Introduction
It has long been suggested that nuclear actin exists
only in a globular form as G-actin (1,2).
However, increasing evidence demonstrates the nuclear localization
of F-actin as part of a large structure in the nucleoplasm and
indicates that it interacts with nuclear matrix proteins (3–8).
The first studies that reported on the presence of nuclear actin,
or its polymerized form, were treated with skepticism and this
issue has been questioned for decades (9). The main problem occurs in detecting
the short polymers of F-actin in the nucleus by fluorescence
staining or at the ultrastructural level. However, newly developed
methods and technological progress allow us to shed new light on
the localization and involvement of actin or its polymerized form
in many cellular compartments. The difficult detection of nuclear
actin filaments is caused by their size (3–5 μm in diameter) and by
the fact that nuclear actin cannot be stained with phalloidin
(10). In the present study, we
used both confocal and transmission electron microscopy (TEM) to
detect actin filaments in the cell nucleus. As previousy
demonstrated Izdebska et al, the combination of pre- and
post-embedding techniques is important in the phalloidin-based
detection of nuclear F-actin by TEM. Moreover, they suggested that
the use of streptavidin-coated quantum dots (QDs), through the
different strategy of their bioconjugation, eliminates weak
labeling problems (11).
Nevertheless, the function and localization of actin in the nucleus
is not yet fully characterized. Undoubtedly, actin exists in
various forms and is involved in distinct functions in the nucleus.
It is also known to play an essential role in the nuclear
architecture and functions (7,12,13). Additionally, actin is localized in
the nucleoli, splicing speckles and Cajal bodies (14–16). It has been suggested that in these
nuclear compartments, actin is associated with gene expression as a
part of chromatin remodeling complexes, linked with the
transcription machineries, as well as with mRNA nuclear export
(17–21). Other nuclear processes in which
actin is involved include the assembly of the nuclear structure
genome organization, and the regulation of transcription factor
activity (17,22–24). Hence, actin seems to be a crucial
protein which interacts with nuclear components, particularly with
matrix proteins and chromatin.
Zimmer and Fabre indicated that spatial chromatin
organization within the highly organized and dynamic structure as
the cell nucleus affects genome expression, stability and
replication (25). As it has been
suggested by Spilianakis et al, it is also possible that
genes on different chromosomes cooperate in a coordinated manner,
thus altering their expression at the same time (26). Moreover, spatial chromatin
organization is directly influenced by the interactions of
chromatin and nuclear matrix proteins accordingly, to control gene
transcription and DNA repair mechanisms (27). One of these identified matrix
proteins is special AT-rich sequence-binding protein 1 (SATB1) that
binds specifically to AT-rich sequences of DNA by recognizing the
base unpairing regions (BURs) (28). The SATB1 protein may coordinate
the control of the expression of various genes even in distant
parts of the genome through the regulation of chromatin-loop
architecture and transcription. It forms three-dimensional
functional cage-like structures inside the cell nucleus which
anchor chromosomes by tethering the BUR regions of the selected
gens (29). SATB1 seems to be of
particular importance to cells which must alter their
functions.
The role of SATB1 in the regulation of apoptotic
cell death process has also been reported (30–33). On the one hand, these studies were
based on the degradation of SATB1 in cells undergoing apoptosis
(31,32,34). On the other hand, there is no
study which describes the link between theapoptotic process and the
high level of SATB1 expression (32). Moreover, to our knowledge, in the
literature, there is no information on its expression during
mitotic catastrophe.
Due to the evidence that the SATB1 protein provides
the structural background for chromatin organization, it can be
significant in the reorganization of chromatin during cell death.
It is also possible that SATB1 and nuclear actin or nuclear F-actin
may jointly act to promote changes in chromatin and nuclear
architecture associated with active cell death process. It has been
demonstrated by a number of authors that nuclear actin is involved
in chromatin remodeling in various nuclear pathways, such as
transcription, RNA export and intra-nuclear transport (7,17,35–40).
In our previous studies, we have also provided
evidence of the polymeric forms of actin in the nucleus of
different cell lines following the induction of active cell death
processes (11,41–44). Continuing with our findings, the
aim of the present study was to evaluate the association between
nuclear F-actin and SATB1 in chromatin remodeling associated with
active cell death and by this means to reveal the functional
association between these two nuclear matrix proteins. These
results extend the knowledge on the role of SATB1 and nuclear actin
in three-dimensional chromatin organization and its influence on
active cell death.
Materials and methods
Cell culture and treatment
The Chinese hamster ovary (CHO AA8) cell line, which
was kindly provided by Professor M.Z. Zdzienicka (Department of
Molecular Cell Genetics, Nicolaus Copernicus University in Toruń,
The Ludwik Rydygier Collegium Medicum in Bydgoszcz, Poland), was
cultured in Eagle’s minimum essential medium (MEM; Sigma-Aldrich,
St. Louis, MO, USA) supplemented with 10% fetal bovine serum
(Gibco, Grand Island, NY, USA) and 50 μg/ml gentamycin
(Sigma-Aldrich) in 5% CO2 at 37°C. For the induction of
cell death, CHO AA8 cells were grown in six-well plates (Falcon; BD
Biosciences, San Jose, CA, USA) and treated with doxorubicin
(Sigma-Aldrich) at a concentration of 2.5 μM for 24 h. The control
cells were grown under the same conditions without doxorubicin
treatment.
Cell death analysis
The analysis of cell death was performed using a
Tali Image-based cytometer and a Tali Apoprosis kit (both from
Invitrogen Life Technologies, Carlsbad, CA, USA) according to the
manufacturer’s instructions. Briefly, following trypsinization, the
suspended cells were resuspended in Annexin binding buffer at a
concentration of approximately 5×105 to
5×106. Subsequently, to each 100 μl of sample, 5 μl of
Annexin V Alexa Fluor 488 were added, mixed and incubated at room
temperature in the dark for 20 min. The cells were then centrifuged
and resuspended in 100 μl of Annexin binding buffer. After the
addition of 1 μl of propidium iodide (PI) to each sample, the cells
were incubated at room temperature in the dark for 3 min. A total
of 25 μl of stained cells were loaded into a Tali Cellular Analysis
Slide (Invitrogen Life Technologies). The data were analyzed using
FCS Express Research Edition software (Ver4.03; De Novo Software,
Los Angeles, CA, USA) on the assumption that viable cells are both
Annexin V Alexa Fluor 488- and PI-negative cells, cells that are in
early apoptosis are Annexin V Alexa Fluor 488-positive and
PI-negative, cells that are in late apoptosis are both Annexin V
Alexa Fluor 488- and PI-positive, whereas necrotic cells are
Annexin V Alexa Fluor 488-negative and PI-positive.
Fluorescence localization of F-actin and
SATB1
For the fluorescence localization of F-actin and
SATB1, the cells grown on sterile glass coverslips were fixed in 4%
(w/v) paraformaldehyde in PBS, pH 7.4 (20 min, room temperature).
After rinsing with PBS (3×5 min), permeabilization using 0.1% (v/v)
Triton X-100 (Serva Electrophoresis GmbH, Heidelberg, Germany) and
washing with PBS (3×5 min), the non-specific protein interactions
were blocked with 1% (w/v) BSA/PBS (Sigma-Aldrich) for 30 min.
Subsequently, SATB1 was labeled by incubating the cells with
primary antibodies diluted in blocking buffer for 1 h at room
temperature [rabbit monoclonal anti-SATB1 (Abcam Cambridge, MA,
USA) or rabbit polyclonal anti-SATB1 (1:50; Sigma-Aldrich)]. After
washing (3X PBS), the cells were incubated with a secondary
antibody [Alexa Fluor 488 goat anti-rabbit IgG (H+L; Invitrogen
Life Technologies)] for 1 h at room temperature, in the dark. For
the localization of F-actin, the cells were incubated with
phalloidin-TRITC conjugate (1:5; Sigma-Aldrich) or Alexa Fluor
488-phalloidin (1:20; Invitrogen Life Technologies) for 20 min in
the dark. After washing (3X PBS), the cell nuclei were
counterstained with DAPI (1:20,000; Sigma-Aldrich). The slides were
mounted in Aqua-Poly/Mount (Polysciences, Inc., Warrington, PA,
USA) and examined using C1 laser-scanning confocal microscope
(Nikon, Tokyo, Japan) with an (×100) oil immersion objective.
Images were captured using Nikon EZ-C1 software (Ver3.80;
Nikon).
5′-Fluorouridine (5-FUrd) incorporation
and its colocalization with F-actin or SATB1
For 5-FUrd incorporation, the cells grown on sterile
glass coverslips were incubated in complete medium containing
5-FUrd for 20 min at 37°C in 5% CO2 and then fixed in 4%
(w/v) paraformaldehyde in PBS, pH 7.4 (20 min, room temperature).
After rinsing with PBS (3×5 min), permeabilization using 0.1% (v/v)
Triton X-100 (Serva Electrophoresis GmbH) and washing with PBS (3×5
min), the non-specific antibody interactions were blocked with 1%
(w/v) BSA/PBS (Sigma-Aldrich) for 30 min. Subsequently, 5′-FUrd was
labeled by incubating the cells with primary mouse monoclonal
anti-BrdU (Sigma-Aldrich) antibody diluted 1:250 in blocking buffer
for 1 h, at room temperature [mouse monoclonal anti-BrdU
(Sigma-Aldrich)]. After washing (3X PBS), the cells were incubated
with Alexa Fluor 555 goat anti-mouse IgG (H+L; 1:500; Invitrogen
Life Technologies) for 1 h at room temperature in the dark. After
washing with PBS (3×5 min), F-actin or SATB1 were localized as
described above. After washing (3X PBS), cell nuclei were
counterstained with DAPI (1:20,000; Sigma-Aldrich). The slides were
mounted in Aqua-Poly/Mount (Polysciences) and examined using a C1
laser-scanning confocal microscope (Nikon) with an (×100) oil
immersion objective. Images were captured using Nikon EZ-C1
software (Ver3.80; Nikon).
Analysis of colocalization between
SATB1/F-actin, F-actin/5-FUrd, and SATB1/5-FUrd
The analysis of the degree of the overlap between
the two channels in the fluorescence images was performed using
imageJ software (Ver1.47i) and the Colocalization Threshold plugin.
Data are presented as a colocalization pixel map and a 2D intensity
histogram with indicated linear regression.
Ultrastructural colocalization of F-actin
and SATB1
For the ultrastructural colocalization of SATB1, a
post-embedding immunogold method was used. The harvested cells were
fixed in 4% (w/v) paraformaldehyde in PBS, pH 7.4 (20 min, room
temperature) and embedded in LR White. The immune-detection of
SATB1 was performed on ultra-thin sections of study material
collected on nickel grids by 1 h of incubation at room temperature
with primary antibodies [anti-SATB1 antibody (1:50;
Sigma-Aldrich)]. After washing in PBS containing 1% (w/v) BSA, the
sections were incubated for 1 h with secondary antibodies tagged
with 5 or 20 nm gold particles [donkey-anti-rabbit IgG (Aurion,
Toowong, Australia)]. In the negative control, the primary antibody
was replaced with PBS containing 1% (w/v) BSA. Following immune
detection, the ultrathin sections were stained with uranyl acetate
and lead citrate and examined under a JEM 100 CX electron
microscope (Jeol, Ltd., Tokyo, Japan) operating at 80 kV.
For the ultrastructural localization of F-actin, the
phalloidin-based method with semiconductor Zn/Se nanocrystals was
used as previously described (44). Briefly, the harvested cells were
fixed in 4% (w/v) paraformaldehyde in PBS, pH 7.4 (20 min, room
temperature). After rinsing with PBS (3×5 min), permeabilization
using 0.1% (v/v) Triton X-100 (Serva Electrophoresis GmbH) and
washing with PBS (3×5 min), the non-specific protein interactions
were blocked with 6% (w/v) BSA/PBS (Sigma-Aldrich) for 1 h.
Subsequently, F-actin was labeled by incubation of the study
material with biotinyled phalloidin (1:85; Sigma-Aldrich) diluted
in blocking buffer for 20 min. In the next step, after rinsing with
PBS (3×5 min), the study material was post-fixed in 1% (w/v)
OsO4 (Serva Electrophoresis GmbH), dehydrated with a
graded series of ethanol, infiltrated and embedded in LR White.
Ultrathin sections were placed on nickel grids (Sigma-Aldrich). The
detection of biotinyled phalloidin (Sigma-Aldrich) was performed by
using Qdot® 525 streptavidin conjugate (1:100;
Invitrogen Life Technologies) diluted in blocking buffer for 1 h.
After staining with uranyl acetate, the preparations were examined
under a JEM 100 CX electron microscope (Jeol, Ltd.) operating at 80
kV.
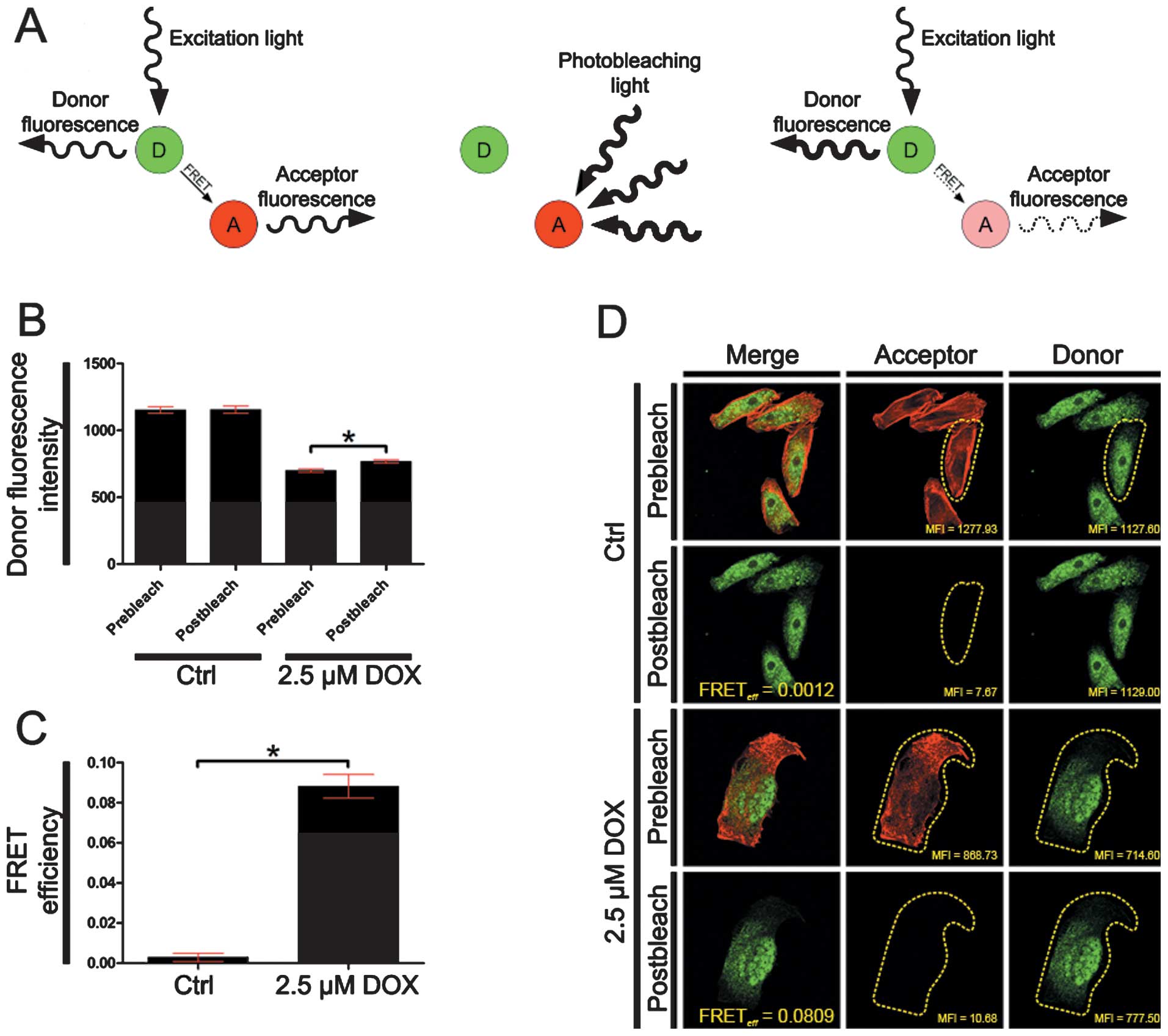
FRET acceptor bleaching (AB) method
For the SATB1 interactions with F-actin, the FRET AB
method was used. The images were collected using a C1
laser-scanning confocal microscope (Nikon) equipped with three
lasers: i) diode 405 nm, ii) diode 488 nm and iii) He-Ne 543 nm.
Images were captured using Nikon EZ-C1 software (Ver3.80; Nikon).
The preparations for FRET analysis were carried out analogically as
for the immunofluorescence experiments, but DAPI counterstaining
was omitted. Briefly, F-actin labeling was performed using
phalloidin-TRITC conjugate (Sigma-Aldrich) and as the secondary
antibody for SATB1 labeling, Alexa Fluor 488 goat anti-rabbit IgG
(H+L; Invitrogen Life Technologies) was used. Alexa Fluor 488
(acceptor) was induced using a diode laser 488 nm, while TRITC
(donor) was induced using a HeNe laser 543 nm. FRET AB method was
performed by comparing donor fluorescence intensity in the same
sample before and after destroying the acceptor by photobleaching.
If FRET was initially present, a resultant increase in donor
fluorescence would occur on photobleaching of the acceptor. The
energy transfer efficiency was calculated using the following
formula: FRETeff = (Dpost-Dpre)/Dpost; where Dpost is the
fluorescence intensity of the Donor after acceptor photobleaching
and Dpre the fluorescence intensity of the Donor before acceptor
photobleaching. The FRET efficiency was considered as positive when
Dpost > Dpre.
Co-immunoprecipitation
For the analysis of the presence of SATB1 in the
protein complex with β-actin, the Direct IP kit (Pierce Thermo
Scientific, Rockford, IL, USA) was used according to the
manufacturer’s instructions. Briefly, the cells were lysed using IP
Lysis buffer (Pierce Thermo Scientific). After normalization of the
protein concentration by the BCA Protein Assay kit (Pierce Thermo
Scientific) using a spectrophotometer, equal amounts of protein
were incubated with 150 μg antibody against β-actin. Subsequently,
the lysates were incubated with protein A/G Plus beads (Pierce
Thermo Scientific). Proteins were analyzed by western blot
analysis.
Pull-down assay
For the analysis of the presence of SATB1 in the
protein complex with F-actin, streptavidin-coated magnetic beads
(Dynabeads M-280 Streptavidin; Invitrogen Life Technologies) were
used according to the manufacturer’s instructions with a number of
key modifications. Briefly, the cells were lysed in RIPA buffer in
the presence of biotinyled phalloidin (1:85) (both from
Sigma-Aldrich). Following lysate clarification by centrifugation at
8 000 × g for 10 min at 4°C, each lysate was divided into two
fractions: ‘supernatant’ and ‘debris’. Following the normalization
of protein concentration in ‘supernatant’ by the BCA Protein Assay
kit (Pierce Thermo Scientific) using a spectrophotometer, equal
amounts of protein were incubated with 0.5 mg of
streptavidin-coated magnetic beads for 30 min at room temperature
with gentle rotation of the tube. ‘Debris’ was resuspended in RIPA
buffer supplemented with biotinyled phalloidin (1:85) and
analogically incubated with streptavidin-coated magnetic beads.
After five cycles of magnetic separation/washing in PBS containing
0.1% BSA, the biotin-streptavidin bonds were broken by boiling the
samples for 5 min in 0.1% SDS. Proteins were analyzed by western
blot analysis.
Western blot analysis
Semi-quantitative analysis of the post-translational
expression of SATB1 or the detection of SATB1 in the protein
complex with β-actin or F-actin was performed by western blot
analysis. Following the normalization of the protein concentration
by the BCA Protein Assay kit (Pierce Thermo Scientific) using a
spectrophotometer, equal amounts of protein (18 μg of total protein
per lane) were separated by 4–12% NuPage Bis-Tris gel (Novex;
Invitrogen Life Technologies) and transferred onto nitrocellulose
membranes using the iBlot dry western blotting system (Invitrogen
Life Technologies). Following co-immunoprecipitation and pull-down,
18 μl of diluted protein complexes were separated. Pre-stained
molecular weight markers (Pierce Thermo Scientific) were used to
estimate the position of the protein bands. Subsequently, the
membranes were processed using a BenchPro 4100 card processing
station (Invitrogen Life Technologies). The membranes were blocked
with 5% non-fat milk in TBS-T for 2 h and then incubated with
primary rabbit monoclonal anti-SATB1 (1:100; Abcam), rabbit
polyclonal anti-SATB1 (1:250) and rabbit anti-GADPH (1:5,000) (both
from Sigma-Aldrich) antibodies diluted TBS-T for 1 h at room
temperature. After washing with TBS-T, the membranes were incubated
with secondary goat anti-rabbit antibodies conjugated with alkaline
phosphatase (1:30,000; Sigma-Aldrich) diluted in TBS-T for 1 h at
room temperature. Protein bands were visualized using NBT/BCIP
ready-to-use tablets (Roche Applied Science, Indianapolis, IN,
USA).
Statistical analysis
The data are presented as the means ± SEM.
Statistical comparisons between two groups of cell death or
fluorescence intensity data were performed using a two-tailed
Mann-Whitney U test. The differences between the groups were
considered significant at P<0.05. The GraphPad Prism 5.0
(GraphPad Software) was used for statistical analyses.
Results
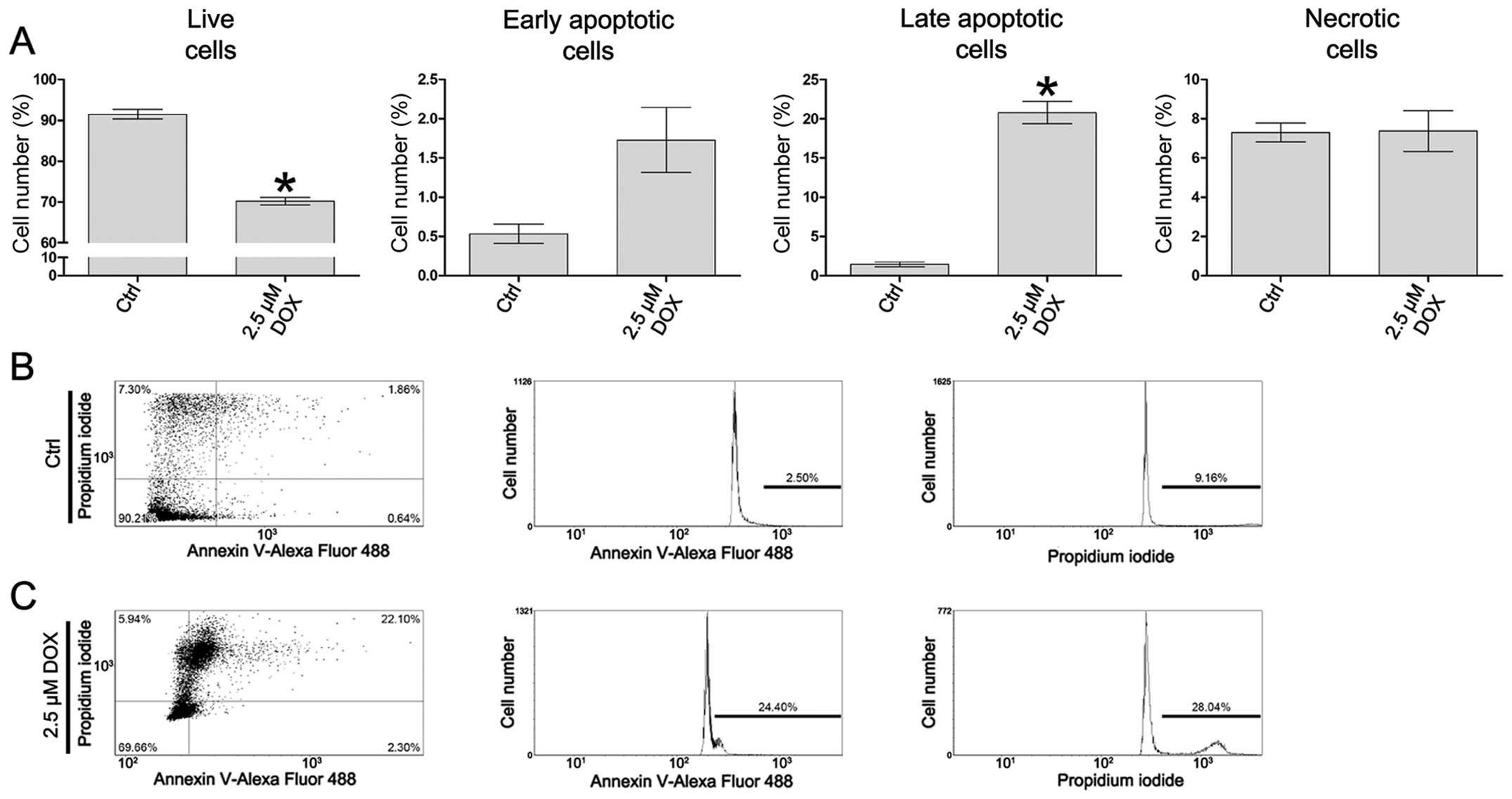
Doxorubicin induces active cell
death
The analysis of cell death was performed using a
Tali Image-based cytometer following Annexin V Alexa Fluor 488 and
PI double staining. In the control cells, the mean percentage of
Annexin V-positive cells was 1.96%, whereas the mean percentage of
PI-positive cells was 7.30%. Following treatment of the cells with
2.5 μM doxorubicin, the mean percentage of Annexin V-positive cells
was 22.49%, whereas the mean percentage of PI-positive cells was
7.37%. Based on the assumption described in ‘Material and methods’,
in the CHO AA8 cells treated with 2.5 μM doxorubicin, a
statistically significant decrease in the percentage of live cells
was observed, as compared to the control (from 91.49 to 70.20%,
P=0.0286). Additionally, following treatment of the cells with
doxorubicin at the concentration of 2.5 μM, a statistically
significant increase in the percentage of late apoptotic cells was
observed (from 1.45 to 20.77%, P=0.0286). The differences between
the populations of early apoptotic and necrotic cells following
treatment with 2.5 μM doxorubicin in comparison to the controls
were statistically insignificant (P=0.2000 and P=0.6631,
respectively) (Fig. 1A–C).
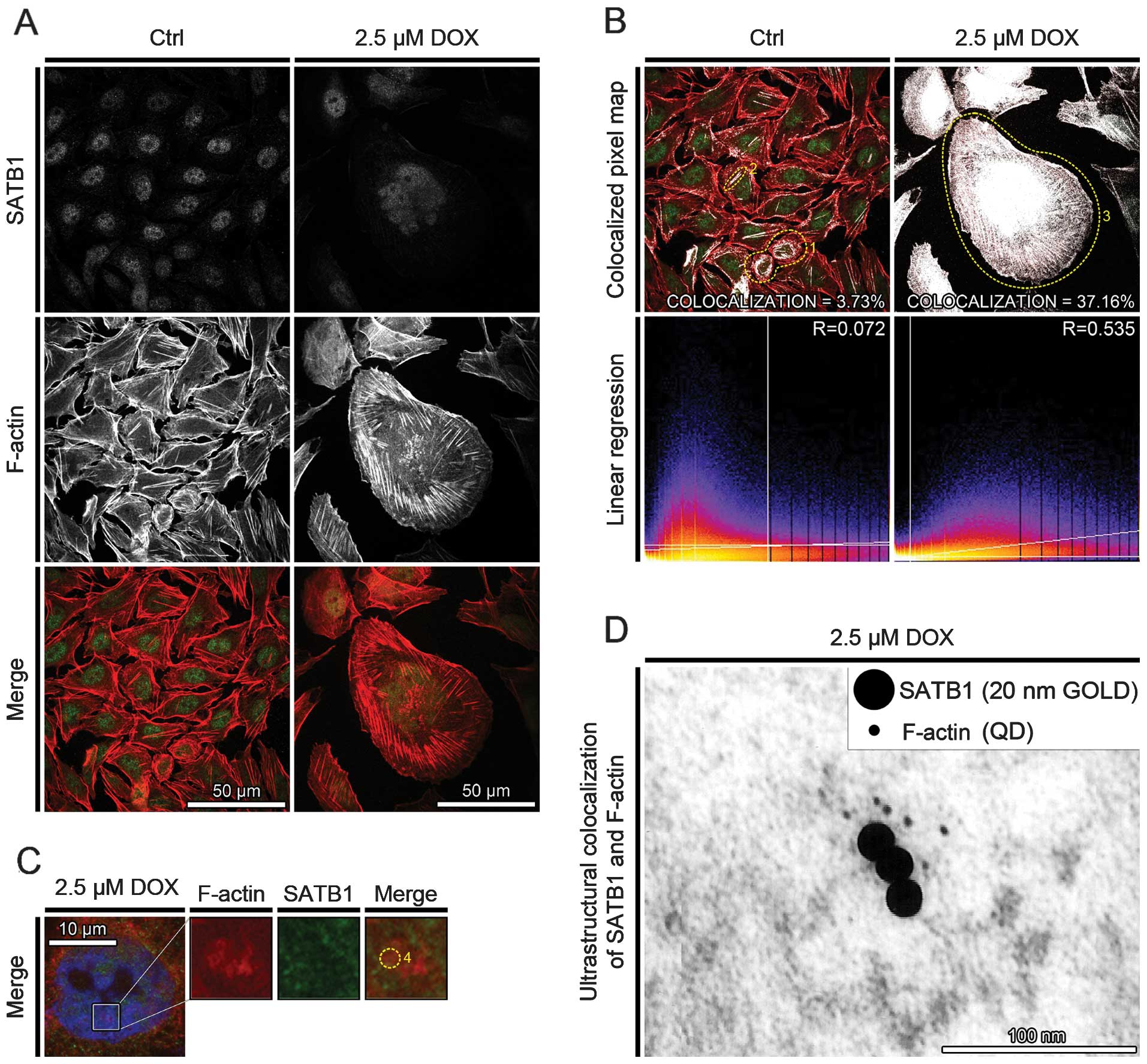
Doxorubicin treatment increases
SATB1/F-actin colocalization
The fluorescence analysis of SATB1 localization in
the control cells revealed its presence in the area of the cell
nucleus in the form of foci and it was scattered in the cytoplasm.
Following the induction of cell death, SATB1 protein was localized
mostly in the cell nucleus. In the cortical cytoplasm of the cells
treated with 2.5 μM doxorubicin, SATB1 formed clearly visible
filament-like structures, associated with F-actin stress fibers
(Fig. 2A).
The analysis of the colocalization between SATB1 and
F-actin indicated its increase after the induction of cell death.
The percentage of colocalized pixels increased from 3.73% (R=0.072)
to 37.16% (R=0.535) for the control cells and the cells treated
with 2.5 μM doxorubicin, respectively. In the controls, the
colocalization of SATB1 and F-actin was observed in association
with actin polymers in freshly divided cells (Fig. 2B, dotted circle 1) or along a few
thick tension cytoplasmic fibers of F-actin (Fig. 2B, dotted circle 1). Following
treatment of the cells with 2.5 μM doxorubicin, SATB1 protein was
colocalized with nuclear F-actin or was associated with F-actin
cytoplasmic stress fibers (Fig.
2B, dotted circle 3). Moreover, in the cell nucleus, SATB1 was
colocalized with more densely organized nuclear F-actin ring-like
structures, at the area of weak DAPI staining (Fig. 2C, dotted circle 4).
Additionally, on the ultrathin sections of the CHO
AA8 cells treated with 2.5 μM doxorubicin labeled for the presence
of SATB1 (20 nm gold) and F-actin (Qdots 525), the double-labeling
was distributed throughout the nucleoplasm. The complexes of SATB1
and F-actin were localized at the border of electron-dense regions
of the cell nuclei, possibly regions between condensed and
decondensed chromatin (Fig.
2D).
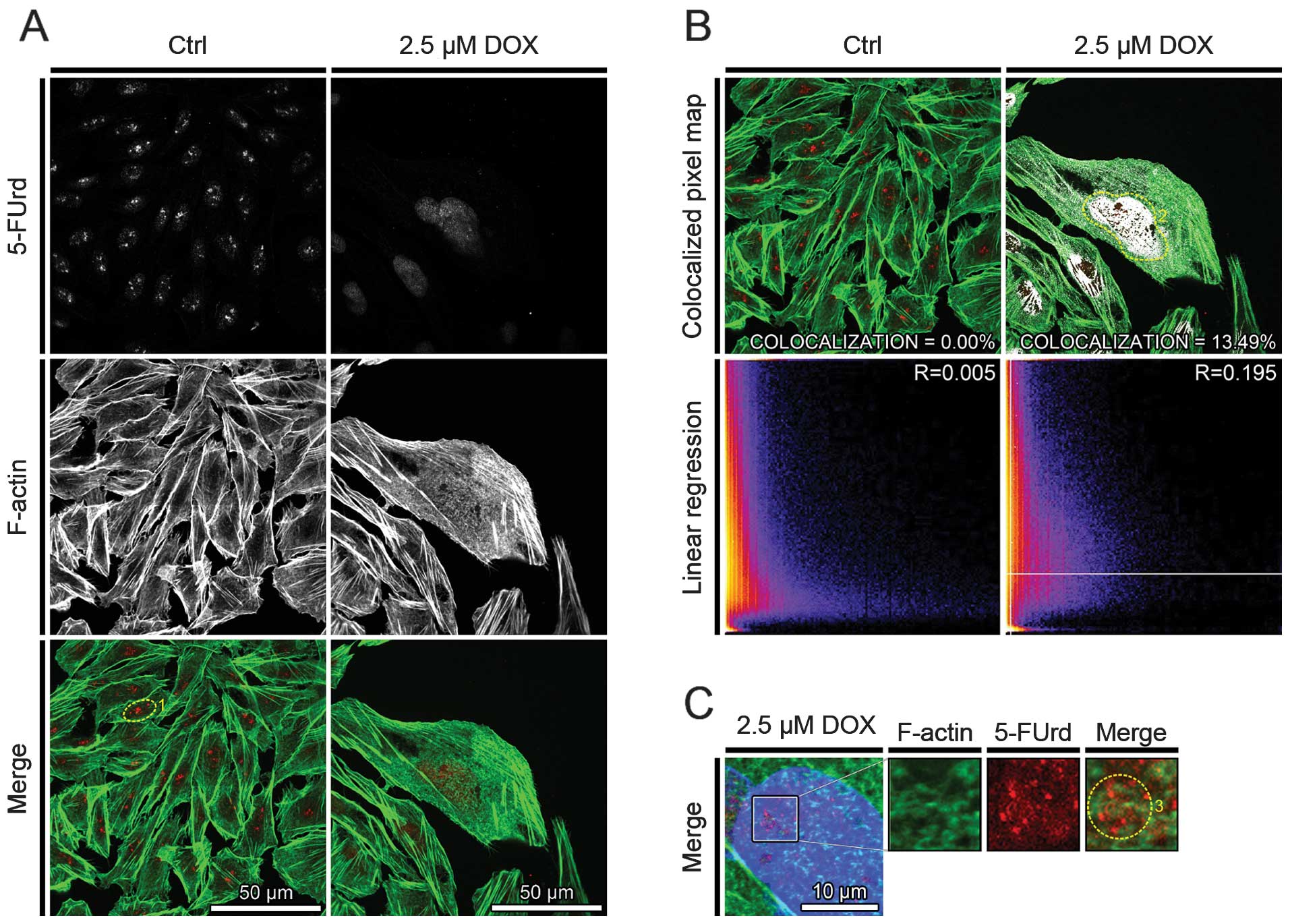
Doxorubicin increases the colocalization
of both SATB1/5FUrd and F-actin/5FUrd
To investigate whether SATB1 or F-actin have
functional consequences in transcriptional processes during active
cell death, the colocalization of the above mentioned proteins with
5-FUrd incorporation into nascent mRNA was performed.
In the controls, 5-FUrd 2–4 large foci, with high
fluorescence intensity (Figs. 3A,
dotted circle 1; and Fig. 4A) and
small foci in the area of the cell nucleus were observed. Following
treatment of the cells with 2.5 μM doxorubicin, the number of
5-FUrd small foci localized in the area of the cell nucleus
increased. Moreover, the overall fluorescence intensity of detected
5-FUrd foci was higher in the cells with the phenotype of mitotic
catastrophe, as compared with the controls (Figs. 3A and 4A).
The analysis of the colocalization between 5-FUrd
and F-actin indicated its absence in the control cells and its
increase after the induction of cell death. The percentage of
colocalized pixels increased from 0.00% (R=0.005) to 13.49%
(R=0.195) for the controls and the cells treated with 2.5 μM
doxorubicin, respectively. Following treatment of the cells with
2.5 μM doxorubicin, 5-FUrd protein was colocalized with nuclear
F-actin (Fig. 3B, dotted circle
2). Moreover, 5-FUrd was colocalized with nuclear F-actin ring-like
structures, at the area of weak DAPI staining (Fig. 3C, dotted circle 3).
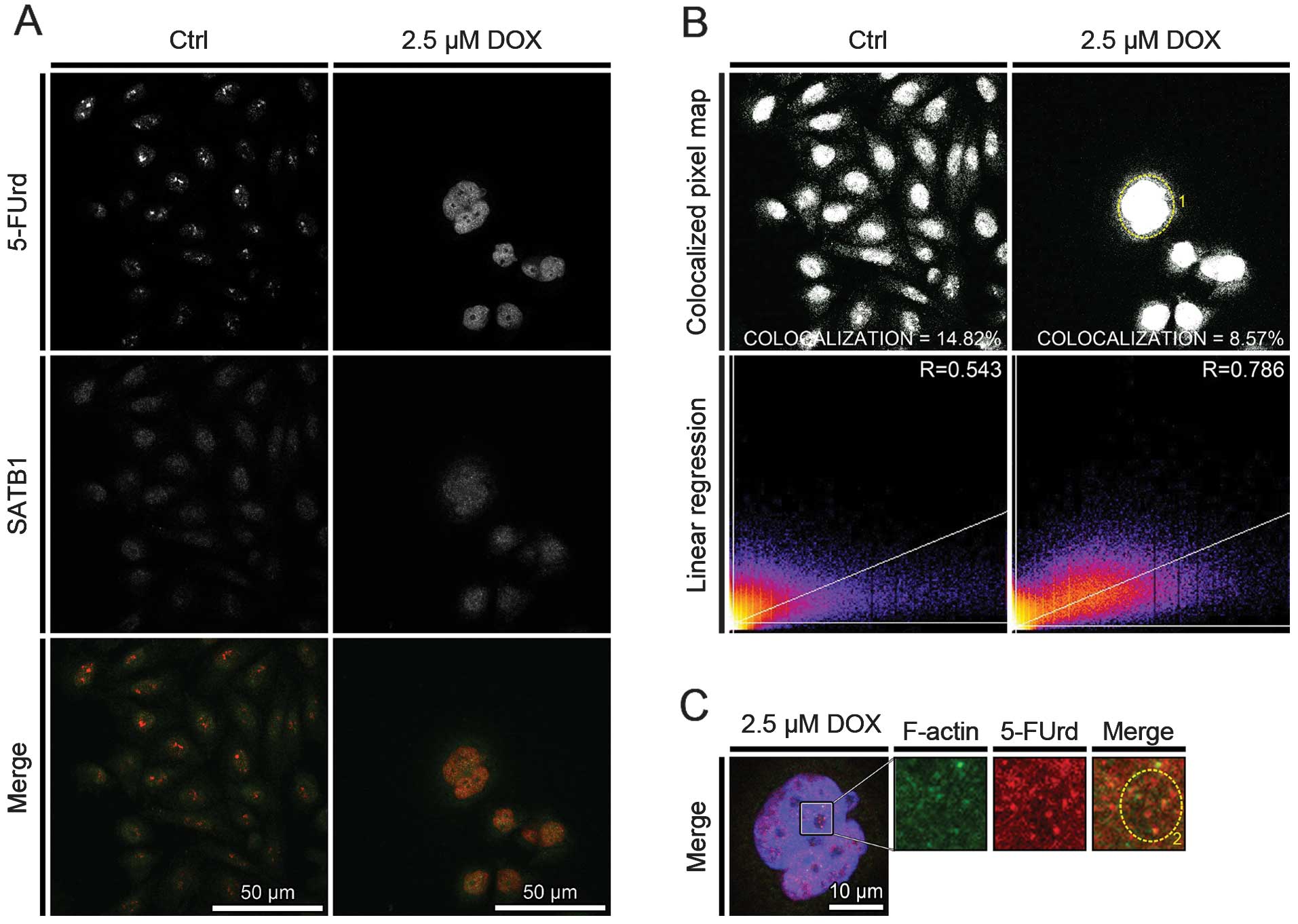
The analysis of the colocalization between 5-FUrd
and SATB1 indicated its decrease following the induction of cell
death. The percentage of colocalized pixels decreased from 14.82
(R=0.543) to 8.57% (R=0.786) for the controls and the cells treated
with 2.5 μM doxorubicin, respectively (Fig. 4B). However, in the cells with the
phenotype of mitotic catastrophe, the nuclear colocalization of
5-FUrd and SATB1 was characterized with a higher regression,
indicating wide-genome transcriptional processes (Fig. 4B, dotted circle 1). Moreover, the
colocalization of 5-FUrd and SATB1 was observed in both the regions
of the cell nucleus stained weakly and intensively with DAPI
(Fig. 4C, dotted circle 2).
Doxorubicin treatment induces
SATB1/F-actin interactions
The fluorescence analysis of SATB1/F-actin
interactions were investigated using the FRET acceptor bleaching
method (Fig. 5A), in the control
cells and cells undergoing mitotic catastrophe following treatment
with 2.5 μM doxorubicin. In the control cells, the mean of donor
fluorescence intensity was statistically insignificant and the
energy transfer efficiency was 0.0029. This confirmed the
examination of the fluorescence colocalization results and indicate
the lack of SATB1/F-actin interactions. Following the induction of
cell death, the mean of donor fluorescence intensity in the cells
undergoing active cell death, was statistically significantly
increased after acceptor photobleaching (from 700.20 to 767.91,
P=0.0286, before and after acceptor photobleaching, respectively)
and the mean energy transfer efficiency was 0.0881 (Fig. 5B–D). These data thus indicate that
SATB1 interacts with F-actin and confirm the colocalizational
resuts that indicate the functional association of both analyzed
nuclear matrix proteins.
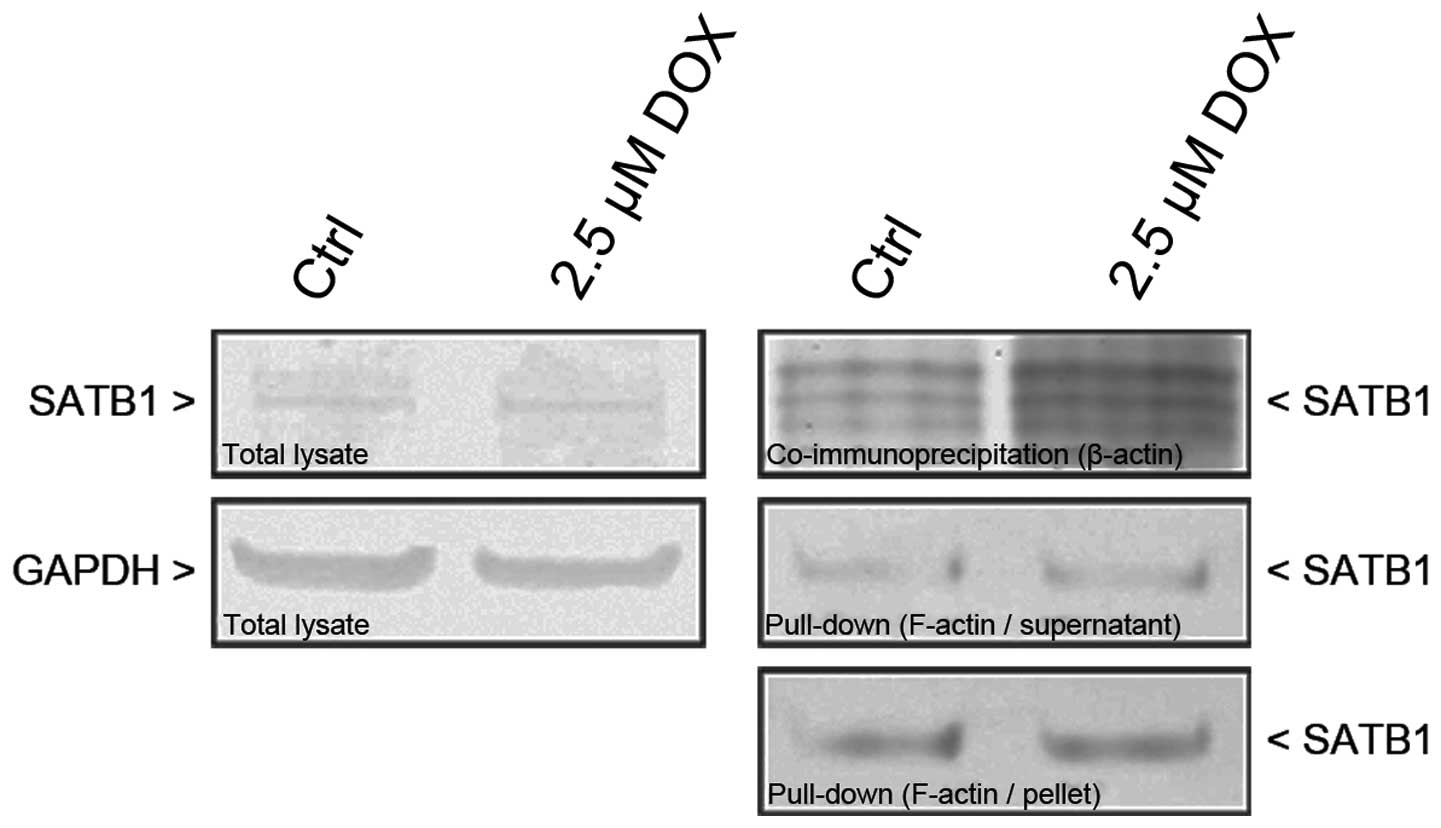
To investigate biochemically whether SATB1 interacts
with actin in vitro, cell extracts from CHO AA8 fibroblasts
were immunoprecipitated with an antibody to β-actin and subjected
to the detection of SATB1 by western blot analysis. The results
revealed significantly elevated levels of SATB1 in the
immunoprecipitates obtained from the cells treated with 2.5 μM
doxorubicin, and indicate that SATB1 interacts with β-actin more
intensely following the induction of active cell death (Fig. 6). Consequently, to confirm
fluorescence-based experiments, pull-down assay using
phalloidin-biotin and streptavidin-coated magnetic beads was used
to investigate whether SATB1 interacts with actin filaments. The
results confirmed those already obtained and indicated the
increased level of SATB1 following the induction of cell death.
Moreover, pull-down assay revealed the interactions of SATB1 not
only with F-actin short polymers present in the supernatant
fractions, but also with more densely organized or
membrane-associated actin filaments in the cell debris (pellet)
(Fig. 6).
Discussion
There are a number of reports linking nuclear actin
with chromatin remodeling during several nuclear processes
(7,17,35–40). Sjölinder et al demonstrated
the association of nuclear actin, chromatin remodeling and RNA
polymerase II transcription (36). Other studies have shown nuclear
actin as a component of chromatin remodeling complexes and its
involvement in the nuclear envelope assembly (17,22,23). It has been also listed that
nuclear actin controls the transcription of its target genes
through different mechanisms: i) specifically binding to a 27-nt
repeat element in intron 4 of the endothelial nitric oxide synthase
gene and regulating its expression (45,46); ii) participation in chromatin
remodeling necessary for gene activation (47–49); iii) direct role in RNA
transcription by being part of the pre-initiation complex with RNA
polymerase II (39); or iv)
participation in transcriptional elongation (50). Moreover, it has been speculated
that under stress conditions, actin may translocate into the cell
nucleus to function as a transcriptional modulator of gene
transcription (51–53). In addition, numerous proteins
which interact with G- and F-actin have been detected in the
nucleus (2,54). Previously, we have provided
evidence of F-actin presence in the nucleus of different cell lines
treated with anti-cancer drugs and speculated that nuclear F-actin
may be involved in chromatin remodeling processes during apoptosis
and mitotic catastrophe (11,41–44). In this study, we confirmed the
involvement of F-actin in nuclear processes accompanied with active
cell death processes. Moreover, we demonstrate that the
SATB1/F-actin complex is localized at the border of condensed and
decondensed chromatin which suggests its involvement in the process
of transcription. Fomproix and Percipalle postulated that the
spatial restriction of actin filaments predominantly to the
interchromatin space argues against their direct involvement of
conventional actin filaments in transcription and chromatin
remodeling (14). Moreover, Belin
et al (8) and Dundr et
al (55) indicated that
nuclear actin filaments are too short to serve as tracks for the
long-range transport of genetic loci cargo through the nucleus. The
authors also referred to the lack of directed motion of nuclear
actin filaments and the little or no colocalization between nuclear
actin filaments and up to now identified nuclear myosins. However,
the conformational differences and some post-translational
modifications in nuclear actin may be responsible for its different
polymerization (56–58).
The matrix associated regions of DNA [MAR-binding
proteins (MARBPs)] are dynamic and their distribution is cell type-
and cell cycle-dependent. Several MARPBs have been characterized as
SATB1, SATB2, BRIGHT, Cux/CDP, Lamin A/B/C, HMG and SMAR1 (59–64). SATB1 is organized into a cage-like
network anchoring loops of heterochromatin and tethering
specialized DNA sequences and serves as a global platform for the
assembly of chromatin remodeling or modifying complexes with the
anchored genomic loci (65). It
has been pointed that depending on its post-translational
modifications, SATB1 activates or represses multiple genes
(66). Furthermore, SATB1 forms a
functional architecture within the cell nucleus, referred to as
‘the SATB1 network’ and functions as a regulatory network of gene
expression (67–69). Although SATB1 function has been
studied mostly using T cells, its expression in the nuclei of other
cell types may exert global gene regulatory activities as well.
In the present study, SATB1 was colocalized with
5-FUrd in both the controls and cells with the phenotype of mitotic
catastrophe, following treatment with doxorubicin. Furthermore, we
observed a reduction in fluorescence intensity of both SATB1 and
5-FUrd. This is consistent with the results obtained in the study
by Chu et al, namely that the downregulation of SATB1
expression is responsible for active cell death initiation
(70).
While the decrease in fluorescence intensity of
SATB1 and 5-FUrd was observed in this study, the colocalization
analysis revealed an increased overlap of their fluorescence signal
in the area of the cell nucleus. This enhances our knowledge on
SATB1-mediated genome-wide transcription and indicates that during
active cell death, SATB1 is involved in the transcription of a
larger number of genes at the same time after the induction of
active cell death, as opposed to cells grown under physiological
conditions.
Similarly, the colocalization analysis of F-actin
and 5-FUrd, performed in the present study, revealed an intense
overlap of the nuclear fluorescence signal following the induction
of active cell death. Our results are in agreement with the
suggestions of Xu et al, that actin translocates into the
cell nucleus to function as a transcriptional modulator under
stress conditions and plays an important role in the regulation of
gene transcription (51,52). Moreover, the comparison of the
above mentioned results with the colocalizational analysis of SATB1
and F-actin, revealed the overlap of their fluorescence,
particularly in the area of the nucleus of in the cells in which
cell death was induced. Additionally, the analysis of the SATB1 and
5-FUrd fluorescence signal in the cell nucleus revealed their
colocalization in more densely organized nuclear F-actin
structures. These results correlate with the analysis of the
colocalization of SATB1 and F-actin shown by TEM. Furthermore, both
the FRET AB technique and phalloidin-based pull-down assay
confirmed that the SATB1/F-actin complex exists. Taken together
with the information that SATB1 was detected in β-actin
immunoprecipited cell lysates, our results are in agreement with
those of other studies, indicating that nuclear actin exists in
multiple forms: monomeric, oligomeric and short-polymeric (3,19,71).
In conclusion, we observed the colocalization of
SATB1 and F-actin in the transcriptional active regions of the cell
nucleus and a functional interaction was observed between SATB1 and
more densely organized nuclear F-actin structures at the border
between condensed and decondensed chromatin. This contributes to
the hypothesis that nuclear actin filaments are involved in
chromatin remodeling associated with transcriptional processes
during active cell death. Our data open the discussion on the
involvement of the SATB1/F-actin functional complex in the
architecture of the cell nucleus of cells undergoing active cell
death and put forward several issues that need be resolved in
future studies: i) the issue of which genes are transcribed by this
complex; ii) the significance of the cooperation between F-actin
and SATB1 in the regulation of active cell death processes; and
iii) the role of F-actin in processes regulated by SATB1.
Acknowledgements
This study was supported by the Polish Ministry of
Science under the Grant no. IP2011016571.
References
|
1
|
Pederson T and Aebi U: Actin in the
nucleus: what form and what for? J Struct Biol. 140:3–9. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pederson T and Aebi U: Nuclear actin
extends, with no contraction in sight. Mol Biol Cell. 16:5055–5060.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
McDonald D, Carrero G, Andrin C, de Vries
G and Hendzel MJ: Nucleoplasmic β-actin exists in a dynamic
equilibrium between low-mobility polymeric species and rapidly
diffusing populations. J Cell Biol. 172:541–552. 2006.
|
|
4
|
Jockusch BM, Schoenenberger CA, Stetefeld
J and Aebi U: Tracking down the different forms of nuclear actin.
Trends Cell Biol. 16:391–396. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ye J, Zhao J, Hoffmann-Rohrer U and Grummt
I: Nuclear myosin I acts in concert with polymeric actin to drive
RNA polymerase I transcription. Genes Dev. 22:322–330. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ferrai C, Naum-Onganía G, Longobardi E, et
al: Induction of HoxB transcription by retinoic acid requires actin
polymerization. Mol Biol Cell. 20:3543–3551. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gieni RS and Hendzel MJ: Actin dynamics
and functions in the interphase nucleus: moving toward an
understanding of nuclear polymeric actin. Biochem Cell Biol.
87:283–306. 2009.PubMed/NCBI
|
|
8
|
Belin BJ, Cimini BA, Blackburn EH and
Mullins RD: Visualization of actin filaments and monomers in
somatic cell nuclei. Mol Biol Cell. 24:982–994. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Grzanka D, Gagat M and Izdebska M: Actin
is required for cellular death. Acta Histochem. 115:775–782. 2013.
View Article : Google Scholar
|
|
10
|
Bettinger BT, Gilbert DM and Amberg DC:
Actin up in the nucleus. Nat Rev Mol Cell Biol. 5:410–415. 2004.
View Article : Google Scholar
|
|
11
|
Izdebska M, Gagat M, Grzanka D and Grzanka
A: Ultrastructural localization of F-actin using phalloidin and
quantum dots in HL-60 promyelocytic leukemia cell line after cell
death induction by arsenic trioxide. Acta Histochem. 115:487–495.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Castano E, Philimonenko VV, Kahle M, et
al: Actin complexes in the cell nucleus: new stones in an old
field. Histochem Cell Biol. 133:607–626. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
De Lanerolle P and Serebryannyy L: Nuclear
actin and myosins: life without filaments. Nat Cell Biol.
13:1282–1288. 2011.PubMed/NCBI
|
|
14
|
Fomproix N and Percipalle P: An
actin-myosin complex on actively transcribing genes. Exp Cell Res.
294:140–148. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gedge LJ, Morrison EE, Blair GE and Walker
JH: Nuclear actin is partially associated with Cajal bodies in
human cells in culture and relocates to the nuclear periphery after
infection of cells by adenovirus 5. Exp Cell Res. 303:229–239.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Saitoh N, Spahr CS, Patterson SD, Bubulya
P, Neuwald AF and Spector DL: Proteomic analysis of interchromatin
granule clusters. Mol Biol Cell. 15:3876–3890. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Olave IA, Reck-Peterson SL and Crabtree
GR: Nuclear actin and actin-related proteins in chromatin
remodeling. Annu Rev Biochem. 71:755–781. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Blessing CA, Ugrinova GT and Goodson HV:
Actin and ARPs: action in the nucleus. Trends Cell Biol.
14:435–442. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen M and Shen X: Nuclear actin and
actin-related proteins in chromatin dynamics. Curr Opin Cell Biol.
19:326–330. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Visa N and Percipalle P: Nuclear functions
of actin. Cold Spring Harb Perspect Biol. 2:a0006202010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kapoor P, Chen M, Winkler DD, Luger K and
Shen X: Evidence for monomeric actin function in INO80 chromatin
remodeling. Nat Struct Mol Biol. 20:426–432. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Krauss SW, Heald R, Lee G, Nunomura W,
Gimm JA, Mohandas N and Chasis JA: Two distinct domains of protein
4.1 critical for assembly of functional nuclei in vitro. J Biol
Chem. 277:44339–44346. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Krauss SW, Chen C, Penman S and Heald R:
Nuclear actin and protein 4.1: essential interactions during
nuclear assembly in vitro. Proc Natl Acad Sci USA. 100:10752–10757.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vartiainen MK, Guettler S, Larijani B and
Treisman R: Nuclear actin regulates dynamic subcellular
localization and activity of the SRF cofactor MAL. Science.
316:1749–1752. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zimmer C and Fabre E: Principles of
chromosomal organization: lessons from yeast. J Cell Biol.
192:723–733. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Spilianakis CG, Lalioti MD, Town T, Lee GR
and Flavell RA: Interchromosomal associations between alternatively
expressed loci. Nature. 435:637–645. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Schneider R and Grosschedl R: Dynamics and
interplay of nuclear architecture, genome organization, and gene
expression. Genes Dev. 21:3027–3043. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
De Belle I, Cai S and Kohwi-Shigematsu T:
The genomic sequences bound to special AT-rich sequence-binding
protein 1 (SATB1) in vivo in Jurkat T cells are tightly associated
with the nuclear matrix at the bases of the chromatin loops. J Cell
Biol. 141:335–348. 1998.
|
|
29
|
Cai S, Lee CC and Kohwi-Shigematsu T:
SATB1 packages densely looped, transcriptionally active chromatin
for coordinated expression of cytokine genes. Nat Genet.
38:1278–1288. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gotzmann J, Meissner M and Gerner C: The
fate of the nuclear matrix-associated-region-binding protein SATB1
during apoptosis. Cell Death Differ. 7:425–438. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Galande S, Dickinson LA, Mian IS, Sikorska
M and Kohwi-Shigematsu T: SATB1 cleavage by caspase 6 disrupts PDZ
domain-mediated dimerization, causing detachment from chromatin
early in T-cell apoptosis. Mol Cell Biol. 21:5591–5604. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mir R, Pradhan SJ and Galande S: Chromatin
organizer SATB1 as a novel molecular target for cancer therapy.
Curr Drug Targets. 13:1603–1615. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Notani D, Ramanujam PL, Kumar PP,
Gottimukkala KP, Kumar-Sinha C and Galande S: N-terminal PDZ-like
domain of chromatin organizer SATB1 contributes towards its
function as transcription regulator. J Biosci. 36:461–469. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu QY, Ribecco-Lutkiewicz M, Carson C, et
al: Mapping the initial DNA breaks in apoptotic Jurkat cells using
ligation-mediated PCR. Cell Death Differ. 10:278–289. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
De Lanerolle P, Johnson T and Hofmann WA:
Actin and myosin I in the nucleus: what next? Nat Struct Mol Biol.
12:742–746. 2005.PubMed/NCBI
|
|
36
|
Sjölinder M, Björk P, Söderberg E, Sabri
N, Farrants AK and Visa N: The growing pre-mRNA recruits actin and
chromatin-modifying factors to transcriptionally active genes.
Genes Dev. 19:1871–1884. 2005.PubMed/NCBI
|
|
37
|
Hofmann WA: Cell and molecular biology of
nuclear actin. Int Rev Cell Mol Biol. 273:219–263. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Louvet E and Percipalle P: Transcriptional
control of gene expression by actin and myosin. Int Rev Cell Mol
Biol. 272:107–147. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hofmann WA, Stojiljkovic L, Fuchsova B, et
al: Actin is part of pre-initiation complexes and is necessary for
transcription by RNA polymerase II. Nat Cell Biol. 6:1094–1101.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Qi T, Tang W, Wang L, Zhai L, Guo L and
Zeng X: G-actin participates in RNA polymerase II-dependent
transcription elongation by recruiting positive transcription
elongation factor b (P-TEFb). J Biol Chem. 286:15171–15181. 2011.
View Article : Google Scholar
|
|
41
|
Grzanka A, Grzanka D and Orlikowska M:
Fluorescence and ultrastructural localization of actin distribution
patterns in the nucleus of HL-60 and K-562 cell lines treated with
cytostatic drugs. Oncol Rep. 11:765–770. 2004.PubMed/NCBI
|
|
42
|
Grzanka D, Domaniewski J and Grzanka A:
Effect of doxorubicin on actin reorganization in Chinese hamster
ovary cells. Neoplasma. 52:46–51. 2005.PubMed/NCBI
|
|
43
|
Grzanka D, Marszałek A, Gagat M, Izdebska
M, Gackowska L and Grzanka A: Doxorubicin-induced F-actin
reorganization in cofilin-1 (nonmuscle) down-regulated CHO AA8
cells. Folia Histochem Cytobiol. 48:377–386. 2010.PubMed/NCBI
|
|
44
|
Izdebska M, Grzanka D, Gagat M, Gackowska
L and Grzanka A: The effect of G-CSF on F-actin reorganization in
HL-60 and K562 cell lines. Oncol Rep. 28:2138–2148. 2012.PubMed/NCBI
|
|
45
|
Ou H, Shen YH, Utama B, Wang J, Wang X,
Coselli J and Wang XL: Effect of nuclear actin on endothelial
nitric oxide synthase expression. Arterioscler Thromb Vasc Biol.
25:2509–2514. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang M-X, Zhang C, Shen YH, et al: Effect
of 27nt small RNA on endothelial nitric-oxide synthase expression.
Mol Biol Cell. 19:3997–4005. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Rando OJ, Zhao K, Janmey P and Crabtree
GR: Phosphatidylinositol-dependent actin filament binding by the
SWI/SNF-like BAF chromatin remodeling complex. Proc Natl Acad Sci
USA. 99:2824–2829. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Song Z, Wang M, Wang X, Pan X, Liu W, Hao
S and Zeng X: Nuclear actin is involved in the regulation of CSF1
gene transcription in a chromatin required, BRG1 independent
manner. J Cell Biochem. 102:403–411. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhao K, Wang W, Rando OJ, Xue Y, Swiderek
K, Kuo A and Crabtree GR: Rapid and phosphoinositol-dependent
binding of the SWI/SNF-like BAF complex to chromatin after T
lymphocyte receptor signaling. Cell. 95:625–636. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Percipalle P, Zhao J, Pope B, Weeds A,
Lindberg U and Daneholt B: Actin bound to the heterogeneous nuclear
ribonucleoprotein hrp36 is associated with Balbiani ring mRNA from
the gene to polysomes. J Cell Biol. 153:229–236. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Xu YZ, Thuraisingam T, Marino R and
Radzioch D: Recruitment of SWI/SNF complex is required for
transcriptional activation of the SLC11A1 gene during macrophage
differentiation of HL-60 cells. J Biol Chem. 286:12839–12849. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Xu T, Wu M, Feng J, Lin X and Gu Z:
RhoA/Rho kinase signaling regulates transforming growth
factor-β1-induced chondrogenesis and actin organization of
synovium-derived mesenchymal stem cells through interaction with
the Smad pathway. Int J Mol Med. 30:1119–1125. 2012.
|
|
53
|
Aoyama K, Yuki R, Horiike Y, et al:
Formation of long and winding nuclear F-actin bundles by nuclear
c-Abl tyrosine kinase. Exp Cell Res. 319:3251–3268. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zheng B, Han M, Bernier M and Wen J:
Nuclear actin and actin-binding proteins in the regulation of
transcription and gene expression. FEBS J. 276:2669–2685. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Dundr M, Ospina JK, Sung M-H, et al:
Actin-dependent intranuclear repositioning of an active gene locus
in vivo. J Cell Biol. 179:1095–1103. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Gonsior SM, Platz S, Buchmeier S, Scheer
U, Jockusch BM and Hinssen H: Conformational difference between
nuclear and cytoplasmic actin as detected by a monoclonal antibody.
J Cell Sci. 112(Pt 6): 797–809. 1999.PubMed/NCBI
|
|
57
|
Schoenenberger CA, Buchmeier S, Boerries
M, Sütterlin R, Aebi U and Jockusch BM: Conformation-specific
antibodies reveal distinct actin structures in the nucleus and the
cytoplasm. J Struct Biol. 152:157–168. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Hofmann WA, Arduini A, Nicol SM, Camacho
CJ, Lessard JL, Fuller-Pace FV and de Lanerolle P: SUMOylation of
nuclear actin. J Cell Biol. 186:193–200. 2009. View Article : Google Scholar
|
|
59
|
Ludérus ME, de Graaf A, Mattia E, den
Blaauwen JL, Grande MA, de Jong L and van Driel R: Binding of
matrix attachment regions to lamin B1. Cell. 70:949–959. 1992.
|
|
60
|
Ludérus ME, den Blaauwen JL, de Smit OJ,
Compton DA and van Driel R: Binding of matrix attachment regions to
lamin polymers involves single-stranded regions and the minor
groove. Mol Cell Biol. 14:6297–6305. 1994.PubMed/NCBI
|
|
61
|
Liu J, Bramblett D, Zhu Q, Lozano M,
Kobayashi R, Ross SR and Dudley JP: The matrix attachment
region-binding protein SATB1 participates in negative regulation of
tissue-specific gene expression. Mol Cell Biol. 17:5275–5287.
1997.PubMed/NCBI
|
|
62
|
Kaplan MH, Zong RT, Herrscher RF,
Scheuermann RH and Tucker PW: Transcriptional activation by a
matrix associating region-binding protein. contextual requirements
for the function of bright. J Biol Chem. 276:21325–21330. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Dobreva G, Dambacher J and Grosschedl R:
SUMO modification of a novel MAR-binding protein, SATB2, modulates
immunoglobulin mu gene expression. Genes Dev. 17:3048–3061. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Kaul-Ghanekar R, Jalota A, Pavithra L,
Tucker P and Chattopadhyay S: SMAR1 and Cux/CDP modulate chromatin
and act as negative regulators of the TCRbeta enhancer (Ebeta).
Nucleic Acids Res. 32:4862–4875. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Han HJ, Russo J, Kohwi Y and
Kohwi-Shigematsu T: SATB1 reprogrammes gene expression to promote
breast tumour growth and metastasis. Nature. 452:187–193. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Pavan Kumar P, Purbey PK, Sinha CK, Notani
D, Limaye A, Jayani RS and Galande S: Phosphorylation of SATB1, a
global gene regulator, acts as a molecular switch regulating its
transcriptional activity in vivo. Mol Cell. 22:231–243.
2006.PubMed/NCBI
|
|
67
|
Alvarez JD, Yasui DH, Niida H, Joh T, Loh
DY and Kohwi-Shigematsu T: The MAR-binding protein SATB1
orchestrates temporal and spatial expression of multiple genes
during T-cell development. Genes Dev. 14:521–535. 2000.PubMed/NCBI
|
|
68
|
Yasui D, Miyano M, Cai S, Varga-Weisz P
and Kohwi-Shigematsu T: SATB1 targets chromatin remodelling to
regulate genes over long distances. Nature. 419:641–645. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Cai S, Han HJ and Kohwi-Shigematsu T:
Tissue-specific nuclear architecture and gene expression regulated
by SATB1. Nat Genet. 34:42–51. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
70
|
Chu SH, Ma YB, Feng DF, Zhang H, Zhu ZA,
Li ZQ and Jiang PC: Upregulation of SATB1 is associated with the
development and progression of glioma. J Transl Med. 10:1492012.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Schuh M and Ellenberg J: Nuclear actin: a
lack of export allows formation of filaments. Curr Biol.
16:R321–R323. 2006. View Article : Google Scholar
|