Top 50 genes altered by treatment
with cobalt chloride.a |
Table I.
Top 50 genes altered by treatment
with cobalt chloride.a
Up-regulated genes
| Down-regulated
genes
|
|---|
| Gene name | F.C | Gene name | F.C | Gene name | F.C | Gene name | F.C |
|---|
| OKL38 | 10.5 | MT1A | 4.19 | HEMGN | 7.56 | XK | 2.69 |
| RN7SK | 8.34 | C17ORF91 | 4.12 | RHOXF2B | 5.29 | EIF2AK2 | 2.66 |
| EMP1 | 7.15 | RAB7B | 4.07 | TSPAN32 | 5.12 | ACSM3 | 2.61 |
| TXNRD1 | 7.14 | MT1X | 4.01 | RHOXF2 | 5.00 | VEGFB | 2.60 |
| CDK5RAP2 | 6.40 | CTH | 3.94 | TESC | 4.52 | SLC10A4 | 2.60 |
| SRXN1 | 6.35 | SLC3A2 | 3.83 | ARHGAP22 | 4.11 | BOLA3 | 2.56 |
| C12ORF48 | 6.33 | UGDH | 3.81 | KISS1R | 3.76 | C5ORF13 | 2.53 |
| AGPAT9 | 6.03 | ARRDC4 | 3.77 | GYPA | 3.75 | GAD1 | 2.48 |
| NDRG1 | 5.90 | SLCO2B1 | 3.74 | GYPE | 3.67 | HADH | 2.48 |
| UPP1 | 5.79 | NOTCH1 | 3.52 | RHAG | 3.32 | MYB | 2.47 |
| CTSL1 | 5.74 | MT1E | 3.48 | GAD1 | 3.24 | SPR | 2.47 |
| SLC7A11 | 5.44 | CD53 | 3.38 | STAT5A | 3.22 | TFDP2 | 2.46 |
| CD44 | 5.32 | SERPINE2 | 3.40 | CA1 | 3.21 | METTL7A | 2.45 |
| FLNB | 5.23 | ITPR1 | 3.37 | SCARB1 | 3.13 | ATP5G1 | 2.45 |
| INHBE | 5.07 | RN5S9 | 3.32 | ELOVL6 | 3.05 | C4ORF18 | 2.43 |
| GCLM | 4.78 | DCUN1D3 | 3.28 | NME4 | 3.05 | NMRAL1 | 2.42 |
| FAM129B | 4.78 | ISG20 | 3.25 | NFE2 | 3.02 | BEX4 | 2.42 |
| BCL6 | 4.74 | CEBPB | 3.24 | CCDC34 | 2.99 | CYP3A5 | 2.42 |
| EID3 | 4.70 | PRSS2 | 3.22 | ATF5 | 2.96 | CFD | 2.42 |
| MT2A | 4.66 | KLF6 | 3.18 | KCNH2 | 2.94 | SLC1A3 | 2.42 |
| DUSP5 | 4.62 | PHLDA1 | 3.16 | STS-1 | 2.83 | SOX21 | 2.41 |
| XIRP1 | 4.59 | PAEP | 3.13 | ACPP | 2.82 | RELN | 2.41 |
| OSGIN1 | 4.31 | KIAA1666 | 3.12 | ZNF121 | 2.73 | SPIN4 | 2.40 |
| SLC3A2 | 4.28 | IER5 | 3.10 | GATA1 | 2.73 | EVL | 2.39 |
| ADM | 4.24 | LGMN | 3.09 | LIPH | 2.71 | PIK3R2 | 2.39 |
 | Table II.Genes sharing a common feature such
as homology or biochemical activity.a |
Table II.
Genes sharing a common feature such
as homology or biochemical activity.a
| Cytokines and
growth factors | Transcription
factors | Homeo-domain
proteins | Cell
differentiation markers | Protein
kinases | Translocated cancer
genes | Oncogenes | Tumor
suppressors |
|---|
| Tumor
suppressor | 0 | 1 (1/0) | 0 | 0 | 0 | 0 | 0 | 2 (1/1) |
| Oncogenes | 0 | 5 (2/3) | 0 | 0 | 1 (1/0) | 8 (6/2) | 9 (6/3) | |
| Translocated cancer
genes | 0 | 4 (2/2) | 0 | 0 | 1 (1/0) | 8 (6/2) | | |
| Protein
kinases | 0 | 0 | 0 | 0 | 10 (6/4) | | | |
| Cell
differentiation markers | 1 (1/0) | 0 | 0 | 18 (14/4) | | | | |
| Homeodomain
proteins | 0 | 0 | 2 (0/2) | | | | | |
| Transcription
factors | 0 | 33 (18/15) | | | | | | |
| Cytokines and
growth factors | 15 (12/3) | | | | | | | |
Although both control BM and myeloma-infiltrated BM
are hypoxic, the hypoxic level in myelomatous BM was revealed to be
much lower than that in native BM, as confirmed by the lower
expression level of HIF-1α in myelomatous BM than in native BM
(29). These results imply that
BM hypoxia is decreased during the progression of myeloma.
Interestingly, using the 5T2 multiple myeloma mouse model, native
BM hypoxia induced both apoptotic cell death of CD45− MM
cells and exerted tumor-initiating effects on CD45+ MM
cells (29). Additionally,
blocking angiogenesis in this model restores the native BM hypoxia
and results in apoptosis and suppressed proliferation of the MM
cells (29). Furthermore, VEGF, a
key angiogenic factor and a HIF-1α-responsive gene, is induced in
CD45− 5T2MM cells but not in CD45+ 5T2MM
cells (29). These data indicate
that although BM hypoxia decreased the proliferation of
CD45− MM cells, angiogenesis was induced, thus
permitting outgrowth of the CD45− MM cells. Similarly,
the microarray data in this study revealed that up-regulated genes
were associated with both apoptosis and development. The MM U266
cells used in this study were CD45−. Therefore, although
hypoxia induces U266 cell death by up-regulating apoptosis-related
gene expression, resistance mechanisms for hypoxia were also
induced via the up-regulation of development-related genes
including VEGF and CD44.
In the microarray data, oxidative stress induced
growth inhibitor 1 (OKL38) was the gene most strongly up-regulated
by CoCl2 treatment (fold change = 10.52). OKL38 is a
tumor suppressor gene, and its overexpression inhibits tumor cell
growth and induces apoptosis (33,34). Although the effect in hypoxia on
OKL38 has not been researched, hypoxia-mediated cell death could be
dependent on OKL38 expression. In addition, CD44 was highly
up-regulated by CoCl2 in U266 cells (fold change =
5.32). CD44 is an antigen glycoprotein that plays an important role
in aspects of cancer progression including growth promotion,
angiogenesis, and metastasis (35); however, its cellular function in
hypoxia has not yet been elucidated. Therefore, the hypoxia-induced
up-regulation of CD44 in U266 cells indicates that CD44 is a novel
key protein controlling hypoxia resistance mechanisms including
angiogenesis in MM cells.
In conclusion, we first examined the diversity of
gene expression responses to hypoxia in MM cells and found
significant differences in gene expression. Additionally, although
hypoxia induced apoptosis, the gene expression patterns indicated
the activation of both antitumor and hypoxia resistance processes.
Therefore, our results may provide a useful approach to
understanding cellular responses to hypoxia.
Acknowledgements
We are grateful to all members of our
research group for their support and advice regarding this study.
This study resulted from the Konkuk University Research Support
Program.
References
|
1.
|
X LuY KangHypoxia and hypoxia-inducible
factors: master regulators of metastasisClin Cancer
Res1659285935201010.1158/1078-0432.CCR-10-136020962028
|
|
2.
|
AL HarrisHypoxia, a key regulatory factor
in tumour growthNat Rev Cancer23847200210.1038/nrc70411902584
|
|
3.
|
GL SemenzaTargeting HIF-1 for cancer
therapyNat Rev Cancer3721732200310.1038/nrc1187
|
|
4.
|
R SullivanCH GrahamHypoxia-driven
selection of the metastatic phenotypeCancer Metastasis
Rev26319331200710.1007/s10555-007-9062-217458507
|
|
5.
|
GP GuptaDX NguyenAC ChiangMediators of
vascular remodelling co-opted for sequential steps in lung
metastasisNature446765770200710.1038/nature0576017429393
|
|
6.
|
KG ShyuFL HsuMJ WangBW WangS
LinHypoxia-inducible factor 1alpha regulates lung adenocarcinoma
cell invasionExp Cell
Res31311811191200710.1016/j.yexcr.2007.01.01317335808
|
|
7.
|
T ImaiA HoriuchiC WangHypoxia attenuates
the expression of E-cadherin via up-regulation of SNAIL in ovarian
carcinoma cellsAm J
Pathol16314371447200310.1016/S0002-9440(10)63501-814507651
|
|
8.
|
MH YangMZ WuSH ChiouDirect regulation of
TWIST by HIF-1alpha promotes metastasisNat Cell
Biol10295305200810.1038/ncb169118297062
|
|
9.
|
B KrishnamacharyD ZagzagH
NagasawaHypoxiainducible factor-1-dependent repression of
E-cadherin in von Hippel-Lindau tumor suppressor-null renal cell
carcinoma mediated by TCF3, ZFHX1A, and ZFHX1BCancer
Res6627252731200610.1158/0008-5472.CAN-05-371916510593
|
|
10.
|
M KoshijiY KageyamaEA PeteI HorikawaJC
BarrettLE HuangHIF-1alpha induces cell cycle arrest by functionally
counteracting MycEMBO
J2319491956200410.1038/sj.emboj.760019615071503
|
|
11.
|
WG AnM KanekalMC SimonE MaltepeMV
BlagosklonnyLM NeckersStabilization of wild-type p53 by
hypoxiainducible factor
1alphaNature392405408199810.1038/329259537326
|
|
12.
|
K GuoG SearfossD KrolikowskiHypoxia
induces the expression of the pro-apoptotic gene BNIP3Cell Death
Differ8367376200110.1038/sj.cdd.440081011550088
|
|
13.
|
Y HuangKM DuZH XueCobalt chloride and low
oxygen tension trigger differentiation of acute myeloid leukemic
cells: possible mediation of hypoxia-inducible
factor-1alphaLeukemia1720652073200310.1038/sj.leu.2403141
|
|
14.
|
JY JungWJ KimInvolvement of mitochondrial-
and Fas-mediated dual mechanism in CoCl2-induced
apoptosis of rat PC12 cellsNeurosci
Lett3718590200410.1016/j.neulet.2004.06.06915519734
|
|
15.
|
Z JiG YangS ShahzidiInduction of
hypoxia-inducible factor-1alpha overexpression by cobalt chloride
enhances cellular resistance to photodynamic therapyCancer
Lett244182189200610.1016/j.canlet.2005.12.01016427735
|
|
16.
|
SG LeeH LeeHM RhoTranscriptional
repression of the human p53 gene by cobalt chloride mimicking
hypoxiaFEBS
Lett507259263200110.1016/S0014-5793(01)02989-111696352
|
|
17.
|
A VengellurBG WoodsHE RyanRS JohnsonJJ
LaPresGene expression profiling of the hypoxia signaling pathway in
hypoxia-inducible factor 1alpha null mouse embryonic
fibroblastsGene
Expr11181197200310.3727/00000000310874906214686790
|
|
18.
|
SJ YangJ PyenI LeeH LeeY KimT KimCobalt
chloride-induced apoptosis and extracellular signal-regulated
protein kinase 1/2 activation in rat C6 glioma cellsJ Biochem Mol
Biol37480486200410.5483/BMBRep.2004.37.4.48015469737
|
|
19.
|
J ArayaM MaruyamaA InoueInhibition of
proteasome activity is involved in cobalt-induced apoptosis of
human alveolar macrophagesAm J Physiol Lung Cell Mol
Physiol283L849858200210.1152/ajplung.00422.200112225962
|
|
20.
|
W ZouM YanW XuCobalt chloride induces PC12
cells apoptosis through reactive oxygen species and accompanied by
AP-1 activationJ Neurosci
Res64646653200110.1002/jnr.111811398189
|
|
21.
|
HJ KimSJ YangYS KimTU KimCobalt
chloride-induced apoptosis and extracellular signal-regulated
protein kinase activation in human cervical cancer HeLa cellsJ
Biochem Mol
Biol36468474200310.5483/BMBRep.2003.36.5.46814536030
|
|
22.
|
JP PiretD MottetM RaesC
MichielsCoCl2, a chemical inducer of hypoxia-inducible
factor-1, and hypoxia reduce apoptotic cell death in hepatoma cell
line HepG2Ann NY Acad Sci973443447200212485908
|
|
23.
|
XH LiuA KirschenbaumS YaoUp-regulation of
vascular endothelial growth factor by cobalt chloride-simulated
hypoxia is mediated by persistent induction of cyclooxygenase-2 in
a metastatic human prostate cancer cell lineClin Exp
Metastasis17687694199910.1023/A:1006728119549
|
|
24.
|
T Van LieshoutJ StaniszV EspirituM
RichardsonG SinghA hypoxic response induced in MatLyLu cells by
cobalt chloride results in an enhanced angiogenic response by the
chick chorioallantoic membraneInt J Oncol237457502003
|
|
25.
|
N LuH ZhouYH LinZQ ChenY PanXJ LiOxidative
stress mediates CoCl(2)-induced prostate tumour cell adhesion: role
of protein kinase C and p38 mitogen-activated protein kinaseBasic
Clin Pharmacol
Toxicol1014146200710.1111/j.1742-7843.2007.00074.x17577315
|
|
26.
|
MH RyuJH ParkJE ParkJ ChungCH LeeHR
ParkCobalt chloride stimulates phosphoinositide 3-kinase/Akt
signaling through the epidermal growth factor receptor in oral
squamous cell carcinomaBiocell341521201020506627
|
|
27.
|
RA KyleSV RajkumarMultiple
myelomaBlood11129622972200810.1182/blood-2007-10-07802218332230
|
|
28.
|
K ParmarP MauchJA VergilioR SacksteinJD
DownDistribution of hematopoietic stem cells in the bone marrow
according to regional hypoxiaProc Natl Acad Sci
USA10454315436200710.1073/pnas.070115210417374716
|
|
29.
|
K AsosinghH De RaeveM de RidderRole of the
hypoxic bone marrow microenvironment in 5T2MM murine myeloma tumor
progressionHaematologica90810817200515951294
|
|
30.
|
J HuDR HandisidesE Van
ValckenborghTargeting the multiple myeloma hypoxic niche with
TH-302, a hypoxia-activated
prodrugBlood11615241527201010.1182/blood-2010-02-26912620530289
|
|
31.
|
SY KimS BaeKH ChoiS AnHydrogen peroxide
controls Akt activity via ubiquitination/degradationOncol
Rep2615611566201121874266
|
|
32.
|
JT ChiZ WangDS NuytenGene expression
programs in response to hypoxia: cell type specificity and
prognostic significance in human cancersPLoS
Med3e47200610.1371/journal.pmed.003004716417408
|
|
33.
|
T WangD XiaN LiBone marrow stromal
cell-derived growth inhibitor inhibits growth and migration of
breast cancer cells via induction of cell cycle arrest and
apoptosisJ Biol
Chem28043744382200510.1074/jbc.M40870820015569677
|
|
34.
|
H HuynhCY NgCK OngKB LimTW ChanCloning and
characterization of a novel pregnancy-induced growth inhibitor in
mammary
glandEndocrinology14236073615200110.1210/endo.142.8.829711459809
|
|
35.
|
R MarhabaM ZollerCD44 in cancer
progression: adhesion, migration and growth regulationJ Mol
Histol35211231200410.1023/B:HIJO.0000032354.94213.6915339042
|