Introduction
Apoptosis represents the key mechanism for the
removal of surplus, damaged and aged cells. It can be activated
under disease conditions, constituting a protective mechanism in a
multi-cellular organism by eliminating potentially dangerous cells
or cells that have lost their functional capabilities. It is an
evolutionarily conserved form of cell death carried out by a highly
complex molecular signaling pathway. In particular, there are two
main pathways which are known to initiate apoptosis: the death
receptor-dependent (extrinsic) pathway and the
mitochondrial-dependent (intrinsic) pathway, both of which converge
on activating the execution caspases (caspase-3 and caspase-7)
(1,2). In the extrinsic pathway, death
receptor ligation leads to the recruitment of adaptor molecules
that activate an initiator caspase (caspase-8), which directly
cleaves and activates the execution caspases. In the intrinsic
pathway, the mitochondria respond to diverse signals (such as DNA
damage, low nutrient levels, increased calcium levels, receptor
signaling, oxidative stress and intracellular aggregation of
misfolded proteins) emanating from other cell compartments. In
response to these signals, a mitochondrial outer membrane
permeabilization (MOMP) occurs, followed by the release of
cytochrome c (Cyt-C) that binds apoptotic
protease-activating factor 1 (APAF1), leading to the formation of a
caspase activation platform (apoptosome). The apoptosome recruits
and activates an initiator caspase (caspase-9), which, in turn,
cleaves and activates the execution caspases. Crosstalk between the
extrinsic and intrinsic pathways occurs through the
caspase-8-mediated cleavage of proteins of the Bcl-2 family,
leading to MOMP (2). Thus, the
opening of channels in the outer mitochondrial membrane, which
induces the release of Cyt-C, represents the most frequent final
step leading to apoptotic death.
In recent years, some proteins have emerged as
important regulators of the apoptotic process. The most well
characterized is the cellular tumor antigen, p53 (3,4).
It has been referred to as the ‘guardian of the genome’ due to its
role in protecting cells from DNA damage, but it is also a key
factor in transmitting programmed cell death signals to the
mitochondrion, which, in turn, regulates the activity of p53
through the generation of reactive oxygen species (4). In addition, increased attention has
been paid to the possible role of neuroglobin (NGB) in the
regulation of the apoptotic process. NGB (5) is a member of the vertebrate globin
superfamily that is mainly expressed in the central and peripheral
nervous system, cerebrospinal fluid, retina and endocrine tissues,
where it exerts a clear-cut neuroprotective role (6). Structural analyses (7) have indicated that human NGB displays
the typical globin fold, comprised of 151 amino acids (molecular
mass, 17 kDa), with only 20–25% of sequence identity with myoglobin
and hemoglobin. It is a particularly highly conserved protein, with
mouse and human NGB differing in only 6% of the amino acid
positions and it has a substitution rate almost four-fold lower
than that of other vertebrate globins. This suggests that its
functions are of basic importance to some types of tissues and
possibly an enriched scenario for NGB functions has been obtained
during evolution. In fact, although NGB reversibly binds oxygen
with an affinity higher than that of hemoglobin, storing and
supplying oxygen to neurons may be one, but not the only one, of
its functions (5).
Of particular interest is the evidence that NGB is
both physically and functionally related to mitochondrial functions
(8,9). NGB may play a role in oxygen sensing
and ATP production (10), and of
particular interest is the ability of the protein to interfere with
the release of Cyt-C from the mitochondria during cell death,
leading to an inhibition of the intrinsic pathway of apoptosis
(1). In addition, NGB scavenges
damaging reactive oxygen or nitrogen species (11), and appears able to regulate G
protein-coupled receptor (GPCR)-triggered signal transduction
pathways, by inhibiting the dissociation of GDP from G protein α
(12). Thus, as Brittain et
al (13) suggest, NGB emerges
as a critical player regulating key mitochondrial events in the
intrinsic pathway of apoptosis, opening new avenues for therapeutic
interventions in a number of disorders. Recent findings identifying
proteins involved in energy metabolism and mitochondrial function
as NGB-interacting proteins (14), provide further experimental
support for this view.
The molecular details of this network of NGB
interactions, however, remain largely undefined. Bioinformatics
methods may provide some clarification to this specific issue and
this type of approach is the focus of the present study. In
particular, the NGB structure is analyzed by well recognized
bioinformatics methods in order to identify the possible
interaction interfaces it can exploit to interact with relevant
proteins of the mitochondrial-dependent pathway of apoptosis.
Materials and methods
NGB datasets
The three-dimensional structure of human NGB, as
determined by X-ray diffraction (7), was obtained from Protein Data Bank
(PDB) and the biological assembly 1 stored in the deposited pdb
file (code: 1oj6) was considered as representative of monomeric
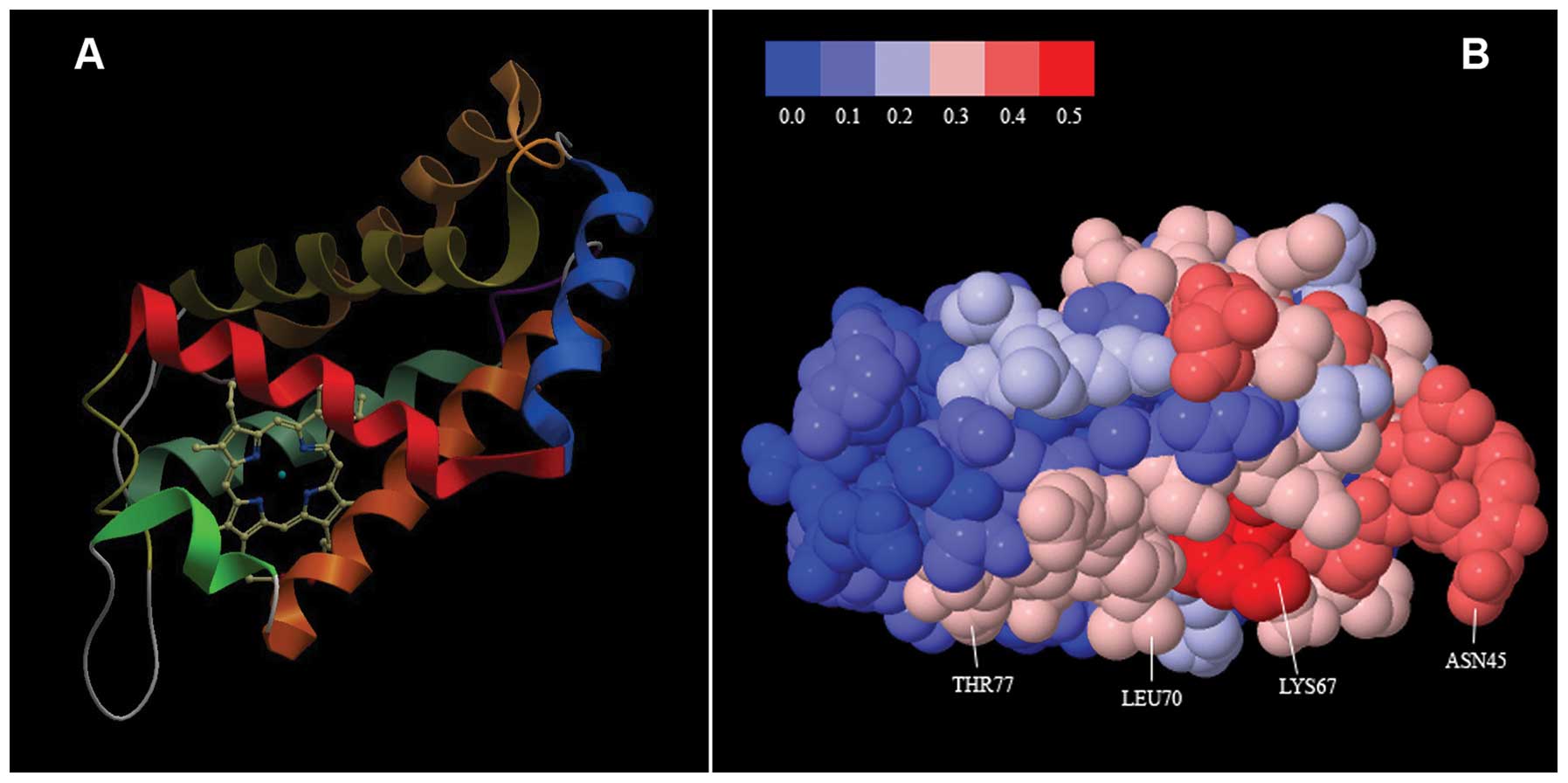
NGB, that is the biologically active form of the protein (15,16). The NGB structure is illustrated in
Fig. 1A.
Protein-protein interaction
propensities
To predict possible protein-protein interaction
sites on the NGB structure, the meta-PPISP (17) and meta-PPI (18) methods were used. Both follow a
consensus strategy (i.e., they combine the results from multiple
predictors) to increase prediction robustness and accuracy.
Meta-PPISP is built on three individual predictors, cons-PPISP
(19), promate (20) and PINUP (21), and a linear regression method,
using the raw scores of the three component methods as input, was
trained on a set of 35 non-homologous proteins to derive the final
predictor. Meta-PPI is based on the individual predictions provided
by the SPIDER (22) and ConSurf
(23) methods, that are combined
into a final confidence score assigned to each residue. In the
present study, a weighted mean of the output scores obtained from
the two abovementioned consensus methods was considered as an index
of the propensity of each NGB residue to take part in
protein-protein interactions. The applied weights were simply given
by the number of individual predictors on which each consensus
method relies (i.e., three for meta-PPISP and two for
meta-PPI).
The network of known and predicted protein-protein
interactions involving human NGB was obtained from the STRING
database (24) and the results it
provided were complemented with information from recent literature
(14). From the set of proteins
identified by this procedure, only those involved in
mitochondrial-dependent apoptosis and characterized by a known
three-dimensional structure were selected for further analysis.
Prediction of interaction interfaces
between NGB and the selected protein set
To predict the most probable interface exploited by
NGB to interact with each of the selected proteins, a docking
analysis was performed. For this purpose, the following docking
methods were applied:
PatchDock (25) is
a geometry-based molecular docking algorithm. It is aimed at
finding docking transformations that yield good molecular shape
complementarity. Such transformations, when applied, induce both
wide interface areas and small amounts of steric clashes.
With GRAMM-X (26), the best surface match between
molecules is determined by a correlation technique (27) using fast Fourier transform (FFT).
It uses a smoothed Lennard-Jones potential on a fine grid during
the global search FFT stage, followed by a refinement optimization
in continuous coordinates. An important feature of GRAMM-X is the
ability to smooth the protein surface representation to account for
possible conformational change upon binding within the rigid body
docking approach.
ZDOCK (28) is
also based on a grid-based representation of two proteins and a
3-dimensional FFT to efficiently explore the rigid-body search
space of docking positions. Its scoring function includes shape
complementarity, electrostatics, and a pairwise atomic statistical
potential developed using contact propensities of transient protein
complexes.
For each of the analyzed interactions the five best
solutions provided by each method were considered, leading to a set
of 15 predictions for each of the selected proteins. They were then
submitted to PDBePISA (29) to
obtain an estimate of their stability, and for each protein the
solution showing the lowest energy docked structure was taken for
the final prediction of the interface with NGB. The main
characteristics of the used predictors (all available as Web
servers) are summarized in Table
I.
 | Table IBioinformatics predictors. |
Table I
Bioinformatics predictors.
Results
Protein-protein interaction propensity of
NGB
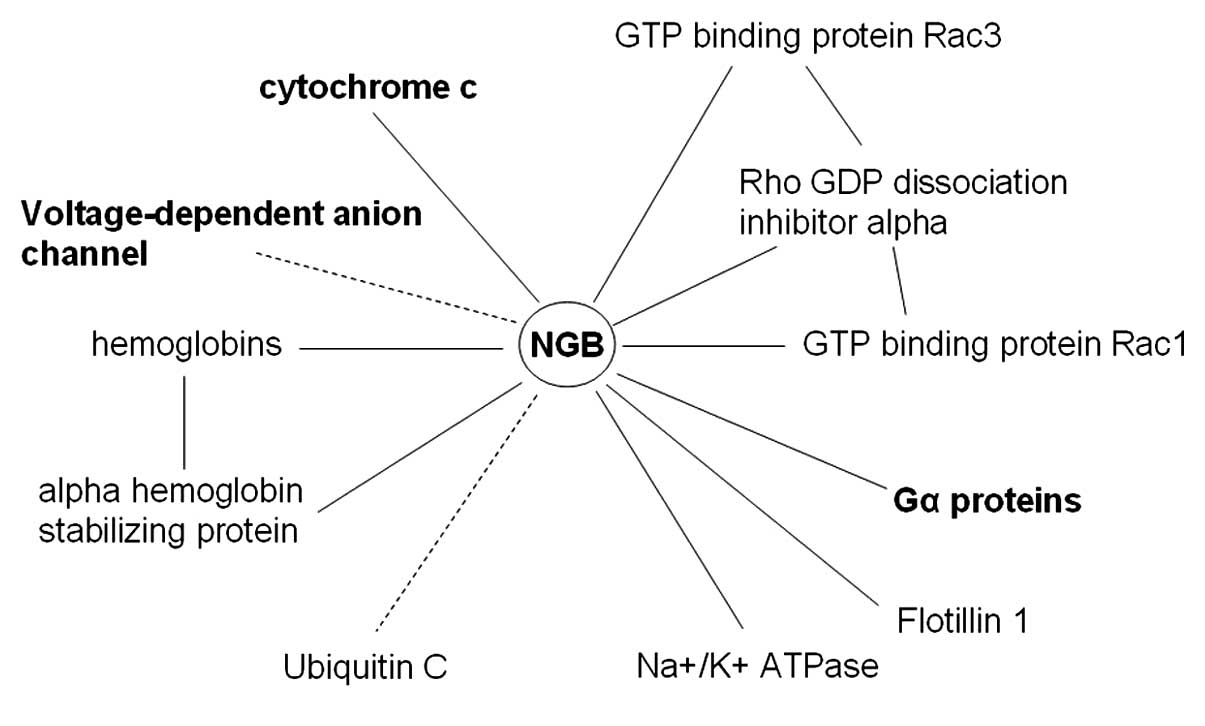
The interaction network provided by the STRING
database for NGB is illustrated in Fig. 2, together with additional
interactions recently identified experimentally (14). The spectrum of proteins that can
interact with NGB appears quite broad and diverse. It involves
plasma membrane proteins (such as flotillin-1) fundamental for the
formation of caveolae or caveolae-like vesicles, globulins involved
in the oxygen transport mechanisms and enzymes. Moreover, of
particular importance to the present study is the possibility for
NGB to interact with a number of proteins involved in the
regulation of the apoptotic process, such as the release of Cyt-C,
voltage-dependent anion channel (VDAC) and elements of signal
transduction mechanisms (such as G proteins). Furthermore, most of
the indicated interactions are predicted by STRING as direct
interactions, involving the binding of the protein to NGB.
The result of the combined use of meta-PPISP and
meta-PPI predictors is consistent and provides further support to
this view. It is summarized in Fig.
1B, where the NGB molecule is shown with the individual amino
acids coded in color according to a score scale ranging from 0 to
0.5 expressing increasing propensity to participate in
protein-protein interaction. As illustrated, >80% of the amino
acids on the molecule surface have non-null propensity, suggesting
that NGB can exploit a number of different sites for interaction
with other proteins.
Prediction of the interaction interfaces
with a selected set of proteins
From the interaction network identified by STRING,
and according to the criteria specified in ‘Materials and methods’,
the set of proteins presented in Table II was selected for docking
analysis. The set included human guanine nucleotide-binding protein
Gi subunit α-1 [Ga(i)], human VDAC and human Cyt-C. The
identified sites of interaction of NGB with each of the
abovementioned proteins identified by the docking analysis are
reported in Table III, together
with some biophysical characteristics (as provided by PDBePISA) of
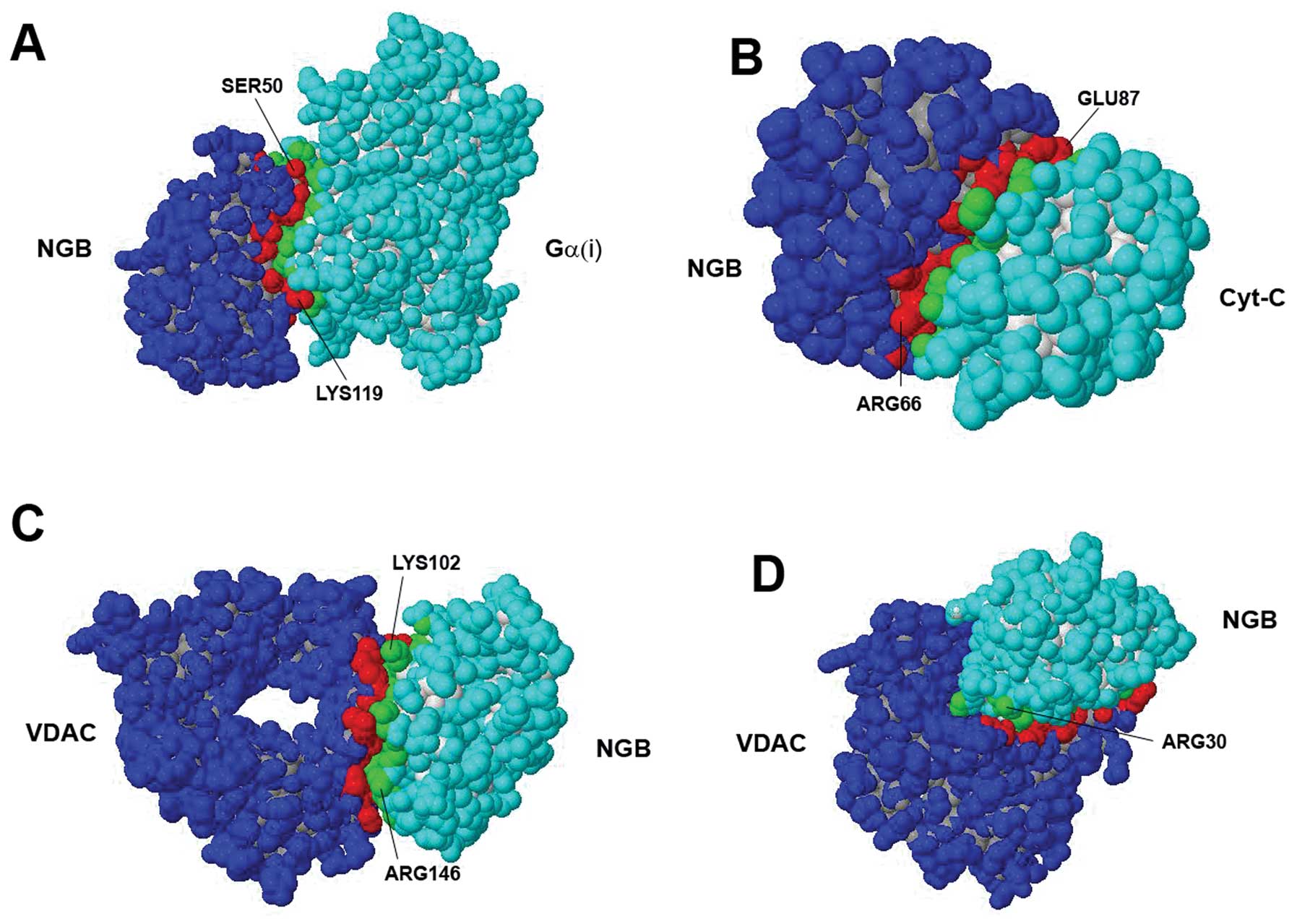
the predicted interface. The identified complexes with NGB are
illustrated in Fig. 3. As far as
the NGB-VDAC complex is concerned, it should be noted that the
analysis identified as the most probable configuration (Fig. 3C), an interaction of NGB with a
region of the VDAC molecule that, however, is usually buried within
the mitochondrial outer membrane. Although less energetically
favored (and likely unstable), an alternative suggested
configuration involved the binding of NGB to residues located at
the boundary of the pore formed by the VDAC molecule. This
assembly, of potential interest from a functional point of view, is
also presented in Table III and
illustrated in Fig. 3D.
 | Table IISet of analyzed proteins. |
Table II
Set of analyzed proteins.
| Protein | PDB code | Refs. |
|---|
| Human
neuroglobin | 1oj6 | (7) |
| Human guanine
nucleotide-binding protein Gi subunit α-1 | 3ums | (41) |
| Human
voltage-dependent anion channel | 2jk4 | (42) |
| Human cytochrome
c | 3nwv | (43) |
 | Table IIIPredicted interaction interfaces of
NGB. |
Table III
Predicted interaction interfaces of
NGB.
| Protein | NGB |
|---|
| Cyt-C | ARG10, TRP13,
ARG18, PRO20, PRO59, GLU60, LEU62, ARG66, LYS67, LEU70, VAL71,
ASP73, ALA74, VAL76, THR77, ASN78, SER84, LEU85, GLU87, TYR88,
ALA90, SER91, LEU92, ARG94, LYS95, HIS96 |
| AA (n) | 26 |
| Surface (%) | 11.3 |
| ΔiG
(p-value) | 0.312 |
| VDAC | ARG3-GLU5, LEU8,
LEU34-ASP37, LYS102, LEU103, SER104, SER107, THR108, GLY110,
GLU111, LEU114, GLU118, PRO127, ALA128, ARG130-ALA132, SER134,
GLN135, TYR137-ALA139, VAL141, GLN142, SER145, ARG146, TRP148,
ASP149 |
| AA (n) | 33 |
| Surface (%) | 14.5 |
| ΔiG
(p-value) | 0.138 |
| ARG3-PRO6, ARG10,
LEU34-ASP37, ARG30, VAL79, GLU80, LEU114, TYR115, GLU118, PRO123,
PRO127, ARG130, ALA131, SER134, GLN135, GLN142 |
| AA (n) | 18 |
| Surface (%) | 7.0 |
| ΔiG
(p-value) | 0.706 |
| Gα(i) | THR25, VAL26,
PHE28-ARG30, PHE32-ASP37, LEU39, PRO40, GLN48-GLU53, LEU56, PHE61,
GLU111, TYR115, GLU118, LYS119 |
| AA (n) | 25 |
| Surface (%) | 11.4 |
| ΔiG
(p-value) | 0.125 |
Discussion
Significant evidence has accumulated, demonstrating
that NGB protects cells from stroke damage, amyloid toxicity and
anoxic injury (32–34). Although the exact mechanisms by
which NGB achieves this protective abiltiy remain controversial, a
major role it plays is certainly in the interception of the
mitochondrial pathway of apoptosis (1). A recent study by Yu et al
(9) suggests that NGB is a
migrating protein capable of moving from the cytoplasm to the
mitochondria under physiological resting conditions, and this
phenomenon is modulated by the oxygen-glucose deprivation condition
of the cell. In both environments, NGB can establish a physical
interaction with mitochondrial proteins, some of which have been
recently identified using yeast two-hybrid screening and confirmed
by co-immunoprecipitation (14).
The bioinformatics analysis performed in the present study provides
further support and adds molecular details to this view. Since, in
general terms, bioinformatics predictions benefit from the combined
use of multiple methods (30,31), all the bioinformatics
investigations were performed not only by using available, well
recognized procedures, but also by organizing them according to a
consensus strategy. They allowed the identification on the globulin
surface of possible interaction interfaces with a set of proteins
relevant for mitochondrial-dependent apoptosis.
The obtained prediction concerning the interaction
of NGB with Cyt-C is consistent with previously reported
experimental and computational data (1), showing that NGB reacts very rapidly
with Cyt-C, and that the interface residues, Glu60 and Glu87, bind
the residues, Lys72 and Lys25, on Cyt-C, that are mandatory for the
binding of Cyt-C to APAF-1 (1,35).
A particularly intriguing suggestion from the analysis presented in
our study, however, concerns a predicted configuration of the
complex NGB-VDAC that involves an interaction at the level of the
channel aperture, opening the possibility for NGB to directly
modulate the permeability of the outer mitochondrial membrane
(43,44), which represents the
key event leading to apoptotic cell death (2). Another possible mechanism through
which NGB regulates apoptosis has been suggested by experimental
data showing that NGB modulates GPCR signaling (6) by interacting with the Gα subunits of
the trimeric G protein. In fact, evidence exists that GPCR-mediated
events can trigger (36) or
inhibit (37) the apoptotic
process. In the present study, the possible docking between NGB and
Ga(i) was analyzed. Interestingly, the Ga(i) molecular regions
involved in the predicted interface appear available when the
molecule is complexed in the GPCR (38), a result in line with available
experimental data showing that NGB binds to the GDP-bound state of
the G protein α subunit, and inhibits the exchange of GDP for GTP
(39).
Deregulated apoptosis has been implicated in the
etiology of diverse pathologies. In a simplified manner, the
diseases in which apoptosis has been involved can be divided into
two categories (40): those in
which there is an increase in cell survival (or diseases associated
with the inhibition of apoptosis, such as cancer and autoimmune
diseases), and those in which there is an excess in cell death (and
hence hyperactive apoptosis, as in neurodegenerative diseases and
ischemic damage). Thus, the search for therapeutic approaches based
on the modulation of apoptosis is of particular importance in
medicine, and NGB may be a target deserving specific attention for
the future development of therapeutic strategies. In this respect,
the results of the present study, based on information from
bioinformatics analyses, can only suggest possibilities and a
theoretical insight. However, the interaction interfaces between
NGB and mitochondrial proteins identified in the present study may
represent a first, guiding step, for the design of strategies aimed
at modulating these interactions, and tune the
mitochondrial-dependent pathway of apoptosis.
Acknowledgements
The study was financially supported by a grant from
the University of Padova (ex 60%) to D.G. and C.T.
References
|
1
|
Raychaudhuri S, Skommer J, Henty K, Birch
N and Brittain T: Neuroglobin protects nerve cells from apoptosis
by inhibiting the intrinsic pathway of cell death. Apoptosis.
15:401–411. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tait SW and Green DR: Mitochondria and
cell death: outer membrane permeabilization and beyond. Nat Rev Mol
Cell Biol. 11:621–632. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Holley AK, Dhar SK and St Clair DK:
Manganese superoxidedismutase versus p53: the mitochondrial center.
Ann NY Acad Sci. 1201:72–78. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Holley AK and St Clair DK: Watching the
watcher: regulation of p53 by mitochondria. Future Oncol.
5:117–130. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Burmester T and Hankeln T: What is the
function of neuroglobin? J ExpBiol. 212:1423–1428. 2009.PubMed/NCBI
|
|
6
|
Yu Z, Liu N, Liu J, Yang K and Wang X:
Neuroglobin, a novel target for endogenous neuroprotection against
stroke and neurodegenerative disorders. Int J Mol Sci.
13:6995–7014. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pesce A, Dewilde S, Nardini M, Moens L,
Ascenzi P, Hankeln T, et al: Human brain neuroglobin structure
reveals a distinct mode of controlling oxygen affinity. Structure.
11:1087–1095. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hankeln T, Ebner B, Fuchs C, Gerlach F,
Haberkamp M, Laufs TL, et al: Neuroglobin and cytoglobin in search
of their role in the vertebrate globin family. J Inorg Biochem.
99:110–119. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yu Z, Xu J, Liu N, Wang Y, Li X, Pallast
S, et al: Mitochondrial distribution of neuroglobin and its
response to oxygen-glucose deprivation in primary-cultured mouse
cortical neurons. Neuroscience. 218:235–242. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu Z, Poppe JL and Wang X: Mitochondrial
mechanisms of neuroglobin's neuroprotection. Oxid Med Cell Longev.
2013:7569892013.
|
|
11
|
Fordel E, Thijs L, Martinet W, Schrijvers
D, Moens L and Dewilde S: Anoxia or oxygen and glucose deprivation
in SH-SY5Y cells: a step closer to the unraveling of neuroglobin
and cytoglobin functions. Gene. 398:114–122. 2007. View Article : Google Scholar
|
|
12
|
Watanabe S and Wakasugi K: Neuroprotective
function of human neuroglobin is correlated with its guanine
nucleotide dissociation inhibitor activity. Biochem Biophys Res
Commun. 369:695–700. 2008. View Article : Google Scholar
|
|
13
|
Brittain T, Skommer J, Raychaudhuri S and
Birch N: An antiapoptotic neuroprotective role for neuroglobin. Int
J Mol Sci. 11:2306–2321. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu Z, Liu N, Wang Y, Li X and Wang X:
Identification of neuroglobin-interacting proteins using yeast
two-hybrid screening. Neuroscience. 200:99–105. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Burmester T, Weich B, Reinhardt S and
Hankeln T: A vertebrate globin expressed in the brain. Nature.
407:520–523. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tiso M, Tejero J, Basu S, Azarov I, Wang
X, Simplaceanu V, et al: Human neuroglobin functions as a
redox-regulated nitrite reductase. J Biol Chem. 286:18277–18289.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Qin S and Zhou HX: meta-PPISP: a meta web
server for protein-protein interaction site prediction.
Bioinformatics. 23:3386–3387. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang B and Schroeder M: Using protein
binding site prediction to improve protein docking. Gene.
422:14–21. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen H and Zhou HX: Prediction of
interface residues in protein-protein complexes by a consensus
neural network method: test against NMR data. Proteins. 61:21–35.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Neuvirth H, Raz R and Schreiber G:
ProMate: a structure based prediction program to identify the
location of protein-protein binding sites. J Mol Biol. 338:181–199.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liang S, Zhang C, Liu S and Zhou Y:
Protein binding site prediction using an empirical scoring
function. Nucleic Acids Res. 34:3698–3707. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Porollo A and Meller J: Prediction-based
fingerprints of protein-protein interactions. Proteins. 66:630–645.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Landau M, Mayrose I, Rosenberg Y, Glaser
F, Martz E, Pupko T and Ben-Tal N: ConSurf 2005: the projection of
evolutionary conservation scores of residues on protein structures.
Nucleic Acids Res. 33:W299–W302. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, et al: STRING v9.1: protein-protein
interaction networks, with increased coverage and integration.
Nucleic Acids Res. 41:D808–D815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schneidman-Duhovny D, Inbar Y, Nussinov R
and Wolfson HJ: PatchDock and SymmDock: servers for rigid and
symmetric docking. Nucleic Acids Res. 33:W363–W367. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tovchigrechko A and Vakser IA: GRAMM-X
public web server for protein-protein docking. Nucleic Acids Res.
34:W310–W314. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Katchalski-Katzir E, Shariv I, Eisenstein
M, Friesem AA, Aflalo C and Vakser IA: Molecular surface
recognition: determination of geometric fit between proteins and
their ligands by correlation techniques. Proc Natl Acad Sci USA.
89:2195–2199. 1992. View Article : Google Scholar
|
|
28
|
Pierce BG, Hourai Y and Weng Z:
Accelerating protein docking in ZDOCK using an advanced 3D
convolution library. PLoS One. 6:e246572011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Krissinel E and Henrick K: Inference of
macromolecular assemblies from crystalline state. J Mol Biol.
372:774–797. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ferron F, Longhi S, Canard B and Karlin D:
A practical overview of protein disorder prediction methods.
Proteins. 65:1–14. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Guidolin D, Agnati LF, Albertin G,
Tortorella C and Fuxe K: Bioinformatics aggregation predictors in
the study of protein conformational diseases of the human nervous
system. Electrophoresis. 33:3669–3679. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Khan AA, Wang Y, Sun Y, et al:
Neuroglobin-overexpressing transgenic mice are resistant to
cerebral and myocardial ischemia. Proc Natl Acad Sci USA.
103:17944–17948. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sun Y, Jin K, Mao XO, Zhu Y and Greenberg
DA: Neuroglobin is up-regulated by and protects neurons from
hypoxicischemic injury. Proc Natl Acad Sci USA. 98:15306–15311.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sun Y, Jin K, Peel A, Mao XO, Xie L and
Greenberg DA: Neuroglobin protects the brain from experimental
stroke in vivo. Proc Natl Acad Sci USA. 100:3497–3500. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kluck RM, Ellerby LM, Ellerby HM, Naiem S,
Yaffe MP, Margoliash E, Bredesen D, Mauk AG, Sherman F and Newmeyer
DD: Determinants of cytochrome c pro-apoptotic activity. The role
of lysine 72 trimethylation. J Biol Chem. 275:16127–16133. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu TR, Yang Y, Ward R, Gao L and Liu Y:
Orexin receptors: Multi-functional therapeutic targets for sleeping
disorders, eating disorders, drug addiction, cancers and other
physiological disorders. Cell Signal. 25:2413–2423. 2013.
View Article : Google Scholar
|
|
37
|
Depoortere I: GI functions of GPR39: novel
biology. Curr Opin Pharmacol. 12:647–652. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lambright DG, Sondek J, Bohm A, Skiba NP,
Hamm HE and Sigler PB: The 2.0 Å crystal structure of a
heterotrimeric G protein. Nature. 379:311–319. 1996.
|
|
39
|
Wakasugi K, Nakano T and Morishima I:
Oxidized human neuroglobin acts as a heterotrimeric Galpha protein
guanine nucleotide dissociation inhibitor. J Biol Chem.
278:36505–36512. 2003. View Article : Google Scholar
|
|
40
|
Carson DA and Ribeiro JM: Apoptosis and
disease. Lancet. 341:1251–1254. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lambert NA, Johnston CA, Cappell SD,
Kuravi S, Kimple AJ, Willard FS and Siderovski DP: Regulators of
G-protein signaling accelerate GPCR signaling kinetics and govern
sensitivity solely by accelerating GTPase activity. Proc Natl Acad
Sci USA. 107:7066–7071. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bayrhuber M, Meins T, Habeck M, Becker S,
Giller K, Villinger S, et al: Structure of the human
voltage-dependent anion channel. Proc Natl Acad Sci USA.
105:15370–15375. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liptak MD, Fagerlund RD, Ledgerwood EC,
Wilbanks SM and Bren KL: The proapoptotic G41S mutation to human
cytochrome c alters the heme electronic structure and increases the
electron self-exchange rate. J Am Chem Soc. 133:1153–1155. 2011.
View Article : Google Scholar : PubMed/NCBI
|