Introduction
Idiopathic pulmonary fibrosis (IPF) is a progressive
interstitial lung disease of unknown etiology, with an appearance
of usual interstitial pneumonia (UIP) on lung biopsy (1). Studies have indicated that the
incidence of IPF is estimated to be 6.8 to 16.3 cases per 100,000
individuals each year in the United States, and the mean survival
rate from the time of diagnosis is 3–5 years, regardless of
treatment, which is a poorer prognosis than lung cancer (2–5).
The distinct characteristics of IPF are identified through clinical
manifestations, pulmonary function tests, radiology and prognosis.
Currently, the diagnosis of IPF relies on the finding of lung mass
in radiology imaging, mainly computed tomography (CT) and pulmonary
function tests; however, the diagnosis of small lesions is
relatively inaccurate. Currently, no known agents have been shown
to reduce the mortality associated with IPF. Clinical trials are
hindered by a dearth of clinically employed biomarkers, despite the
establishment of some clinical characteristics and the fact that
some peripheral blood proteins, such as the mucin-like
glycoprotein, KL-6 (6),
CC-chemokine ligand 18 (CCL-18) (7), surfactant protein A (SP)-A and D
(SP-D) (8) and matrix
metalloproteinase (MMP)-7 (9) are
currently being investigated as potential biomarkers in patients
with IPF. Therefore, seeking informative diagnostic markers with
greater clinical significance is essential for the successful
treatment and improved survival of patients with IPF.
microRNAs (miRNAs or miRs) are a novel class of
naturally occurring, short, non-coding, single-stranded RNAs, that
play key roles in various cellular processes commonly implicated in
cancer, such as cell differentiation, cell growth, angiogenesis,
epithelial-mesenchymal transition (EMT) and invasion (10–14). Following the release from
apoptotic or necrotic cells as a result of tissue damage or chronic
inflammation (15,16), miRNAs are highly stable and
abundant in biological fluids, such as blood, serum, plasma and
cerebrospinal fluid (CSF) (17–20). With an increasing number of
studies analyzing circulating miRNA expression profiles in plasma
or serum, miRNAs have become an attractive source of new nucleic
acid-based biomarkers. We previously demonstrated that miRNAs can
be detected in the serum of patients with IPF (21). This finding raised the possibility
that assaying miRNAs in serum may serve as a novel approach for the
blood-based detection of IPF. In this study, we focused on the
circulating miRNA expression profiles in serum samples of patienits
with IPF, validated our microarray results through quantitative
reverse transcription-polymerase chain reaction (qRT-PCR), and
performed a preliminary analysis of the biological functions of the
most differentially expressed miRNAs. These results may provide a
basis for further study of the molecular markers associated with
IPF for its early diagnosis.
Materials and methods
Clinical samples and serum
collection
Following approval by the Ethics Committee of
Zhengzhou University, Zhengzhou, China and after written informed
consent was obtained, serum samples were collected from 149
subjects, including 76 patients with IPF and 73 age- and
gender-matched healthy donors who served as the controls for this
study. All samples were collected from consenting individuals at
the First Affiliated Hospital of Zhengzhou University (FAHZZU).
Patients were diagnosed with IPF using a multidisciplinary approach
involving clinicians, radiologists and pathologists, along with
high-resolution computed tomography (HRCT), and the relevant
demographic and clinicopathological characteristics of the subjects
were obtained. Control subjects were recruited from individuals who
sought a routine health check-up at the Healthy Physical
Examination Centre of FAHZZU.
For serum collection, venous blood (2 ml/subject)
was collected in the morning before breakfast from the patients
with IPF and the control subjects, via a direct venous puncture,
into tubes containing ethylenediaminetetraacetic acid (EDTA). The
tubes were centrifuged at 2,500 × g for 10 min at 4°C to completely
remove cellular components. Tthe serum was then collected gently
and transferred into an RNase-free tube for the extraction of RNA
and stored at −80°C.
RNA extraction
Total RNA, which included miRNAs, was isolated from
the serum using a total RNA extraction kit (Shanghai Novland Co.,
Ltd., Shanghai, China) following the manufacturer’s instructions.
The quality of the total RNA was verified using a NanoDrop ND-1000
spectrophotometer (Thermo Scientific, Waltham, MA, USA). We defined
an OD260/280 value of approximately 1.8 as a criterion of
acceptable purity for further microarray analysis.
microRNA microarray analysis
To assess the labeling and hybridization
efficiencies, the total RNA samples were spiked with the microRNA
Spike-In kit (Agilent Technologies, Santa Clara, CA, USA).
Following treatment with calf intestine phosphatase (CIP), a
labeling reaction was initiated with 100 ng of total RNA per
sample. A T4 RNA ligase that incorporates cyanine 3-cytidine
bisphosphate (miRNA Complete Labeling and Hyb kit; Agilent
Technologies) was used to label the dephosphorylated RNA. The
cyanine-3-labeled miRNA samples were then prepared for
one-color-based hybridization (Complete miRNA Labeling and Hyb kit;
Agilent Technologies). Hybridization was performed at 55°C for 20 h
using a human miRNA microarray kit, Release 16.0, 8×60K format
(Agilent Technologies). The microarray slides were washed (Gene
Expression Wash Buffers; Agilent Technologies) and dried with
acetonitrile (Sigma-Aldrich, St. Louis, MO, USA). Fluorescent
signal intensities were detected using an Agilent DNA Microarray
Scanner and Scan Control A.8.4.1 software (both from Agilent
Technologies). The data were extracted using Feature Extraction
10.7.3.1 software (Agilent Technologies). Predicted target genes
were analyzed with Gene Ontology (GO) enrichment analysis and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway mapping.
qRT-PCR
The microarray data was validated by qRT-PCR. U6
snRNA served as an endogenous control for normalization. A
High-Specificity miRNA qRT-PCR Detection kit (Stratagene Corp., La
Jolla, CA, USA) was used in conjunction with an ABI 7500 thermal
cycler, according to the manufacturer’s instructions. The
correlating primers are shown in Table I. The PCR reaction was conducted
at 95°C for 180 sec, followed by 40 cycles of 95°C for 15 sec and
65°C for 30 sec. The results were analyzed using ABI Prism 7500 SDS
software V1.3.1 (Applied Biosystems, Bedford, MA, USA) and
expressed as relative quantification (2−ΔCt) mode with
triplicate measurements, as previously described (22), where the change in the threshold
cycle (ΔCt) = Ct (miRNA) − Ct (U6). Fold change values of the
selected miRNAs were transformed to log10 values, both for the
qRT-PCR and the microarray results.
 | Table IList of primers used for qRT-PCR. |
Table I
List of primers used for qRT-PCR.
| Gene name | RT primers | PCR primers |
|---|
| miR-101-3p |
5′-GTCGTATCCAGTGCAGGGTCCGAGG
TATTCGCACTGGATACGACATGTCAT-3′ | F:
5′-TCCGAAAGTCAATAGTGTC-3′
R: 5′-GTGCAGGGTCCGAGGT-3′ |
| miR-142-5p |
5′-GTCGTATCCAGTGCAGGGTCCGAGG
TATTCGCACTGGATACGACGTATTTC-3′ | F:
5′-TCCGATCATCACGAAAGAT-3′
R: 5′-GTGCAGGGTCCGAGGT-3′ |
| miR-557 |
5′-GTCGTATCCAGTGCAGGGTCCGAGG
TATTCGCACTGGATACGACCAAACGT-3′ | F:
5′-TCCGATCTGTTCCGGGTGGGC-3′
R: 5′-GTGCAGGGTCCGAGGT-3′ |
| miR-187-5p |
5′-GTCGTATCCAGTGCAGGGTCCGAGG
TATTCGCACTGGATACGACCCGATGTT-3′ | F:
5′-TCCGACGGGCCCAGGACAC-3′
R: 5′-GTGCAGGGTCCGAGGT-3′ |
| miR-484 |
5′-GTCGTATCCAGTGCAGGGTCCGAGG
TATTCGCACTGGATACGACAGTCCGAG-3′ | F:
5′-TCCGATAGCCCTCCCCTGA-3′
R: 5′-GTGCAGGGTCCGAGGT-3′ |
| miR-21 |
5′-GTCGTATCCAGTGCAGGGTCCGAGG
TATTCGCACTGGATACGACATCGAAT-3′ | F:
5′-TCCGAAGTTGTAGTCAGACT-3′
R: 5′-GTGCAGGGTCCGAGGT-3′ |
| miR-155 |
5′-GTCGTATCCAGTGCAGGGTCCGAGG
TATTCGCACTGGATACGACAATTACG-3′ | F:
5′-TCCGATGGGGATAGTGCTAAT-3′
R: 5′-GTGCAGGGTCCGAGGT-3′ |
| miR-3675-3p |
5′-GTCGTATCCAGTGCAGGGTCCGAGG
TATTCGCACTGGATACGACGTAGAGA-3′ | F:
5′-TCCGAAACCCCCTCAAGGAA-3′
R: 5′-GTGCAGGGTCCGAGGT-3′ |
| miR-1229 |
5′-GTCGTATCCAGTGCAGGGTCCGAGG
TATTCGCACTGGATACGACGAGAGTG-3′ | F:
5′-TCCGAGACACCCTCCCGTCAC-3′
R: 5′-GTGCAGGGTCCGAGGT-3′ |
| U6 snRNA |
5′-GTCGTATCCAGTGCAGGGTCCGAGG
TATTCGCACTGGATACGACAAAATA-3′ | F:
5′-TCCGATCGTGAAGCGTTC-3′
R: 5′-GTGCAGGGTCCGAGGT-3′ |
Statistical analysis
SPSS version 17.0 software was used for statistical
analysis (SPSS, Inc., Chicago, IL, USA). The difference in miRNA
expression levels between 2 serum samples was determined by the
Mann-Whitney U test. The Mann-Whitney U test and Kruskal-Wallis
test were used to compare the differences among clinicopathological
parameters. For multiple comparisons among groups (functional and
radiographical variables), either one-way analysis of variance
(ANOVA) followed by Tukey’s test, or Kruskal-Wallis tests, where
appropriate, were performed. A value of P<0.05 was considered to
indicate a statistically significant difference. Quantitative data
are presented as the means ± standard deviation (SD).
Results
Differential miRNA expression in serum
from patients with IPF
Comparing the miRNA expression profiles in serum
from patients with IPF and the healthy controls on the basis of the
miRNA array results, we found that 60 miRNAs were differentially
expressed in the serum of patients with IPF (P<0.05), including
8 upregulated and 52 downregulated miRNAs. Among the 60
differentially expressed miRNAs, all of the upregulated miRNAs
(Table III) and 21 of the 52
downregulated miRNAs (Table II)
showed at least a 2.5-fold change in expression in the patients
with IPF.
 | Table IIIUpregulated miRNAs with a ≥2.5-fold
change in expression in serum from patients with IPF. |
Table III
Upregulated miRNAs with a ≥2.5-fold
change in expression in serum from patients with IPF.
| Systematic
name | FC | Log FC | Regulation |
|---|
|
hsa-miR-3675-3p | 74.52153 | 6.2195854 | Up |
| hsa-miR-21 | 54.23154 | 5.7234412 | Up |
| hsa-miR-1229 | 32.81375 | 5.0362287 | Up |
| hsa-miR-155 | 3.2157842 | 1.6084715 | Up |
| hsa-miR-18b-3p | 2.9419162 | 1.5567561 | Up |
| hsa-miR-636 | 2.8691778 | 1.5206374 | Up |
| hsa-miR-3646 | 2.8469105 | 1.5093971 | Up |
| hsa-miR-574-5p | 2.7281206 | 1.4479074 | Up |
 | Table IIDownregulated miRNAs with a ≥2.5-fold
change in expression in serum from patients with IPF. |
Table II
Downregulated miRNAs with a ≥2.5-fold
change in expression in serum from patients with IPF.
| Systematic name | FC | Log FC | Regulation |
|---|
| hsa-miR-142-5p | −142.85098 | −7.158367 | Down |
| hsa-miR-3652 | −71.64325 | −6.162759 | Down |
| hsa-miR-4322 | −58.7685 | −5.8769712 | Down |
| hsa-miR-557 | −55.494648 | −5.7942767 | Down |
| hsa-miR-187-5p | −54.88799 | −5.7784185 | Down |
|
hsa-miR-1224-5p | −51.036926 | −5.6734695 | Down |
| hsa-miR-484 | −49.851433 | −5.639563 | Down |
|
hsa-miR-3682-3p | −48.302345 | −5.5940213 | Down |
| hsa-miR-3131 | −48.020832 | −5.5855885 | Down |
|
hsa-miR-148b-3p | −47.77412 | −5.5781574 | Down |
| hsa-miR-874 | −44.02572 | −5.4602747 | Down |
| hsa-miR-150-5p | −42.34698 | −5.404187 | Down |
| hsa-miR-1305 | −41.710274 | −5.382331 | Down |
| hsa-miR-663a | −36.454872 | −5.18804 | Down |
| hsa-miR-101-3p | −34.896984 | −5.1250305 | Down |
|
hsa-miR-516a-5p | −34.492382 | −5.108206 | Down |
| hsa-miR-328 | −33.15238 | −5.0510406 | Down |
| hsa-miR-877-5p | −32.35714 | −5.016012 | Down |
| hsa-miR-3137 | −31.168226 | −4.962004 | Down |
| hsa-miR-483-3p | −30.725616 | −4.94137 | Down |
| hsa-miR-23b-3p | −30.128986 | −4.91308 | Down |
Relevant GO enrichment analysis of miRNAs
in serum from patients with IPF
To determine the effects of the differentially
expressed miRNAs on IPF and to better understand the functions of
two types of genes in different biological processes, which provide
more information on the miRNA expression profiles in serum from
patients with IPF at the cellular level, we performed GO enrichment
analysis with FunNet software. GO enrichment analysis includes
molecular functions, cellular components and biological processes.
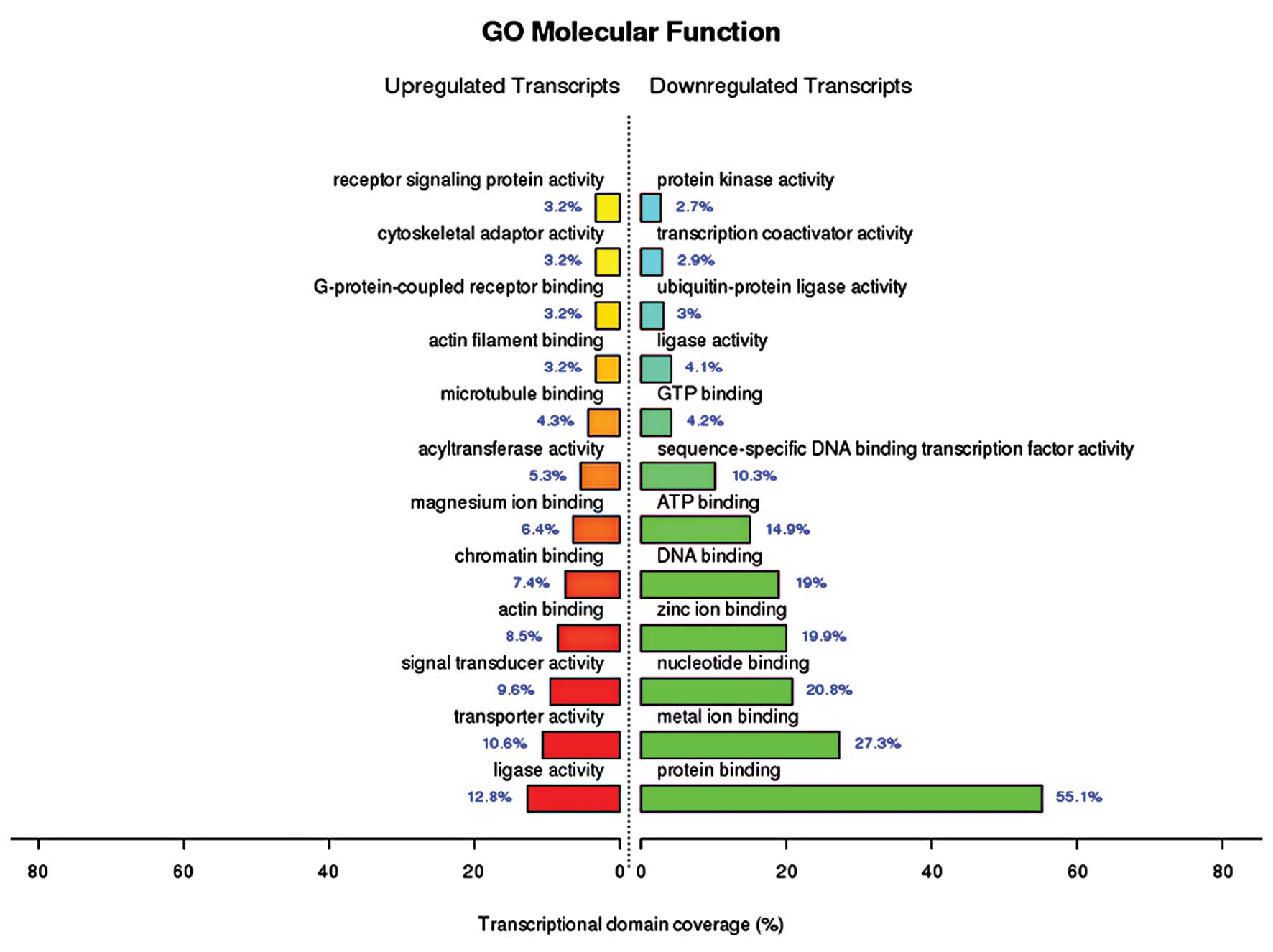
The molecular functions of the upregulated genes identified by GO
enrichment analysis included (among others) ligase activity,
transporter activity, signal transducer activity and actin binding,
while the molecular functions of the downregulated genes included
(among others) protein binding, mental ion binding, nucleotide
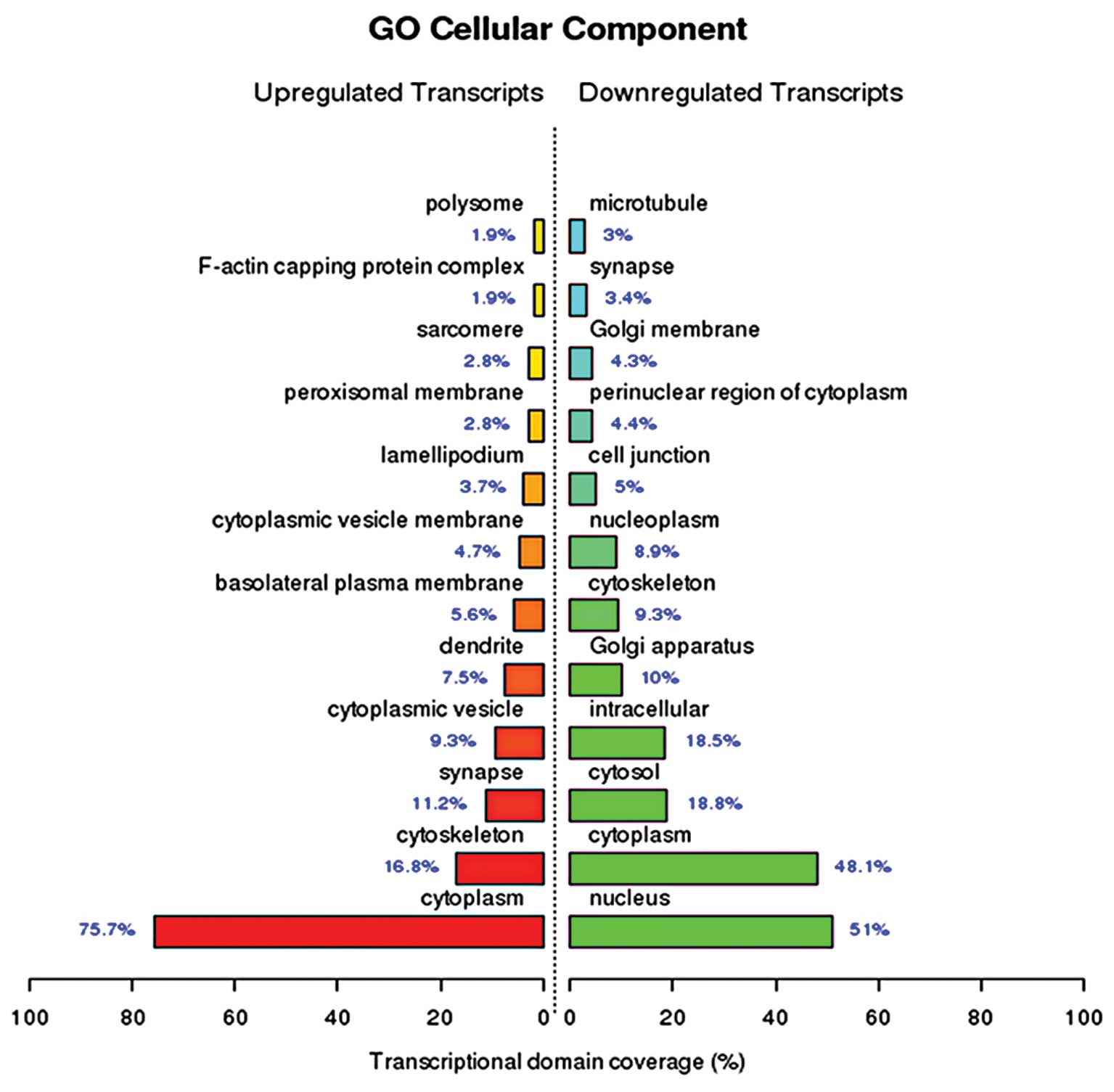
binding, zinc ion binding, DNA binding and ATP binding (Fig. 1). The cellular components of the
upregulated genes identified by GO enrichment analysis included
(among others) the cytoplasm, cytoskeleton, synapses and
cytoplasmic vesicles, while the cellular components of the
downregulated genes included (among others) the nucleus, cytoplasm,
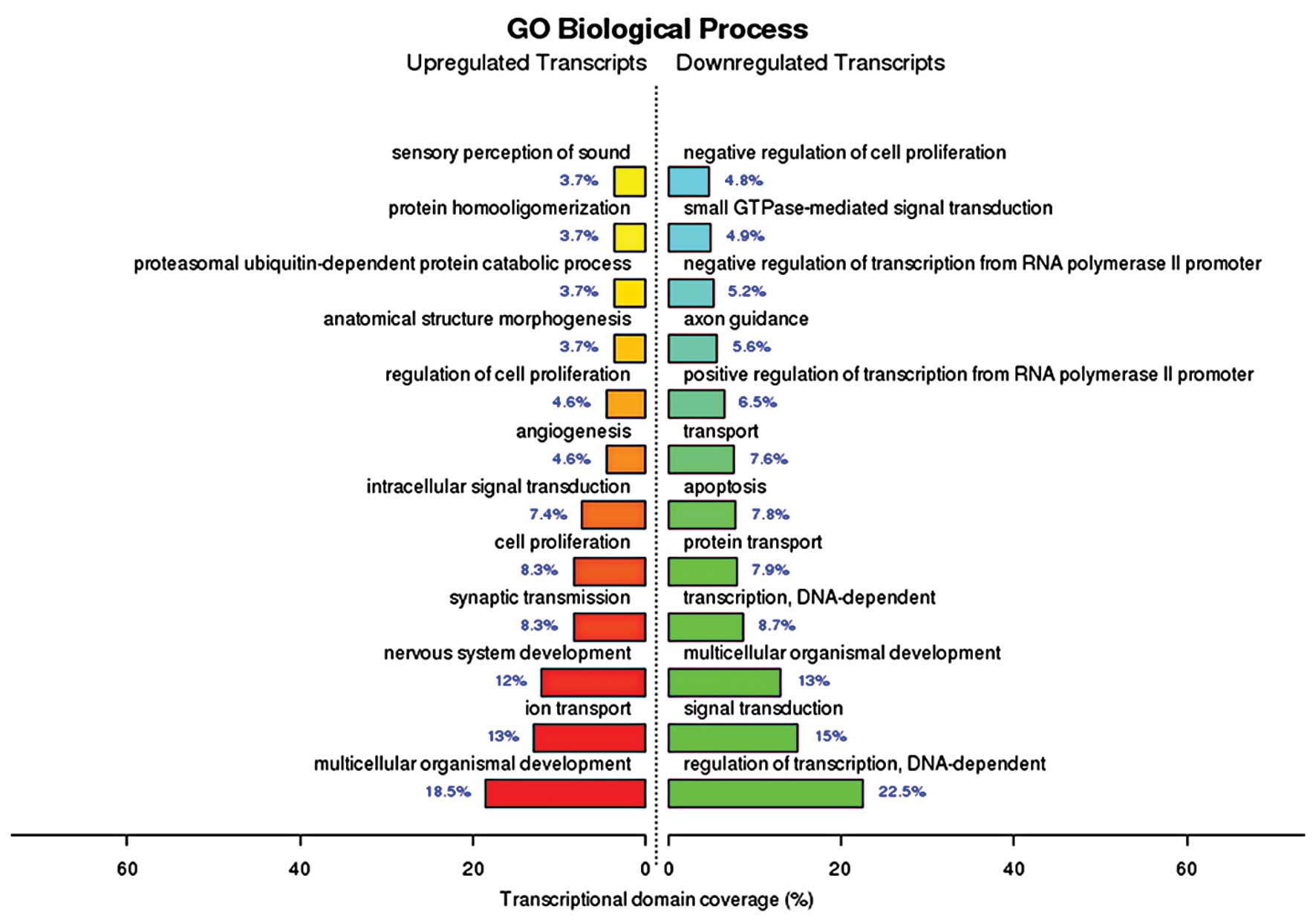
cytosol and intracellular compartments (Fig. 2). The biological processes of the
upregulated genes identified by GO enrichment analysis included
(among others) multicellular organismal development, ion transport
and nervous system development, while the biological processes of
the downregulated genes included (among others) the regulation of
transcription, DNA-dependent signal transduction and multicellular
organismal development (Fig. 3).
All of the results showed that two types of genes exhibited
variable distribution of some functions.
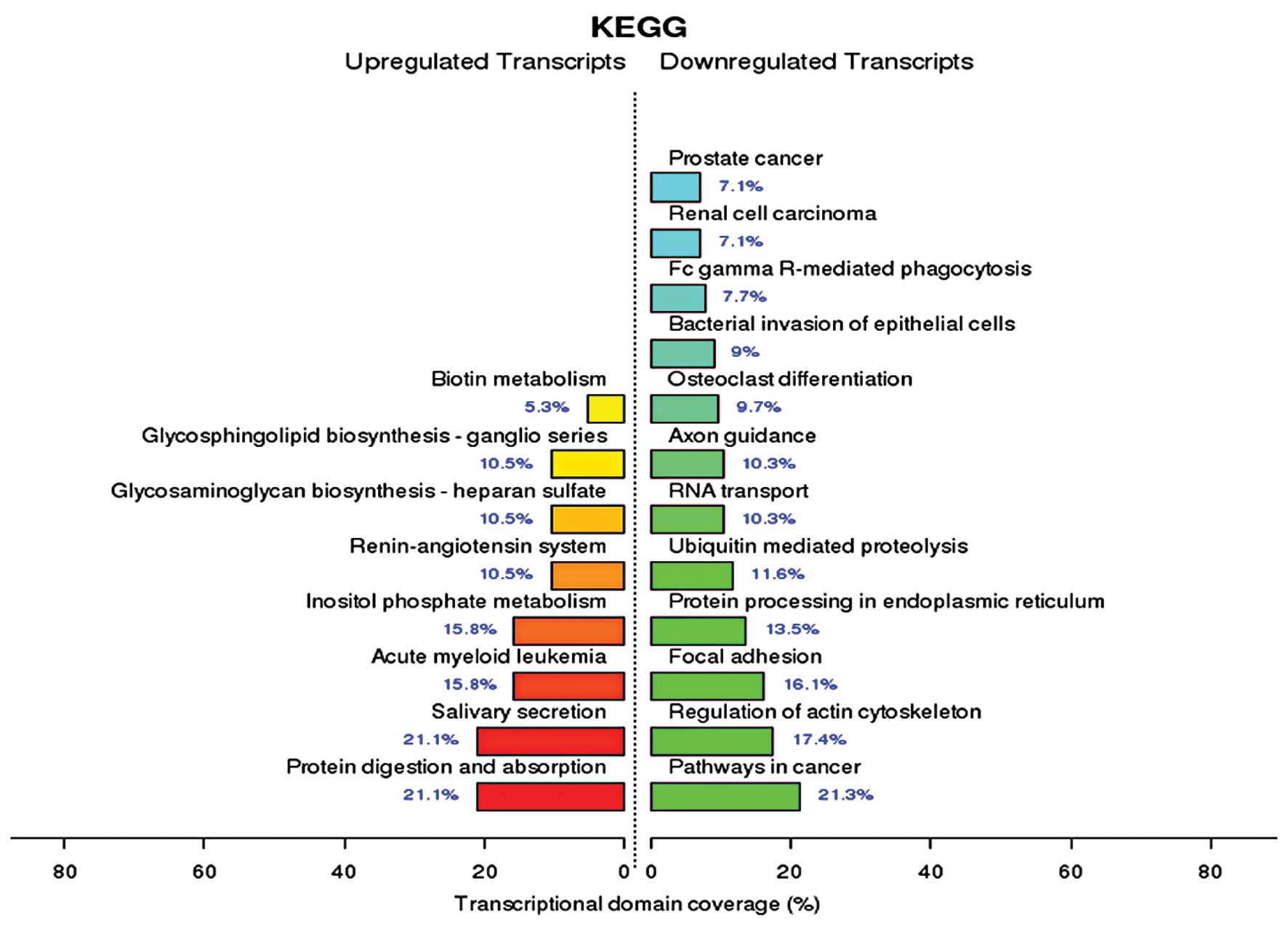
KEGG enrichment analysis of miRNAs in
serum from patients with IPF
KEGG is a knowledge base for the systematic analysis
of gene functions, in terms of the networks of genes and molecules.
The major component of KEGG is the Pathway database, which consists
of graphical diagrams of biochemical pathways including most of the
known metabolic pathways and some of the known regulatory pathways.
Upregulated transcripts included those involved in protein
digestion and absorption, salivary secretion, acute myeloid
leukemia, inositol phosphate metabolism, as well as others.
Downregulated transcripts included those involved in cancer
pathways, such as the regulation of the actin cytoskeleton, focal
adhesion, protein processing in endoplasmic reticulum, as well as
others (Fig. 4).
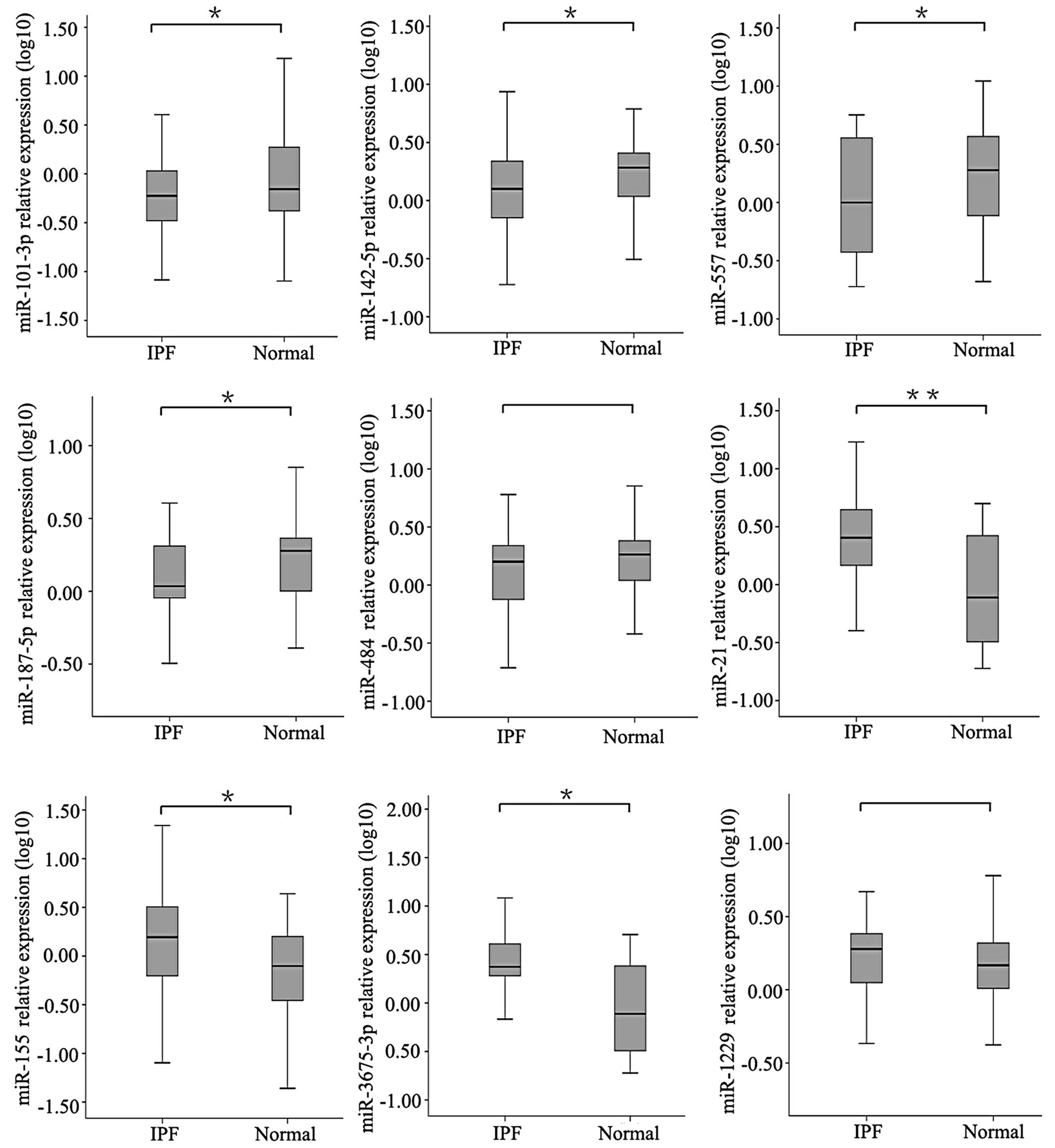
Validation of selected miRNA expression
profiles in serum by qRT-PCR
On the basis of the microarray results, we selected
9 miRNAs, including 4 upregulated miRNAs (miR-3675-3p, miR-21,
miR-1229 and miR-155) and 5 downregulated miRNAs (miR-142-5p,
miR-557, miR-484, miR-187-5p and miR-101-3p) for further validation
in IPF and in healthy serum by qRT-PCR. The expression of
miR-142-5p, miR-557, miR-187-5p and miR-101-3p was found to be
significantly reduced in the serum from patients with IPF
(P<0.05). The expression of miR-3675-3p, miR-21 and miR-155 was
found to be significantly increased in the serum of patients with
IPF (P<0.05). However, the expression of miR-1229 and miR-187-5p
did not differ significantly between the IPF and control serum
samples (P>0.05). Generally, the results of the both methods
(qRT-PCR and microarray) were quite similar (Fig. 5).
Expression levels of miRNAs in serum from
patients with IPF are associated with clinicopathologic
characteristics of IPF
Combining the results of qRT-PCR and Agilent
microarray, we found that the changes in the expression of miR-21
and miR-101-3p were more obvious than those of the other miRNAs.
For all the samples, clinicopathological information (gender, age,
FVC and HRCT) were available (Table
IV). With further analysis, we also found that the expression
levels of miR-21, miR-155 and miR-101-3p were associated with FVC
and radiological features (P<0.05 or P<0.01) (Table IV); however, no significant
differences were observed between miR-21, miR-155 and miR-101-3p
expression and gender and age (P>0.05) (Table IV).
 | Table IVmiRNA expression levels associated
with clinicopathological characteristics of patients with IPF. |
Table IV
miRNA expression levels associated
with clinicopathological characteristics of patients with IPF.
| Clinicopathological
characteristic | n | miRNA-21 expression
(2−ΔCt) | miR-155 expression
(2−ΔCt) | miR-101-3p
expression (2−ΔCt) |
|---|
|
|
|
|---|
| Median ± SD | P-value | Median ± SD | P-value | Median ± SD | P-value |
|---|
| Age (years) | | | 0.670 | | 0.509 | | 0.218 |
| >52 | 37 | 4.33±3.44 | | 3.70±4.65 | | 0.89±1.17 | |
| ≤52 | 39 | 3.88±2.64 | | 2.89±3.95 | | 0.78±0.43 | |
| Gender | | | 0.763 | | 0.597 | | 0.059 |
| Male | 46 | 4.28±3.22 | | 3.61±4.21 | | 0.76±0.94 | |
| Female | 30 | 3.97±1.99 | | 2.77±2.86 | | 0.95±0.69 | |
| FVC (%) | | | 0.016 | | 0.042 | | 0.037 |
| Mild (80) | 18 | 2.20±0.98a | | 1.43±1.16a | | 1.21±1.43a | |
| Moderate
(50–80) | 22 | 3.45±2.89 | | 2.70±2.70 | | 0.95±0.88 | |
| Severe (50) | 36 | 4.60±3.36a | | 3.98±4.51a | | 0.57±0.40a | |
| HRCT | | | 0.002 | | 0.005 | | 0.021 |
| GGO | 17 | 1.90±0.78b | | 1.51±1.15b | | 1.17±1.04b | |
| Reticulation | 26 | 3.20±2.13 | | 2.57±2.68 | | 0.92±0.80 | |
| Honeycombing | 33 | 4.72±3.43b | | 4.79±4.71b | | 0.59±0.34b | |
Discussion
The diagnostic criteria for IPF are presented in the
ATS/ERS statement (23). In the
appropriate clinical setting, the presence of a UIP pattern on HRCT
is sufficient for the diagnosis of IPF. However, some cases of UIP
are difficult to differentiate from fibrotic non-specific
interstitial pneumonia, which may exhibit honeycombing. Thus, the
major and minor criteria for the clinical diagnosis of IPF have
been eliminated in this new ATS/ERS statement. The diagnosis of IPF
can be achieved with certainty only after the pulmonologist,
radiologist and pathologist have reviewed all of the clinical,
radiological and pathological data.
Increasing evidence indicates that serum miRNAs may
offer an interesting new avenue for diagnostic development in a
variety of diseases. In the field of cancer, miR-21 was the first
serum miRNA to be identified, and was associated with large B-cell
lymphoma (24). Serum miRNAs have
also been suggested to be novel biomarkers for ovarian cancer
(25), lung cancer (26,27) and colorectal cancer (28). Furthermore, miRNAs have been
implicated as circulating biomarkers in tissue injury, including
liver, muscle and brain (29,30). The interest in serum miRNAs lies
in their central roles in the regulation of gene expression and the
implication of miRNA-specific aberrant expression in the
pathogenesis of a number of diseases.
In the present study, we obtained a total of 76
serum samples from patients with IPF to study changes in miRNA
expression compared to serum samples obtained from healthy
subjects. The microarray data showed that 60 important miRNAs were
differentially expressed in patients with IPF. The varying
expression of miRNAs provides a useful clue for the in-depth
research of serological diagnosis in IPF. With the results of
microarray analysis, we selected 9 miRNAs that exhibited marked
variations in expression for validation by qRT-PCR, including 4
upregulated miRNAs and 5 downregulated miRNAs. We found that the
qRT-PCR and Agilent microarray miRNA profiles significantly
correlated. Among the 9 miRNAs examined, miR-3675-3p, miR-21,
miR-155, miR-142-5p, miR-557, miR-187-5p and miR-101-3p showed
significant changes in expression (P<0.05). We also found that
the expression of miR-21 was markedly altered, and that the
upregulation of miR-21 was consistent with its expression level in
IPF tissue, as previously demonstrated (31). These observations demonstrate the
importance of investigating the roles of the remaining miRNAs in
the pathogenesis of IPF and analyzing the biologically relevant
molecular mechanisms in future studies. Furthermore, the specific
functions of serum miRNAs, such as those identified by GO
enrichment and KEGG enrichment analysis, may also provide some
guidance for further verification through functional studies, which
may have ties with the biological processes of IPF at the cellular
level. Additionally, we demonstrated that the expression levels of
miR-21, miR-155 and miR-101-3p in serum were associated with the
degree of FVC and HRCT in IPF. However, no correlation was observed
between these miRNAs and other clinicopathological characteristics,
including gender and age. These findings suggest that miR-21,
miR-155 and miR-101-3p play an important role in the progression
and development of IPF. Multi-analysis showed that serum miRNA
expression levels were the potential prognostic factors for
patients with IPF. Of course, more extensive data and studies with
a larger sample number are required to confirm the diagnostic
significance of miRNA expression levels in patients with IPF.
In conclusion, our study established differential
expression profiles of miRNAs, which seem to be involved in
different biological processes in IPF. Additionally, we found that
the altered expression levels of miR-21, miR-155 and miR-101-3p
were associated with the degree of FVC and HRCT in IPF. Our data
may serve as a basis for further investigation, preferably in large
prospective studies, before miRNAs can be used as a non-invasive
screening tool for IPF in routine clinical practice.
Acknowledgements
The authors are grateful to all staff at the study
centre who contributed to this study. This study was supported by a
grant from the Ministry of Major Science and Technology of Henan
(20130205).
References
|
1
|
Maher TM, Wells AU and Laurent GJ:
Idiopathic pulmonary fibrosis: multiple causes and multiple
mechanisms? Eur Respir J. 30:835–839. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fernández Pérez ER, Daniels CE, Schroeder
DR, St Sauver J, Hartman TE, Bartholmai BJ, Yi ES and Ryu JH:
Incidence, prevalence, and clinical course of idiopathic pulmonary
fibrosis: a population-based study. Chest. 137:129–137.
2010.PubMed/NCBI
|
|
3
|
Navaratnam V, Fleming KM, West J, Smith
CJ, Jenkins RG, Fogarty A and Hubbard RB: The rising incidence of
idiopathic pulmonary fibrosis in the U.K. Thorax. 66:462–467. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nalysnyk L, Cid-Ruzafa J, Rotella P and
Esser D: Incidence and prevalence of idiopathic pulmonary fibrosis:
review of the literature. Eur Respir Rev. 21:355–361. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ley B, Collard HR and King TE Jr: Clinical
course and prediction of survival in idiopathic pulmonary fibrosis.
Am J Respir Crit Care Med. 183:431–440. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yokoyama A, Kondo K, Nakajima M,
Matsushima T, Takahashi T, Nishimura M, Bando M, Sugiyama Y, Totani
Y, Ishizaki T, Ichiyasu H, Suga M, Hamada H and Kohno N: Prognostic
value of circulating KL-6 in idiopathic pulmonary fibrosis.
Respirology. 11:164–168. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Prasse A, Probst C, Bargagli E, Zissel G,
Toews GB, Flaherty KR, Olschewski M, Rottoli P and Muller-Quernheim
J: Serum CC-chemokine ligand 18 concentration predicts outcome in
idiopathic pulmonary fibrosis. Am J Respir Crit Care Med.
179:717–723. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kinder BW, Brown KK, McCormack FX, Ix JH,
Kervitsky A, Schwarz MI and King TE Jr: Serum surfactant protein-A
is a strong predictor of early mortality in idiopathic pulmonary
fibrosis. Chest. 135:1557–1563. 2009. View Article : Google Scholar
|
|
9
|
Rosas IO, Richards TJ, Konishi K, Zhang Y,
Gibson K, Lokshin AE, Lindell KO, Cisneros J, Macdonald SD, Pardo
A, Sciurba F, Dauber J, Selman M, Gochuico BR and Kaminski N: MMP1
and MMP7 as potential peripheral blood biomarkers in idiopathic
pulmonary fibrosis. PLoS Med. 5:e932008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin KY, Zhang XJ, Feng DD, Zhang H, Zeng
CW, Han BW, Zhou AD, Qu LH, Xu L and Chen YQ: miR-125b, a target of
CDX2, regulates cell differentiation through repression of the core
binding factor in hematopoietic malignancies. J Biol Chem.
286:38253–38263. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Han BW, Feng DD, Li ZG, Luo XQ, Zhang H,
Li XJ, Zhang XJ, Zheng LL, Zeng CW, Lin KY, Zhang P, Xu L and Chen
YQ: A set of miRNAs that involve in the pathways of drug resistance
and leukemic stem-cell differentiation is associated with the risk
of relapse and glucocorticoid response in childhood ALL. Hum Mol
Genet. 20:4903–4915. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hwang MS, Yu N, Stinson SY, Yue P, Newman
RJ, Allan BB and Dornan D: miR-221/222 targets adiponectin receptor
1 to promote the epithelial-to-mesenchymal transition in breast
cancer. PLoS One. 8:e665022013. View Article : Google Scholar
|
|
13
|
Paterson EL, Kazenwadel J, Bert AG,
Khew-Goodall Y, Ruszkiewicz A and Goodall GJ: Down-regulation of
the miRNA-200 family at the invasive front of colorectal cancers
with degraded basement membrane indicates EMT is involved in cancer
progression. Neoplasia. 15:180–191. 2013.PubMed/NCBI
|
|
14
|
Cheng CW, Wang HW, Chang CW, Chu HW, Chen
CY, Yu JC, Chao JI, Liu HF, Ding SL and Shen CY: MicroRNA-30a
inhibits cell migration and invasion by downregulating vimentin
expression and is a potential prognostic marker in breast cancer.
Breast Cancer Res Treat. 134:1081–1093. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bala S, Tilahun Y, Taha O, Alao H, Kodys
K, Catalano D and Szabo G: Increased microRNA-155 expression in the
serum and peripheral monocytes in chronic HCV infection. J Transl
Med. 10:1512012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
van der Meer AJ, Farid WR, Sonneveld MJ,
de Ruiter PE, Boonstra A, van Vuuren AJ, Verheij J, Hansen BE, de
Knegt RJ, van der Laan LJ and Janssen HL: Sensitive detection of
hepatocellular injury in chronic hepatitis C patients with
circulating hepatocyte-derived microRNA-122. J Viral Hepat.
20:158–166. 2013.PubMed/NCBI
|
|
17
|
Haghikia A, Haghikia A, Hellwig K,
Baraniskin A, Holzmann A, Décard BF, Thum T and Gold R: Regulated
microRNAs in the CSF of patients with multiple sclerosis: a
case-control study. Neurology. 79:2166–2170. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mitchell PS1, Parkin RK, Kroh EM, Fritz
BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J,
O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS,
Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB and
Tewari M: Circulating microRNAs as stable blood-based markers for
cancer detection. Proc Natl Acad Sci USA. 105:10513–10518. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu R, Chen X, Du Y, Yao W, Shen L, Wang
C, Hu Z, Zhuang R, Ning G, Zhang C, Yuan Y, Li Z, Zen K, Ba Y and
Zhang CY: Serum microRNA expression profile as a biomarker in the
diagnosis and prognosis of pancreatic cancer. Clin Chem.
58:610–618. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang C, Wang C, Chen X, Chen S, Zhang Y,
Zhi F, Wang J, Li L, Zhou X, Li N, Pan H, Zhang J, Zen K, Zhang CY
and Zhang C: Identification of seven serum microRNAs from a
genome-wide serum microRNA expression profile as potential
noninvasive biomarkers for malignant astrocytomas. Int J Cancer.
132:116–127. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li P, Zhao GQ, Chen TF, Chang JX, Wang HQ,
Chen SS and Zhang GJ: Serum miR-21 and miR-155 expression in
idiopathic pulmonary fibrosis. J Asthma. 50:960–964. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Raghu G, Collard HR, Egan JJ, et al: An
official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis:
evidence-based guidelines for diagnosis and management. Am J Respir
Crit Care Med. 183:788–824. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lawrie CH, Gal S, Dunlop HM, Pushkaran B,
Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J,
Wainscoat JS, Hatton CS and Harris AL: Detection of elevated levels
of tumour-associated microRNAs in serum of patients with diffuse
large B-cell lymphoma. Br J Haematol. 141:672–675. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zheng H, Zhang L, Zhao Y, Yang D, Song F,
Wen Y, Hao Q, Hu Z, Zhang W and Chen K: Plasma miRNAs as diagnostic
and prognostic biomarkers for ovarian cancer. PLoS One.
8:e778532013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu XG, Zhu WY, Huang YY, Ma LN, Zhou SQ,
Wang YK, Zeng F, Zhou JH and Zhang YK: High expression of serum
miR-21 and tumor miR-200c associated with poor prognosis in
patients with lung cancer. Med Oncol. 29:618–626. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang Y, Gu J, Roth JA, Hildebrandt MA,
Lippman SM, Ye Y, Minna JD and Wu X: Pathway-based serum microRNA
profiling and survival in patients with advanced stage non-small
cell lung cancer. Cancer Res. 73:4801–4809. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang Q, Huang Z, Ni S, Xiao X, Xu Q, Wang
L, Huang D, Tan C, Sheng W and Du X: Plasma miR-601 and miR-760 are
novel biomarkers for the early detection of colorectal cancer. PLoS
One. 7:e443982012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Laterza OF, Lim L, Garrett-Engele PW,
Vlasakova K, Muniappa N, Tanaka WK, Johnson JM, Sina JF, Fare TL,
Sistare FD and Glaab WE: Plasma MicroRNAs as sensitive and specific
biomarkers of tissue injury. Clin Chem. 55:1977–1983. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tryndyak VP, Latendresse JR, Montgomery B,
Ross SA, Beland FA, Rusyn I and Pogribny IP: Plasma microRNAs are
sensitive indicators of inter-strain differences in the severity of
liver injury induced in mice by a choline- and folate-deficient
diet. Toxicol Appl Pharmacol. 262:52–59. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu G, Friggeri A, Yang Y, Milosevic J,
Ding Q, Thannickal VJ, Kaminski N and Abraham E: miR-21 mediates
fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J
Exp Med. 207:1589–1597. 2010. View Article : Google Scholar : PubMed/NCBI
|