Introduction
Human cytomegalovirus (HCMV) is an ubiquitous
β-herpesvirus, which is able to maintain a persistent or latent
infection during the host’s lifetime. It always displays
asymptomatic infection in healthy adults, and also causes
significant morbidity and mortality in newborn and
immunocompromised patients (1).
HCMV genomic DNA comprises ~230 kb and potentially encodes ~165
open reading frames (ORFs) (2).
Compared with the genome of the prototype laboratory strain, AD169,
genomes of low-passage HCMV clinical isolates contain the UL/b′
region, including ORFs UL133-UL151, considered as a critical
candidate cluster to clinical pathogenesis (3). Although the UL/b′ region is not
essential to viral growth or replication (4), products of this region, including
UL141, UL142 and UL144 have been experimentally identified to aid
in viral escape from immune surveillance through interactions with
cellular molecules (5–10).
A number of previous studies have demonstrated that
a species of regulatory RNA molecules, known as microRNAs (miRNAs),
are encoded in non-coding regions and are involved in the
regulation of diverse cellular processes such as development,
differentiation, cell cycle, apoptosis and immune responses
(11–13). miRNAs display their
post-transcriptional regulation through RNA interference by 2
different mechanisms (14–17).
Binding within the first 10 bases of a miRNA, particularly within
bases 2 to 7 at the 5′ end of the miRNA known as the seed region,
is considered of particular importance (18–20).
HCMV encodes at least 14 miRNAs expressed in 11
unique RNA structures (14,21). Unlike other miRNAs expressed in
herpesviruses in the clustered form, 14 miRNAs encoded by the HCMV
are scattered in the whole genome. The target transcripts and
regulatory functions of HCMV miRNAs remain to be elucidated. Only
some HCMV miRNAs, such as hcmv-miR-UL112 (22,23) and hcmv-miR-US4-1 (24) have been verified experimentally to
be involved in viral pathogenesis by inhibiting host immune
molecules or regulating viral proteins. As the only miRNA expressed
in the HCMV UL/b′ region, identifying the target genes and
regulatory functions of hcmv-miR-UL148D may provide further insight
into the viral pathogenesis of clinical isolates (25,26).
In this study, we identified and characterized the
functional targets of hcmv-miR-UL148D using a hybrid-PCR approach
in combination with luciferase report assays and western blot
analysis. The cellular target gene, human immediate early gene X-1
(IEX-1), was identified as the target of hcmv-miR-UL148D. The
suppression of IEX-1 expression by hcmv-miR-UL148D in an in
vitro system was primarily investigated for the anti-apoptotic
effect on host cells.
Materials and methods
Cell line and cell culture
Human embryonic lung fibroblast (HELF) and human
embryonic kidney 293 (HEK293) cells were obtained from Shanghai
Biology Institution (Shanghai, China). The HELF and HEK293 cells
were cultured in RPMI-1640 or DMEM supplemented with L-glutamine
and pennicilin/streptomycin with 10% FBS at 37°C and 5%
CO2 in a humidified incubator.
Virus, RNA and complementary DNA (cDNA)
preparation
A HCMV clinical strain, termed Han, was isolated
from a urine sample of an infant (<5 months old) hospitalized in
Shengjing Hospital of China Medical University, Shenyang, China.
The strain has been passaged 6 times in HELF cells before being
used in this study.
For preparation of immediate early (IE) RNA,
cycloheximide (100 μg/ml) was added to the culture medium 1 h prior
to infection. The cells were harvested at 24 h post-infection
(hpi). For early (E) RNA, phosphonoacetic acid (100 μg/ml) was
added immediately after infection and the cells were harvested at
48 hpi. Late (L) RNA and mock-infected cellular RNA were derived
from infected and uninfected cells, respectively, cultured in
parallel and harvested at 96 hpi.
Total RNA was isolated from ~1×107
HCMV-infected HELF cells by TRIzol reagent (Invitrogen/Life
Technologies, Shanghai, China) using the classic phenol/chloroform
methods, and then dissolved in 50 μl RNase-free H2O,
using the TURBO DNA-free™ kit (Ambion/Life Technologies, Austin,
TX, USA). The integrity of the RNA was analyzed by 1% agarose gel
electrophoresis.
cDNA preparations of HCMV non-infected HELF cells
and HCMV infected HELF cells from the IE, E and L phases were
carried out using the 3′-Full RACE Core Set kit (Takara, Dalian,
China). Reverse transcription was performed with 1 μg RNA and an
oligo(dT)3 site adaptor, which was provided in the kit, introducing
a special sequence into the 5′-terminal of the cDNA, according to
instructions provided by the manufacturer.
Hybrid-PCR and BLAST for candidate target
genes
Target genes of hcmv-miR-UL148D were screened using
the hybrid-PCR method as described in a previous study of ours
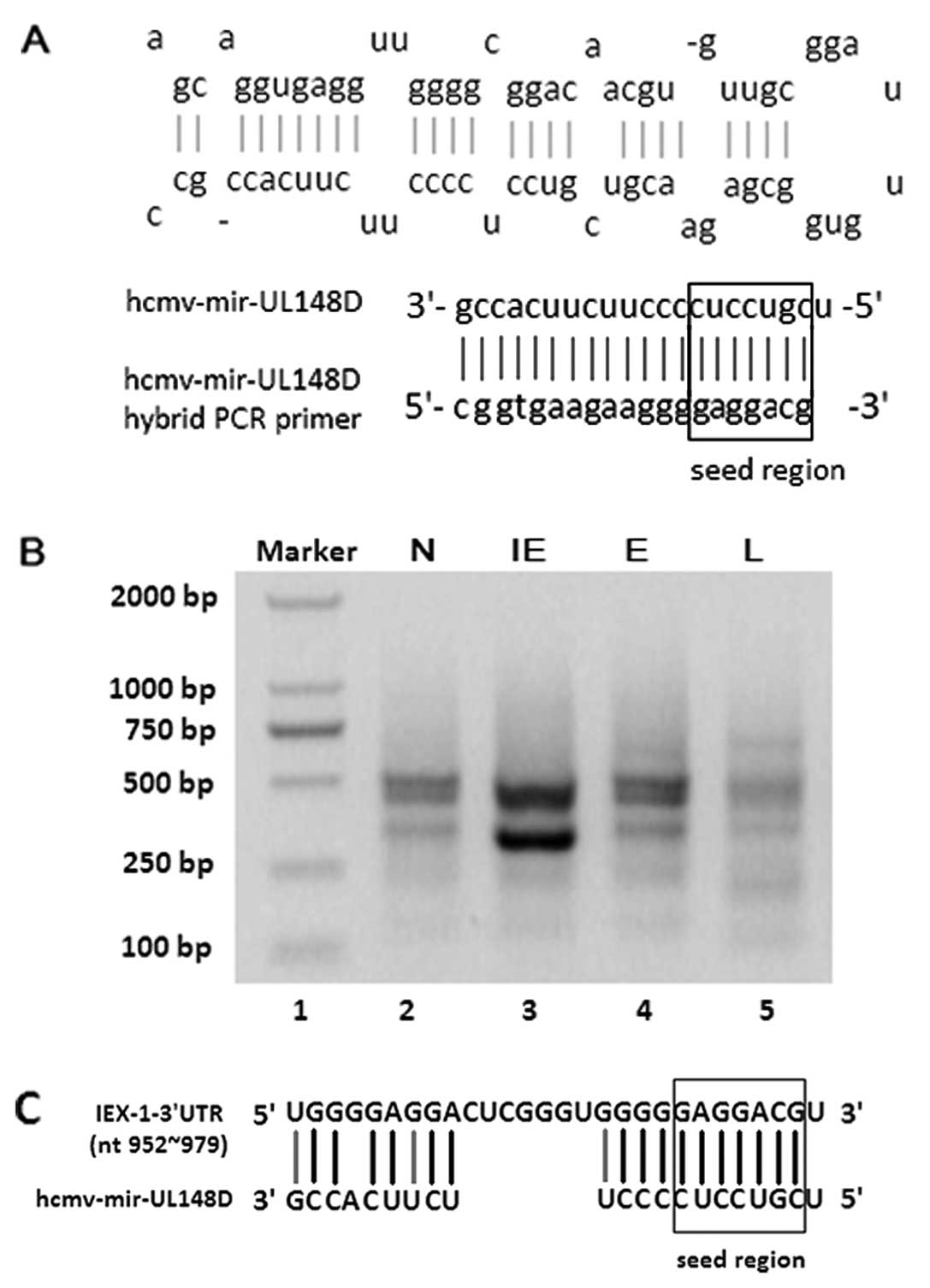
(27). The hybrid-PCR primer of
hcmv-miR-UL148D, A1, was designed according to the reverse sequence
of hcmv-miR-UL148D (Fig. 1A). A
semi-nested PCR was performed using primer A1 as the forward primer
and the primer A2/A3 provided in the 3′-Full RACE Core Set kit
(Takara) as the reverse primer. All primers used in this study are
listed in Table I. The purified
PCR products were T-A cloned and the clones were randomly selected
for sequencing analysis using the ABI PRISM 3700 DNA sequencer
(Applied Biosystem/Life Technologies, Carlsbad, CA, USA).
 | Table INames and sequences of primers used
in this study. |
Table I
Names and sequences of primers used
in this study.
| Primer names | Primer
sequences | Use of the
primers |
|---|
| A1 |
5′-gcggtgaagaaggggaggacg-3′, | Hybrid-PCR |
| A2 |
5′-taccgtcgttccactagtgattt-3′ | |
| A3 |
5′-cgcggatcctccactagtgatttcactatacg-3′ | |
| B1 |
5′-gcggatccgattctccagggaacgacag-3′ |
pSilence-hcmv-mir-UL148D |
| B2 |
5′-gcaagcttacaaccgccgctattcttt-3′ | |
| C1 |
5′-ggcggtgaagaagggagacggcacgttctcgccacga-3′ |
pSilence-hcmv-mir-UL148D-mutant |
| C2 |
5′-tcgtggcgagaacgtgccgtctcccttcttcaccgcc-3′ | |
| D1 |
5′-gcactagtctgtgactccccgcactc-3′ |
pMIR-IEX-1-3′UTR |
| D2 |
5′-gcaagcttcacagtagacagacggagttga-3′ | |
| E1 |
5′-aggactcgggtggggagacggctcccggctgggatga-3′ |
pMIR-IEX-1-3′UTR-mutant |
| E2 |
5′-tcatcccagccgggagccgtctccccacccgagtcct-3′ | |
| F1 |
5′-cgcggatccatgtgtcactctcgcagctgcc-3′ |
pBI-IEX-1-3′UTR |
| F2 |
5′-cccaagctttgtgttcacagaacatactaggc-3′ | |
| G1 |
5′-cccaaggcttatgtgtcactctcgcagctgcc-3′ |
pcDNA3.1-IEX-1-3′UTR |
| G2 |
5′-cgcggatcctgtgttcacagaacatactaggc-3′ | |
The candidate target sequences of hcmv-miR-UL148D
were blasted on line (http://www.ncbi.nlm.nih.gov/blast) using the mRNA
specific sequences located between primer A1 (i.e., the binding
site) and poly A obtained from hybrid-PCR. Putative target genes
were identified according to the following sequence criteria: i)
containing a sequence either completely complementary to the
hcmv-miR-UL148D seed region or with only one base unpaired; ii) the
hcmv-miR-UL148D binding site is located within the 3′ untranslated
region (3′UTR) or coding domains (CDS) of the putative target
genes; and iii) containing a poly A structure, which indicates that
the sequences originate from mRNA.
Plasmid construction and site-directed
mutagenesis
According to the sequence of the Han strain, the
primers for construction of the hcmv-miR-UL148D expression plasmid
(pSilence-hcmv-mir-UL148D) were designed as primer-B1/B2. The
purified PCR products of the hcmv-mir-UL148D encoding sequence were
inserted into the pSilencer 4.1 vector (Ambion/Life Technologies)
between the BamHI and the HindIII sites. The selected
clone was confirmed by sequencing analysis.
As the control of the hcmv-miR-UL148D expression
plasmid, the plasmid with a full-length mutation at the seed region
of hcmv-miR-UL148D was obtained using site-directed mutagenesis.
The mutant plasmid was firstly synthesized based on a PCR
amplification with 2 primers (C1/C2) designed for the specific
nucleotide mutation, pfu DNA polymerase (Takara), and the
wild-type pSilence-hcmv-mir-UL148D as the template. The PCR
products were then treated with methylase DpnI at 37°C for 1
h to exclude the original plasmid of pSilence-hcmv-mir-UL148D,
methylated during replication in Escherichia coli. Finally,
the mutant plasmid was transformed into Escherichia coli
DH5a to repair the nicks in the mutant plasmid resulting from the
synthesized step. Clones were randomly selected, and the mutant
plasmid, pSilence-hcmv-mir-UL148D-mutant, was confirmed by
sequencing analysis.
For dual luciferase report assays, the full-length
3′UTR sequence of IEX-1 was inserted into the pMIR vector at the
SpelI and the HindIII sites to construct
pMIR-IEX-1-3′UTR. The primers for amplification of the IEX-1-3′UTR
sequence were D1/D2. As an inner control, the
pMIR-IEX-1-3′UTR-mutant with the whole mutant binding site of the
hcmv-miR-UL148D seed region in the IEX-1-3′UTR was obtained by
site-directed mutageneisis as described above. The primer sequences
for the mutagenesis of the IEX-1-3′UTR sequence were E1/E2.
For western blot analysis, the expression plasmid,
pBI-IEX-1-3′UTR, was constructed by inserting the full-length IEX-1
cDNA with 3′UTR sequence into the pBI vector at the BamHI
and HindIII sites. The primer sequences for amplification of
the IEX-1 cDNA were F1/F2. The expression plasmid containing the
mutant 3′UTR sequence of IEX-1, pBI-IEX-1-3′UTR-mutant, was
obtained by site-directed mutagenesis using the same primers as
those used for the construction of the pMIR-IEX-1-3′UTR-mutant
plasmid.
For the apoptosis assay, the expression plasmid,
pcDNA3.1-IEX-1-3′UTR, was constructed by inserting the full-length
of IEX-1 cDNA with the 3′UTR sequence into the pcDNA3.1+
vector at the HindIII and BamHI sites. The primer
sequences were G1/G2. The control plasmid,
pcDNA3.1-IEX-1-3′UTR-mutant, was obtained by site-directed
mutagenesis using the primers, E1/E2, which were the same as those
used for the construction of the pMIR-IEX-1-3′UTR-mutant
plasmid.
Dual luciferase reporter assays
To evaluate the binding effect of hcmv-miR-UL148D to
the 3′UTR of IEX-1, a dual-luciferase reporter assay (Promega,
Madison, WI, USA) was carried out, with a pRL-TK plasmid as the
control for transfection efficiency.
A group of plasmids, pSilence-hcmv-miR-UL148D (600
ng), pMIR-IEX-1-3′UTR (100 ng) and pRL-TK (100 ng), was
co-transfected into the HEK293 cells using Lipofectamine 2000
(Invitrogen/Life Technologies) according to the manufacturer’s
instructions in a 24-well culture plate. The miRNA control,
consisting of pSilence-hcmv-miR-UL148D-mutant (600 ng),
pMIR-IEX-1-3′UTR (100 ng) and pRL-TK (100 ng), and the IEX-1-3′UTR
control, containing pSilence-hcmv-miR-UL148D (600 ng),
pMIR-IEX-1-3′UTR-mutant (100 ng) and pRL-TK (100 ng), were
transfected at the same time. After 48 h, the cells were harvested
for the detection of the luciferase activity using a luminometer
(Berthold Technologies, Oak Ridge, TN, USA) according to the
manufacturer’s instructions. The normalized firefly luciferase
activity to that of Renilla was used to evaluate the binding effect
of hcmv-miR-UL148D with the 3′UTR of IEX-1. The experiments were
carried out at least 3 times.
Western blot analysis
To detect the expression kinetics of the IEX-1
protein during HCMV infection, the HELF cells were incubated with
the Han strain and treated by cycloheximide and phosphonoacetic
acid, as described above. Cellular proteins were extracted from the
mock-infected and HCMV-infected cells at the IE, E and L stages
using cytoplasmic protein lysis buffer (50 mM Tris, PH 7.5, 10%
glycerol, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100 and protease and
phosphatase inhibitors). Protein concentrations were determined
using the BCA Protein Assay kit (P0012; Beyotime, Nantong, China).
Equal amounts of proteins were separated by sodium dodecyl
sulfate-polyacrylamide gel electrophoresis and transferred onto a
polyvinylidene fluoride membrane at 4°C. The membranes were
subsequently incubated with primary antibody to IEX-1 (1:1,000;
ab65152; Abcam, Cambridge, UK) or β-actin (1:1,000; SC-1615; Santa
Cruz Biotechnology, Inc., Santa Cruz, CA, USA), followed by
incubation with HRP-conjugated secondary antibody (1:2,000; Beijing
Zhongshan Biotechnology Co., Beijing, China). Luminescence was
visualized on a Bio-Rad image station.
In order to detect the effects of hcmv-mir-UL148D on
IEX-1 expression, the HEK293 cells in 60-mm culture plate were
co-transfected using Lipofectamine 2000 with the miRNA expression
plasmid, pSilence-hcmv-mir-UL148D, and the IEX-1 expression
plasmid, pBI-IEX-1-3′UTR, or its mutant plasmid,
pBI-IEX-1-3′UTR-mutant. At the same time, the HEK293 cells were
co-transfected with the IEX-1 expression plasmid, pBI-IEX-1-3′UTR,
and the miRNA expression plasmid, pSilence-hcmv-mir-UL148D, or its
mutant plasmid, pSilence-hcmv-mir-UL148D-mutant. The cells were
harvested at 48 h after transfection. The expression of IEX-1 was
detected by western blot analysis using primary antibodies to IEX-1
and green fluorescent protein (GFP), which was expressed by the pBI
vector and served as the internal control, separately.
Apoptosis assay
HEK293 cells cultured in a 6-well culture plate were
first transfected using Lipofectamine 2000 with the miRNA
expression plasmid, pSilence-hcmv-mir-UL148D (3,600 ng), or its
mutant plasmid, pSilence-hcmv-mir-UL148D-mutant (3,600 ng). After
24 h, the cells were further transfected with the IEX-1 expression
plasmid, pcDNA3.1-IEX-1-3′UTR (400 ng). The HEK293 cells
transfected with the miRNA expression plasmid,
pSilence-hcmv-mir-UL148D, or its mutant plasmid,
pSilence-hcmv-mir-UL148D-mutant, were further transfected with the
IEX-1 mutant expression plasmid, pcDNA3.1-IEX-1-3′UTR-mutant. Cells
were harvested at 48 h after transfection, and washed with PBS
buffer. The recovered cells were then analyzed for apoptosis using
the Annexin V-FITC apoptosis detection kit (c1063; Beyotime)
following the procedures outlined by the manufacturer on a flow
cytometer (FACSCalibur; BD Biosciences, Franklin Lakes, NJ,
USA).
Prediction of binding sites of HCMV
encoding miRNAs in IEX-1 using Biosoftware
RNAhybrid-Submission
To analyze whether IEX-1 can be regulated by other
miRNAs encoded by HCMV, a Biosoftware RNAhybrid-Submission
(http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/submission.html)
was used. The binding sites of HCMV encoding miRNAs in IEX-1 were
identified according to the following miRNA selection criteria: i)
>6 nts in its seed region were complementary to the IEX-1-3′UTR;
ii) the minimum free energy (mfe) of the binding was <-20
kcal/mol.
Statistical analysis
Data are expressed as the means ± SD. Statistical
analysis was carried out using the two-tailed, unpaired Student’s
t-test. A level of P<0.05 was considered to indicate a
statistically significant difference.
Results
Putative target genes of hcmv-miR-UL148D
screened by hybrid-PCR
Since miRNAs exhibit post-trancriptional regulatory
effects by binding to target mRNAs, the target mRNA sequences were
screened within mRNA-derived cDNA from HCMV-infected cells using
hybrid-PCR, which has been experimentally demonstrated to be
efficient for screening target genes of any known miRNAs (27). Two main products of
hcmv-mir-UL148D obtained from the HCMV-infected cells by hybrid-PCR
were ~300 and 500 bp in length. Of note, the ~300 bp fragment was
stronger in the IE phase of the HCMV-infected cells than in the E
and L phase cells, as well as in the non-infected cells (Fig. 1B).
The ~300 bp fragment was subsequently cloned by the
T-A clone method, and the inserts of 11 clones were successfully
sequenced. Using BLAST analysis, 9 of them contained sequences
homologous to the cellular gene, IEX-1 with the length of 297 bp
from 968 nt to the polyA signal of IEX-1 mRNA. The sequence of the
3′UTR of IEX-1 mRNA from 968 to 978 nt was completely complementary
to the 11 nt sequence of the hcmv-miR-UL148D seed region; while its
upward sequence was partially complementary to the other 10 nt
sequence in the 3′-terminal of hcmv-mir-UL148D (Fig. 1C). Therefore, IEX-1 was
preferentially considered to be the candidate target gene of
hcmv-miR-UL148D.
Binding ability of hcmv-mir-UL148D to
3′UTR of the IEX-1 mRNA
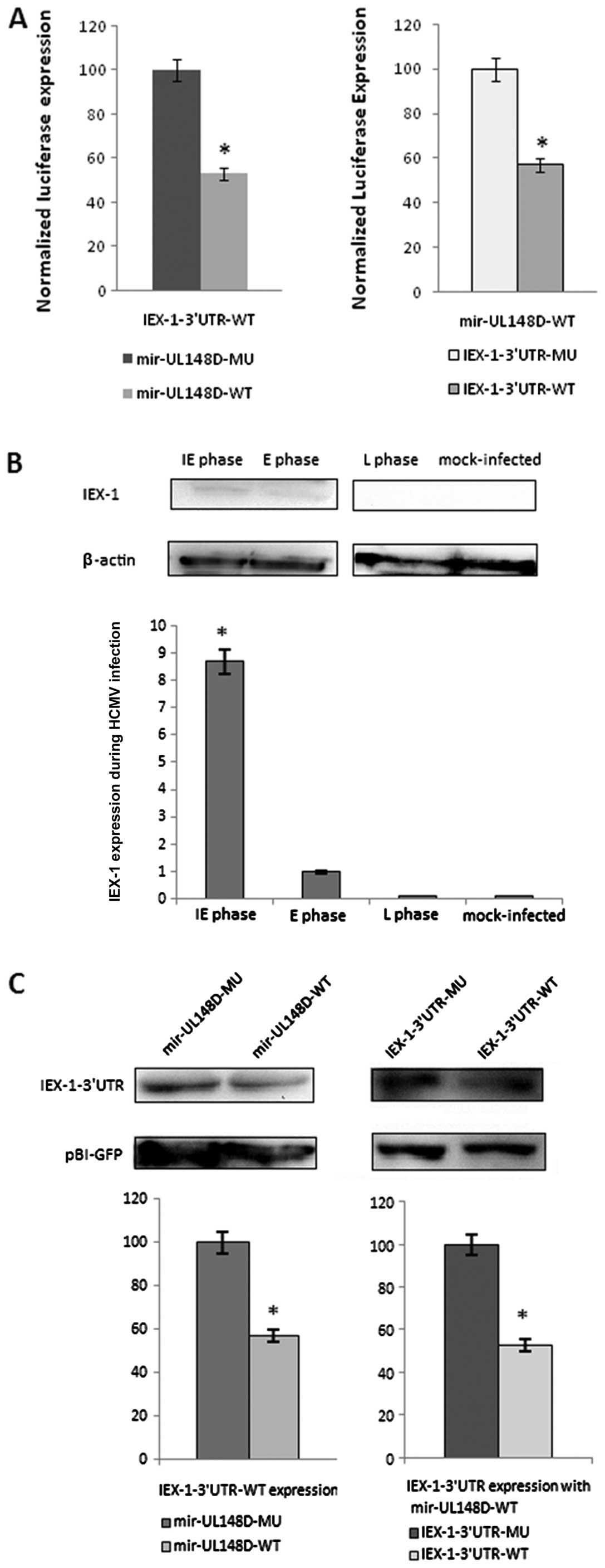
The entire wild-type and mutant 3′UTR of IEX-1 were
inserted downstream of the luciferase gene. The results of
dual-luciferase reporter assays showed that compared with the
mutant hcmv-miR-UL148D, hcmv-miR-UL148D suppressed the relative
luciferase activity of pMIR-IEX-1-3′UTR by ~47%; the relative
luciferase activity of pSilence-hcmv-miR-UL148D was suppressed ~40%
by the wild-type plasmid compared to the control
pMIR-IEX-1-3′UTR-mutant plasmid (Fig.
2A). These results suggest that the binding site at 952 to 979
nt of IEX-1-3′UTR may be specifically affected by
hcmv-miR-UL148D.
Expression kinetics of IEX-1 during HCMV
infection
To examine the downregulatory effect of
hcmv-mir-UL148D on the cellular gene, IEX-1, the expression
kinetics of IEX-1 during HCMV infection were primarily detected
using proteins from cells infected with the Han strain by western
blot analysis. In cells collected at the IE and E phase, a slight
IEX-1 expression band was detected; while in cells at the L phase
cells or mock-infected HELF cells, no IEX-1 expression was
detected. The expression level of IEX-1 in cells at the E phase was
only 10% of that in cells at the IE phase. This divergence in
expression levels suggested that IEX-1 expression was elevated
during HCMV infection at the IE phase, and was downregulated
gradually at the E and L phase (Fig.
2B).
Suppression of IEX-1 gene expression by
hcmv-miR-UL148D
In order to further evaluate the specific effect of
hcmv-mir-UL148D on the expression of IEX-1, an in vitro
system was utilized. HEK293 cells were transfected with IEX-1 and
hcmv-mir-UL148D. The western blot analysis results showed that
compared with mutant hcmv-miR-UL148D, hcmv-miR-UL148D suppressed
the expression of pBI-IEX-1-3′UTR ~43%; the expression was
suppressed ~47% compared with the expression of
pBI-IEX-1-3′UTR-mutant together with pSilence-hcmv-mir-UL148D
(Fig. 2C). These results suggest
that hcmv-miR-UL148D can functionally downregulate IEX-1
expression.
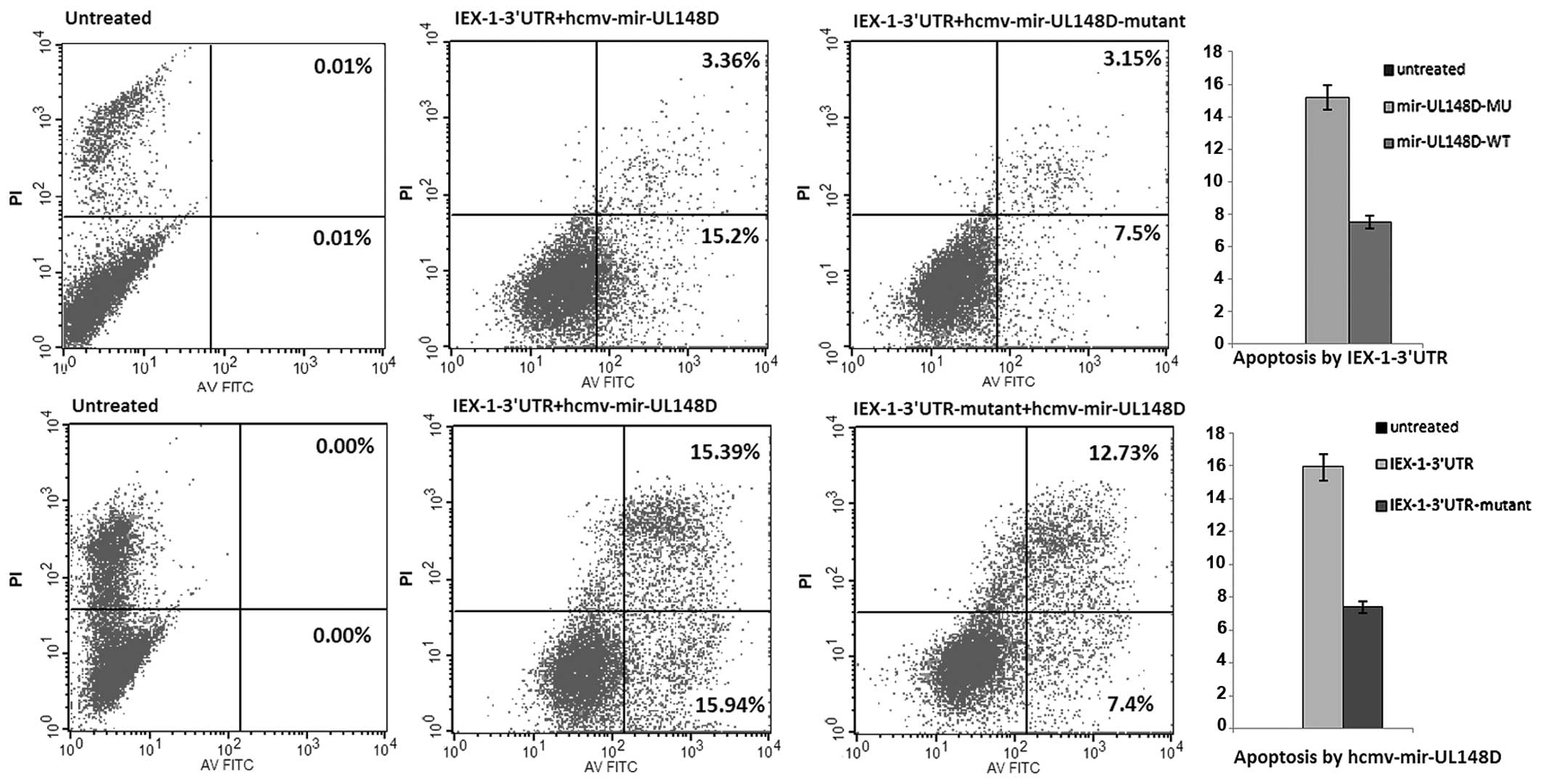
hcmv-miR-UL148D exerts anti-apoptotic
effects by downregulating IEX-1 expression
In order to identify the effects of hcmv-mir-UL148D
on cell apoptosis caused by ectopically expressed IEX-1, HEK293
cells were first transfected with pSilence-hcmv-mir-UL148D followed
by the IEX-1 expression plasmid, pcDNA3.1-IEX-1-3′UTR. The
percentage of apoptotic cells in the cells transfected with the
expression plasmids of hcmv-mir-UL148D and IEX-1 was decreased ~51%
compared with the control cells expressing mutant hcmv-mir-UL148D
and IEX-1 with a normal 3′UTR; the percentage was decreased ~54%
compared with the cells expressing hcmv-mir-UL148D and IEX-1 with a
mutant 3′UTR (Fig. 3). These
results indicate that hcmv-mir-UL148D exerts anti-apoptotic effects
by downregulating the elevated expression of the cellular gene,
IEX-1, during HCMV infection.
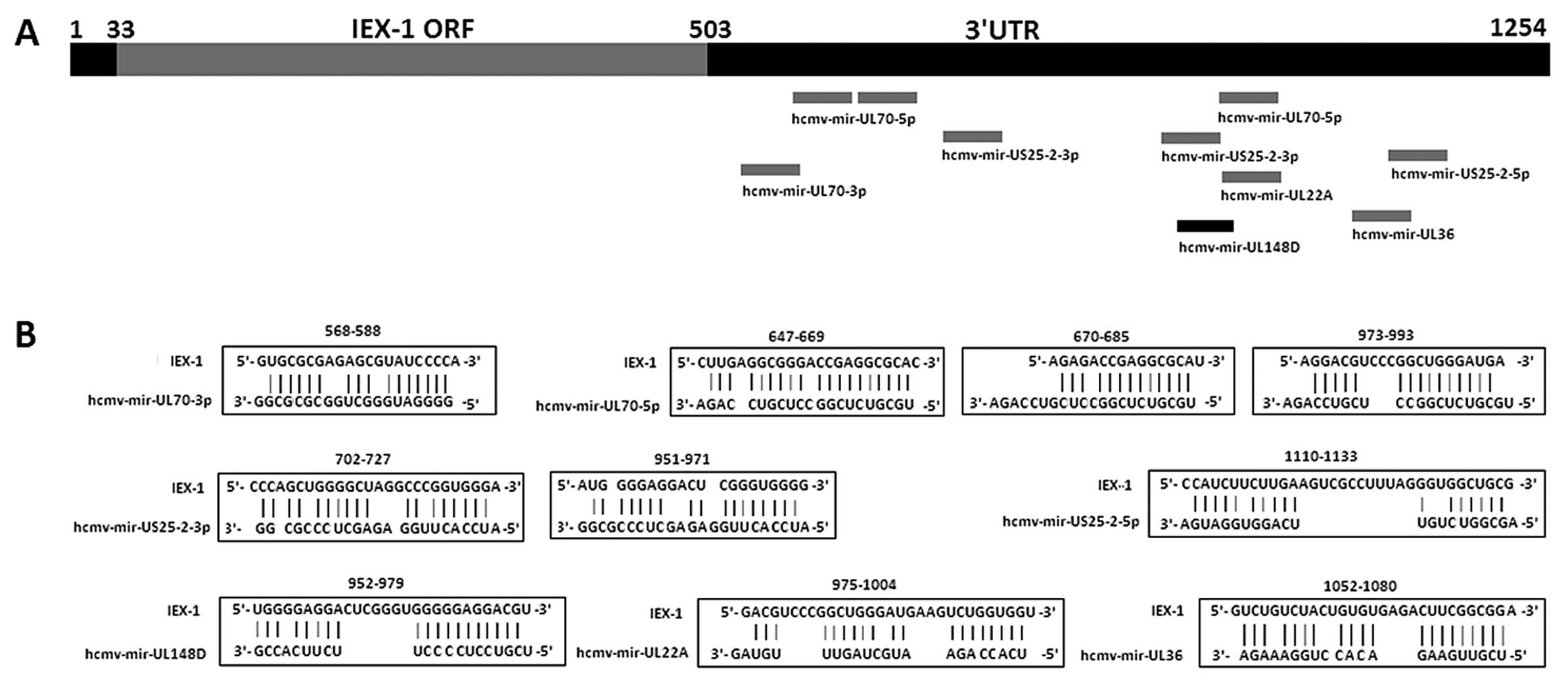
Prediction of binding sites of HCMV
encoding miRNAs in IEX-1 by Biosoftware RNAhybrid-Submission
Multiple HCMV miRNAs were predicted to be partially
complementary to IEX-1-3′UTR with the mfe <−20 kcal/mol
(Fig. 4). The hcmv-mir-UL148D
binding site at 952–979 nt in the IEX-1 gene is located 23 nt
upstream of the predicted binding site of hcmv-mir-UL22A (975–1004
nt), and 21 nt upstream of one of the binding sites of
hcmv-mir-UL70 (973–993 nt) predicated by Biosoftware
RNAhybrid-Submission. The binding sites in IEX-1-3′UTR of these
miRNA were located separately >21 nt from each other.
Discussion
Since miRNAs have been found to be expressed in a
number of viruses and exert downregulatory effects on target gene
expression, the study of miRNAs may provide further insight into
the mechanisms of viral pathogenesis and host defense. The
determination of target genes and the relative functions of a given
miRNA have been the new research highlights. A recent study showed
that hcmv-mir-UL148D, concomitant with other miRNAs encoded by
HCMV, is expressed in low-passage clinical strains recovered from
the amniotic fluid and urine of congenitally-infected infants
(26). In the present study, the
cellular target gene, IEX-1, of hcmv-mir-UL148D, which resides in
the HCMV UL/b′ region only found in clinical isolates, was
identified by a series of experiments.
To date, the prediction of viral miRNA targets has
been particularly challenging due to the complex prediction
algorithms (28) and the high
false-positive rate (29). The
hybrid-PCR method used in our study has proven to be a useful
experimental method for screening putative target genes of any
known miRNA (27). It conforms to
the idea that only the seed region of the miRNA is completely
complementary to and/or other sequences partially complementary to
the target mRNA, and that this miRNA can exhibit a translational
inhibitory effect. In fact, the more completely complementary the
nucleotides (nts) in the 3′-terminal sequence of a hybrid-PCR
primer, the stronger the binding of the miRNA to the target mRNA,
and the easier it will be for the targets to be amplified by the
hybrid-PCR method.
The main hybrid-PCR product of hcmv-mir-UL148D in
the IE, E and L phases of the HCMV infected cells was ~300 bp in
length. This band was confirmed to be derived from the cellular
gene, IEX-1, by sequencing analysis. A total of 11 nts around the
hcmv-mir-UL148D seed region were complementary to the IEX-1-3′UTR.
It has been reported that the complementary sequence between 6 nt
of the miRNA seed region and its target sequence is enough to form
stable bindings (30). The
prediction results by Biosoftware RNAhybrid-Submission in the
present study showed that only one binding site from 952 to 979 nt
of IEX-1 was satisfactory with consecutive 11 nt pairs completely
complementary to hcmv-mir-UL148D 5′ end including the entire seed
region, and the mfe of this binding was −35.0 kcal/mol. This
predicted binding site was the same as that screened by hybrid-PCR.
The high thermodynamic stability of hcmv-mir-UL148D:IEX-1-3′UTR
suggests that the sequence from 952 to 979 nt of IEX-1 should be
the real binding site of hcmv-mir-UL148D. As shown by luciferase
assay, the relative luciferase activity of pMIR-IEX-1-3′UTR was
suppressed ~47% by hcmv-miR-UL148D, compared to the mutant control
plasmids of hcmv-mir-UL148D and IEX-1-3′UTR. This evidence further
demonstrates the binding ability of hcmv-mir-UL148D to
IEX-1-3′UTR.
The coding sequence of hcmv-mir-UL148D resides in
the antisense strand of UL150, and is completely complementary to
the UL150 sequence. Unfortunately, the exact function of the UL150
gene remains unknown, which hinders the determination of the exact
bio-behavioral effects of hcmv-mir-UL148D interference. However,
the UL150 gene was not identified as a candidate target of
hcmv-mir-UL148D in our screening tests by hybrid-PCR. This may due
to a number of resons: i) The number of selected clones was too
limited to contain all the putative target genes. ii) The extracted
RNAs used in hybrid-PCR contained two genomes, Homo sapiens
and HCMV. The higher amount of Homo RNAs than HCMV RNAs in
the hybrid-PCR samples made cellular RNAs easier to be amplified.
iii) Residing in the high GC region, the UL150 gene is more
difficult to be amplified with hybrid-PCR primer and oligo(dT)-3′
site adaptor primer at the annealing temperature of 37°C (27).
Although the 3′UTR sequence of IEX-1 mRNA from 952
to 979 nt was not completely complementary to the whole sequence of
hcmv-mir-UL148D, it was completely complementary to the entire seed
region. To determine whether the binding of hcmv-mir-UL148D to the
3′UTR sequence of IEX-1 mRNA has functional effects on IEX-1
expression, western blot analysis was performed. As shown by
western blot analysis, the inhibitory effect of ectopically
expressed IEX-1 was similar to that of the suppression of relative
luciferase activity as shown by luciferase assay. These results
further suggests that the cellular gene, IEX-1, is one of the
targets of hcmv-mir-UL148D.
Previous studies have shown that a miRNA may exert
synergistic post-regulatory inhibitory effects on a target gene
with multiple binding sites (31–33). As shown by western blot analysis,
the similar expression levels of IEX-1 in the parallel tests of
mutant hcmv-mir-UL148D and wild-type IEX-1 with that in tests of
wild-type hcmv-mir-UL148D and mutant IEX-1 suggest that the binding
site in the 3′UTR of IEX-1 mRNA from 952 to 979 nt is an exclusive
site.
IEX-1 plays a role in controling apoptosis and
cellular growth. IEX-1 expression can be rapidly induced in various
cells by irradiation, viral infection, inflammatory cytokines,
chemical carcinogens, growth factors and hormones. The increased
expression of IEX-1 has been associated with an increase in the
growth rate of keratinocytes and HeLa cells, and the disruption of
IEX-1 expression in HeLa and 293 cells has been associated with a
decrease in cellular proliferation (34–37). A previous study on the
hcmv-mir-UL148D expression kinetics showed that the expression is
first evident at the IE phase, and gradually increases at the E
phase and reaches maximum levels at the L phase during HCMV
infection (25). In our study,
the weak expression of IEX-1 was detected in the IE phase and the
expression was even weaker in the E phase of HCMV infection, but
not in cells in the L phase and mock-infected cells, as shown by
western blot analysis. The elevated expression of IEX-1 at the IE
phase of HCMV infection is likely due to viral invasion. The
expression level of IEX-1 in HCMV-infected cells negatively
correlates with that of hcmv-mir-UL148D. This may partially due to
the specific downregulatory effect hcmv-mir-UL148D on IEX-1
expression, which was identified in our in vitro system.
Apoptosis is an antiviral defense mechanism by which
the host can eliminate infected cells and restrict viral
propagation. It has been demonstrated that HCMV-encoded proteins,
such as UL37, an immediately early gene product, can prevent or
attenuate apoptosis in infected cells (38–40). By utilizing an in vitro
apoptosis assay we demonstrated primarily that hcmv-mir-UL148D
suppresses the apoptotic cells ratio by downregulating IEX-1
expression. The downregulation rate of IEX-1 expression by
hcmv-mir-UL148D in this in vitro system was ~50%, which was
lower than the decreased level of IEX-1 expression from the IE to E
phase during HCMV infection (90%). Certain studies have shown that
multiple miRNAs may display cooperative effects on one target
(31–33). To explain this result, the binding
sites of HCMV other miRNAs in the IEX-1 sequence were analyzed
using Biosoftware RNAhybrid-Submission. The prediction result
showed that multiple miRNAs were partially complementary to
IEX-1-3′UTR with the mfe <−20 kcal/mol (Fig. 4). The binding sites in the
IEX-1-3′UTR of these miRNA were located separately >21 nt from
each other. This result suggests that multiple miRNAs may
cooperatively suppression on IEX-1 expression during HCMV
infection. These results may explain the manifestation that the
downregulation rate of ectopically expressed IEX-1 by
hcmv-mir-UL148D is lower than the decreased level of IEX-1
expression from the IE to the E phase during HCMV infection. The
cooperative effect on IEX-1 expression exerted by hcmv-mir-UL148D
and other miRNAs encoded by HCMV and their anti-apoptotic effects
require further study.
In conclusion, in the current study, we demonstrate
that the expression of the cellular gene, IEX-1, increased at the
IE phase, and rapidly decreased at the E and L phases during HCMV
infection. Hcmv-mir-UL148D, a miRNA residing in the HCMV UL/b′
region, was identified to downregulate IEX-1 expression through
only one binding site within the 3′UTR, and to contribute to the
anti-apoptotic effects caused by ectopically expressed IEX-1 in an
in vitro system. These findings provide new insight into
IEX-1 expression kinetics during HCMV infection, as well as the
interactions between hcmv-miR-UL148D and IEX-1 expression.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (30672248, 30901625, 81171580 and
81171581) and the Specialized Research Fund for the Doctoral
Program of Higher Education (20112104110012) and the Outstanding
Scientific Fund of Shengjing Hospital.
References
|
1
|
Griffiths PD, Cope AV, Hassan-Walker AF
and Emery VC: Diagnostic approaches to cytomegalovirus infection in
bone marrow and organ transplantation. Transpl Infect Dis.
1:179–186. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dolan A, Cunningham C, Hector RD, et al:
Genetic content of wild-type human cytomegalovirus. J Gen Virol.
85:1301–1312. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cha TA, Tom E, Kemble GW, Duke GM,
Mocarski ES and Spaete RR: Human cytomegalovirus clinical isolates
carry at least 19 genes not found in laboratory strains. J Virol.
70:78–83. 1996.PubMed/NCBI
|
|
4
|
Dunn W, Chou C, Li H, et al: Functional
profiling of a human cytomegalovirus genome. Proc Natl Acad Sci
USA. 100:14223–14228. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ashiru O, Bennett NJ, Boyle LH, Thomas M,
Trowsdale J and Wills MR: NKG2D ligand MICA is retained in the
cis-Golgi apparatus by human cytomegalovirus protein UL142. J
Virol. 83:12345–12354. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bennett NJ, Ashiru O, Morgan FJ, et al:
Intracellular sequestration of the NKG2D ligand ULBP3 by human
cytomegalovirus. J Immunol. 185:1093–1102. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Prod’homme V, Sugrue DM, Stanton RJ, et
al: Human cytomegalovirus UL141 promotes efficient downregulation
of the natural killer cell activating ligand CD112. J Gen Virol.
91:2034–2039. 2010.PubMed/NCBI
|
|
8
|
Poole E, Groves I, MacDonald A, Pang Y,
Alcami A and Sinclair J: Identification of TRIM23 as a cofactor
involved in the regulation of NF-kappaB by human cytomegalovirus. J
Virol. 83:3581–3590. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Poole E, Atkins E, Nakayama T, Yoshie O,
Groves I, Alcami A and Sinclair J: NF-kappaB-mediated activation of
the chemokine CCL22 by the product of the human cytomegalovirus
gene UL144 escapes regulation by viral IE86. J Virol. 82:4250–4256.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Montag C, Wagner JA, Gruska I, Vetter B,
Wiebusch L and Hagemeier C: The latency-associated UL138 gene
product of human cytomegalovirus sensitizes cells to tumor necrosis
factor alpha (TNF-alpha) signaling by upregulating TNF-alpha
receptor 1 cell surface expression. J Virol. 85:11409–11421. 2011.
View Article : Google Scholar
|
|
11
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao Y and Srivastava D: A developmental
view of microRNA function. Trends Biochem Sci. 32:189–197. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Grey F, Antoniewicz A, Allen E, Saugstad
J, McShea A, Carrington JC and Nelson J: Identification and
characterization of human cytomegalovirus-encoded microRNAs. J
Virol. 79:12095–12099. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hutvagner G and Zamore PD: A microRNA in a
multiple-turnover RNAi enzyme complex. Science. 297:2056–2060.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hammond SM, Bernstein E, Beach D and
Hannon GJ: An RNA-directed nuclease mediates post-transcriptional
gene silencing in Drosophila cells. Nature. 404:293–296.
2000. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mourelatos Z, Dostie J, Paushkin S, et al:
miRNPs: a novel class of ribonucleoproteins containing numerous
micro-RNAs. Genes Dev. 16:720–728. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Brennecke J, Stark A, Russell RB and Cohen
SM: Principles of microRNA-target recognition. PLoS Biol.
3:e852005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Doench JG and Sharp PA: Specificity of
microRNA target selection in translational repression. Genes Dev.
18:504–511. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tuddenham L and Pfeffer S: Roles and
regulation of microRNAs in cytomegalovirus infection. Biochim
Biophys Acta. 1809:613–622. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Stern-Ginossar N, Elefant N, Zimmermann A,
et al: Host immune system gene targeting by a viral miRNA. Science.
317:376–381. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Grey F, Meyers H, White EA, Spector DH and
Nelson J: A human cytomegalovirus-encoded microRNA regulates
expression of multiple viral genes involved in replication. PLoS
Pathog. 3:e1632007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kim S, Lee S, Shin J, et al: Human
cytomegalovirus microRNA miR-US4–1 inhibits CD8(+) T cell responses
by targeting the aminopeptidase ERAP1. Nat Immunol. 12:984–991.
2011.PubMed/NCBI
|
|
25
|
Kim Y, Lee S, Kim S, Kim D, Ahn JH and Ahn
K: Human cytomegalovirus clinical strain-specific microRNA
miR-UL148D targets the human chemokine RANTES during infection.
PLoS Pathog. 8:e10025772012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Stern-Ginossar N, Saleh N, Goldberg MD,
Prichard M, Wolf DG and Mandelboim O: Analysis of
humancytomegalovirus-encoded microRNA activity during infection. J
Virol. 83:10684–10693. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang Y, Qi Y, Ruan Q, Ma Y, He R, Ji Y
and Sun Z: A rapid method to screen putative mRNA targets of any
known microRNA. Virol J. 8:82011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rajewsky N: microRNA target predictions in
animals. Nat Genet. 38:S8–S13. 2006. View
Article : Google Scholar
|
|
29
|
Baek D, Villén J, Shin C, Camargo FD, Gygi
SP and Bartel DP: The impact of microRNAs on protein output.
Nature. 455:64–71. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bentwich I: Prediction and validation of
microRNAs and their targets. FEBS Lett. 579:5904–5910. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tirabassi R, Hook L, Landais I, Grey F,
Meyers H, Hewitt H and Nelson J: Human cytomegalovirus US7 is
regulated synergistically by two virally encoded microRNAs and by
two distinct mechanisms. J Virol. 85:11938–11944. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hon LS and Zhang Z: The roles of binding
site arrangement and combinatorial targeting in microRNA repression
of gene expression. Genome Biol. 8:R1662007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Saetrom P, Heale BS, Snøve O Jr, Aagaard
L, Alluin J and Rossi JJ: Distance onstraints between microRNA
target sites dictate efficacy and cooperativity. Nucleic Acids Res.
35:2333–2342. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Arlt A, Grobe O, Sieke A, Kruse ML, Folsch
UR, Schmidt WE and Schafer H: Expression of the NF-kappa B target
gene IEX-1 (p22/PRG1) does not prevent cell death but instead
triggers apoptosis in HeLa cells. Oncogene. 20:69–76. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kumar R, Kobayashi T, Warner GM, Wu Y,
Salisbury JL, Lingle W and Pittelkow MR: A novel immediate early
response gene, IEX-1, is induced by ultraviolet radiation in human
keratinocytes. Biochem Biophys Res Commun. 253:336–341. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Arlt A, Kruse ML, Breitenbroich M, et al:
The early response gene IEX-1 attenuates NF-kappaB activation in
293 cells, a possible counter-regulatory process leading to
enhanced cell death. Oncogene. 22:3343–3351. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Schafer H, Arlt A, Trauzold A,
Hunermann-Jansen A and Schmidt WE: The putative apoptosis inhibitor
IEX-1L is a mutant nonspliced variant of p22(PRG1/IEX-1) and is not
expressed in vivo. Biochem Biophys Res Commun. 262:139–145. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Skaletskaya A, Bartle LM, Chittenden T,
McCormick AL, Mocarski ES and Goldmacher VS: A
cytomegalovirus-encoded inhibitor of apoptosis that suppresses
caspase-8 activation. Proc Natl Acad Sci USA. 98:7829–7834. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hayajneh WA, Colberg-Poley AM, Skaletskaya
A, et al: The sequence and antiapoptotic functional domains of the
human cytomegalovirus UL37 exon 1 immediate early protein are
conserved in multiple primary strains. Virology. 279:233–240. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Goldmacher VS: vMIA, a viral inhibitor of
apoptosis targeting mitochondria. Biochimie. 84:177–185. 2002.
View Article : Google Scholar : PubMed/NCBI
|