Introduction
In the latest guidelines by the National
Comprehensive Cancer Network (NCCN), epidermal growth factor
receptor (EGFR)-tyrosine kinase inhibitor (TKI) is recommended as
first-line therapy for patients with advanced non-small cell lung
cancer (NSCLC) harboring activating EGFR mutations.
EGFR mutations occur more frequently in
patients of Asian ethnicity, females, individuals with no smoking
history, and in patients diagnosed with adenocarcinoma. Treatment
with EGFR-TKI manifests the greatest efficacy in these patients,
with no smoking history being the best predictor of good response
to TKI. Mutations are associated with an enhanced sensitivity to an
EGFR-TKI located in EGFR exons 18–21, which encode the
tyrosine kinase domain. In-frame deletions in exon 19 and a point
mutation in exon 21 (p.L858R) are the most prevalent EGFR
mutations. Mutations associated with resistance to TKI include a
point mutation (p.T790M) and insertions (e.g., p.D770_N771insNPG)
in exon 20, and a point mutation (p.D761Y) in exon 19 (1–3).
Although the clinical relevance of activating
EGFR mutations with TKI response in advanced NSCLC is
well-addressed, a standardized and commonly accepted approach in
terms of optimal sensitivity, specificity, reproducibility and
accuracy in detecting EGFR mutations has not been adopted to
date (4–6). A variety of techniques for mutation
analysis of the EGFR gene exist. These are classified into
screening methods that identify all mutations and targeted methods
that distinctively detect known and pre-determined mutations.
Among diverse screening methods, the direct
sequencing of polymerase chain reaction (PCR) products is still
widely used, despite its low sensitivity. Direct sequencing does
not require sample batching, while it provides better contamination
control since the exact, specific mutation is presented. However,
direct sequencing is time-consuming and successful only when viable
tumor cells constitute at least 25% of the tissues (7,8).
Alternative screening methods include high resolution melting
(HRM), pyrosequencing and denaturing high pressure liquid
chromatography (dHPLC) analysis. HRM is an in-tube, fast method
that detects sequence variation by monitoring the melting curve of
PCR amplicons. HRM is able to detect mutant genes at levels of
1–10% (9–11). Nevertheless, the requirement for
sequencing validation increases the turn-around time and reduces
the value of high sensitivity.
The limited sensitivity of conventional sequencing
necessitates the adoption of more sensitive approaches. Scorpion
amplification refractory mutation system (ARMS) falls into the
targeting method category and has been successfully used to analyze
the EGFR mutation status in the phase III Iressa Pan-Asia
Study (IPASS) clinical trial (12). ARMS detects mutations in samples
with a mutation frequency as low as 0.1 to 1%, but detects known
mutations only. Additionally, ARMS requires the batching of samples
and the reagents required are expensive.
Samples used for analyzing EGFR mutations
differ between laboratories. Samples can be obtained either at the
stage of diagnosis (biopsy) or at the stage of surgical
intervention (resections). Large samples from surgical intervention
are preferred, but small biopsy samples are also regularly used.
Unfortunately, tissue samples are not always available; therefore,
cytological materials, including pleural effusion, bronchial
scraping and bronchofiberscopic brushing are being increasingly
used. In fact, the mutation detection rate achieved with
cytologyical material is comparable with that achieved with tissue
samples obtained by biopsy or resection (5).
In the present study, we analyzed the EGFR
mutation status of 356 patients with advanced NSCLC and
systemically compared the mutation detection rate of direct
sequencing, the gold standard, with more sensitive methods, i.e.,
ARMS and HRM, in different tissue types. The overall mutation rate
of the EGFR gene was 44.10% and the rate of activating
EGFR mutations was 39.04%. The activating EGFR
mutations occurred more frequently in females and patients
diagnosed with adenocarcinoma. The EGFR mutation frequency
identified from the bronchoscopic biopsies was lower than that from
surgical resections, regardless of the method used. The mutation
rate detected from the cytological sample was similar to that
achieved with surgical resections and a sensitive method for
detecting mutations in the cytological samples was required. Each
method has advantages and disadvantages; it was thus suggested that
the choice of method in clinical practice should be made based on
the sample type. ARMS was recommended when mutations were detected
in bronchoscopic biopsies and cytological samples. Direct
sequencing was recommended when mutations were identified in
surgical resections. However, the lack of EGFR mutations
tested by direct sequencing is possibly due to the limited
sensitivity of the method. The absence of EGFR mutations,
determined by methods that detect known mutations, such as ARMS,
cannot be the exclusion criterion for TKI treatment. To reduce
false positives and false negatives caused by the limitations of
each method, the combination of direct sequencing and a more
sensitive technique, such as ARMS, is recommended for identifying
EGFR mutations in clinical practice.
Patients and methods
Subjects
This study was approved by the Institutional Review
Board, where samples were collected from and analyzed (the First
Affiliated Hospital of Soochow University, Suzhou, China). A total
of 356 patients diagnosed with NSCLC were included in the study.
The pleural effusion cytological samples were collected from
patients diagnosed with NSCLC and confirmed by a pathologist to
contain tumor cells.
Sensitivity determination
Two lung cancer cell lines, PC-9 and A549, were
used. The A549 cell line was purchased from the Shanghai Institute
for Biological Sciences (Shanghai, China). PC-9 cells harbor
in-frame deletions in exon 19 of the EGFR gene (heterozygous
for c.2235_2249del15). A549 cells are wild-type for the EGFR
gene. Serial dilutions of the EGFR mutant PC-9 cells with
A549 cells were used to determine the sensitivity of direct
sequencing, ARMS and HRM.
Genomic DNA (gDNA) extraction
Before the extraction of gDNA, representative
formalin-fixed, paraffin-embedded (FFPE) sections were stained with
hematoxylin and eosin (H&E) and diagnosed by pathologists. At
least 20% of tumor cells was observed in the FFPE sections. DNA
extraction was performed usng the QIAamp™DNA FFPE Tissue kit
(Qiagen, Shanghai, China) according to the manufacturer’s
instructions. To obtain DNA fromthe cell lines, the cells were
harvested by trypsinization when grown to confluence. To obtain DNA
from pleural effusion, the cells were collected by centrifugation.
DNA was extracted using gDNA isolation kits (Omega BioTek
Guangzhou, Ltd., Guangzhou, China) according to the manufacturer’s
instructions. DNA was quantified using a NanoDrop ND-1000
fluorospectrometer (ThermoFisher Scientific, Shanghai, China), and
the A260/280 value was ensured between 1.8–2.0.
Direct sequencing
Mutation screening of EGFR exons 18–21 was
carried out by PCR amplification as previously described (13). The primers for PCR amplification
were as follows: EGFR exon 18 forward,
GCATGGTGAGGGCTGAGGTGAC and reverse, TATACAGCTTGCAAGGACT CTG; exon
19 forward, GTGCATCGCTGGTAACATCCA and reverse, GGAGAT
GAGCAGGGTCTAGAGCA; exon 20 forward, GATCGC ATTCATGCGTCTTCACC and
reverse, TTGCTATCCCAGG AGCGCAGACC; exon 21 forward, TCAGAGCCTGGCAT
GAACATGACCCTG and reverse, GGTCCCTGGTGTCAGG AAAATGCTGG. PCR
reaction was amplified using Platinum Taq DNA polymerase
(Invitrogen, Beijing, China) and conducted under the following
conditions: 94°C 5 min, (94°C, 30 sec, 60°C, 30 sec, 72°C, 45 sec)
×40 cycles, 72°C 10 min. The PCR products were checked on 2%
agarose gels. PCR products were purified and followed by
bi-directional sequencing using an ABI 3730 DNA analyzer (Applied
Biosystems, Inc., Beijing, China). Sequencing chromatograms were
analyzed using DNA Baser 3.0. Nucleotide changes detected by
sequencing were all checked in Sanger’s COSMIC database (http://cancer.sanger.ac.uk/cosmic/gene/analysis?ln=EGFR#histo),
and diagnosed as mutations accordingly.
ARMS assay
The presence of EGFR mutations was determined
using the AmoyDx™ EGFR 29 Mutations Detection kit, (Amoy
Diagnostics Co., Ltd., Xiamen, China). The kit, which has been
approved for clinical use by the State Food and Drug Administration
(SFDA) in China, detects the most commonly reported 29 somatic
mutations (both activating and TKI resistance-related) in the
EGFR gene: 19 deletions in exon 19, three insertions in exon
20 and point mutations p.G719X (exon 18), p.S768I and p.T790 M
(exon 20), p.L858R and p.L861Q (exon 21). The test detects the
presence of these mutations, but does not distinguish between them.
The analysis was performed according to the manufacturer’s
instructions using a LightCycler 480 (Roche Diagnostic, Ltd.,
Shanghai, China).
HRM assay
HRM assay was performed using an EGFR gene
mutation detection kit, detecting mutations in EGFR exons
18–21 (Suzhou MicroDiag Biomedicine Co., Suzhou, China) on a
LightCycler 480. The melting profiles of the amplicons were
analyzed using gene scanning software to detect wild-type and
mutations. To affirm the gene scanning results, the amplicons were
sequenced after HRM assay.
Statistical analysis
The association of the mutation status of the
EGFR gene with any of the clinicopathological characeristics
was evaluated. Frequencies were compared using two-tailed Pearson’s
Chi-square or Fisher’s exact test. The difference was considered
significant when the P-value was P<0.05. Analyses were performed
using the GraphPad Prism 5 program.
Results
Patient characteristics
A total of 356 samples (from August 2010 to December
2012) were collected and successfully evaluated for EGFR
mutations. The clinical characteristics of the patients, such as
age, gender, smoking history and pathological evaluation, are
summarized in Table I. A total of
210 patients (58.99%) were male and 146 (41.01%) were female, with
a median age of 57.5 years (range, 27–88 years). Of these patients,
162 (45.51%) were current smokers, 173 (48.60%) were non-smokers,
and 21 (5.89%) had an unknown smoking history. All samples were
confirmed to contain malignant cells and the pathological and
cytological diagnosis revealed that 173 (48.60%) of the samples
contained adenocarcinoma cells, 56 (15.73%) contained squamous cell
carcinoma cells and 127 (35.67%) contained other types of carcinoma
cells.
 | Table IClinical characteristics of the lung
cancer patients. |
Table I
Clinical characteristics of the lung
cancer patients.
|
Characteristics | No. of patients
(total, 356) | Frequency (%) |
|---|
| Age (years) |
| Median
(range) | 57.5 (27–88) | |
| >60 | 195 | 54.77 |
| ≤60 | 161 | 45.23 |
| Gender |
| Male | 210 | 58.99 |
| Female | 146 | 41.01 |
| Smoking
history |
| Current
smoker | 162 | 45.51 |
| Non-smoker | 173 | 48.60 |
| Unknown | 21 | 5.89 |
| Histological
subtype |
|
Adenocarcinoma | 173 | 48.60 |
| Squamous | 56 | 15.73 |
| Other | 127 | 35.67 |
EGFR mutations status
The presence of mutations in EGFR exons 18–21
was analyzed, and the overall mutation rate was 44.10% (157/356).
The mutation frequencies in the males and females were 37.61
(79/210) and 53.42% (78/146), respectively. There was a significant
difference (P=0.0034) between males and females as regards the
EGFR mutation frequency (Table II).
 | Table IIMutation rates between male and
female NSCLC patients. |
Table II
Mutation rates between male and
female NSCLC patients.
| Gender | All
n (%) | Wild-type
n (%) | Mutation
n (%) | P-value |
|---|
| Male | 210 (58.99) | 131 (65.83) | 79 (50.32) | 0.0034a |
| Female | 146 (41.01) | 68 (34.17) | 78 (49.68) | |
The EGFR mutations detected were further
classified into three types, i.e., activating (TKI-sensitive)
mutations, TKI-resistant mutations and mutations that were not
associated with TKI response. The frequencies of the three types of
mutations were 88.54 (139/157), 3.18 (5/157) and 8.28% (13/157),
respectively.
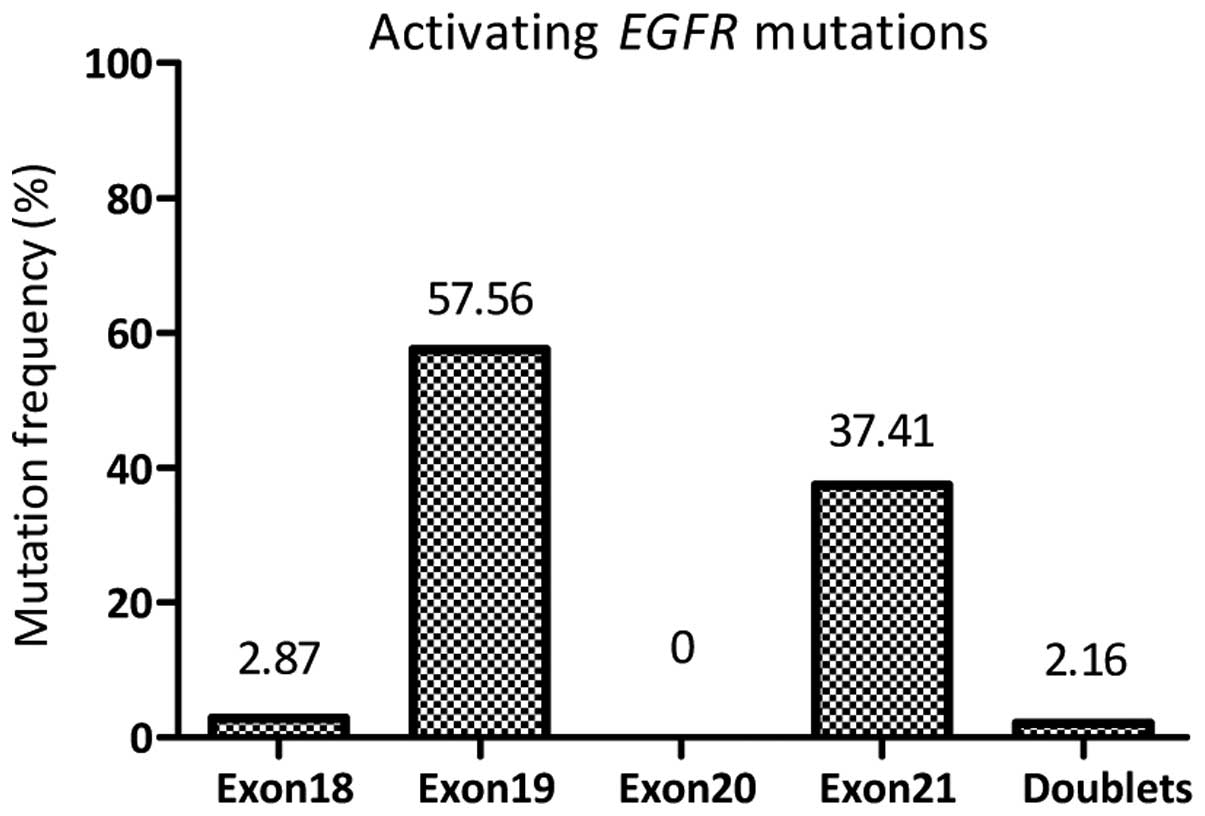
The activating EGFR mutations most frequently
occurred in exon 19, comprising 57.56% (80/139) of all the
activating mutations, followed by 35.25% (49/139) point mutations
in exon 21 (p.L858R and p.L861Q). Ten types of deletion/insertion
in exon 19 detected in the study are summarized in Table III. Of note, three patients
harboring specific mutations of p.H835L and p.H838V in EGFR
exon 21 were detected. Four patients (2.87%, 4/139) with a point
mutation (p.G719S or p.G719A) in exon 18 and three patients (2.16%,
3/139) with double mutations in exons 19 and 21 (p.E746_A750del
& p.L858R) were found as activating EGFR mutations
(Fig. 1 and Table III).
 | Table IIIMolecular characteristics of the 157
NSCLC patients with EGFR mutations. |
Table III
Molecular characteristics of the 157
NSCLC patients with EGFR mutations.
| Exon | Mutation type | Nucleotide
change | Amino acid
change | No. | TKI response |
|---|
| 18 | Missense |
c.2152_2153CT>TA |
L718* | 1 | U |
| | c.2155G>A | p.G719S | 2 | S |
| | c.2156G>C | p.G719A | 2 | S |
|
Deletion/insertion |
c.2127_2129delAAC | p.E709_T710del | 1 | U |
| 19 | Missense | c.2186G>A | p.G729E | 1 | U |
| | c.2227G>A | p.A743T | 1 | U |
| |
c.2239_2240TT>CC | p.L747P | 1 | U |
| | c.2255C>T | p.S752F | 1 | U |
|
Deletion/insertion |
c.2235_2249del15 | p.E746_A750del | 56 | S |
| |
c.2236_2250del15 | p.E746_A750del | 13 | S |
| |
c.2237_2253del17insTTGCT |
p.E746_T751delinsVA | 1 | S |
| |
c.2237_2255del19insT |
p.E746_S752delinsV | 1 | S |
| |
c.2238_2248del11insGC |
p.E746_A750delinsEP | 1 | S |
| |
c.2239_2250del12insCCG |
P.L747_A750delinsP | 1 | S |
| |
c.2239_2248del10insC |
p.L747_A750delinsP | 1 | S |
| |
c.2240_2248del9 | p.L747_A750del | 1 | S |
| |
c.2240_2254del15 | p.L747_T751del | 2 | S |
| |
c.2240_2257del18 | p.L747_T753del | 3 | S |
| 20 | Missense | c.2230T>A | p.L777Q | 1 | U |
| |
c.2345_2346CT>AA | p.L782N | 1 | U |
| | c.2369C>T | p.T790M | 1 | R |
|
Deletion/insertion |
c.2310_2311insGGT |
p.D770_N771insG | 1 | R |
| |
c.2319_2320insCCCCAC |
p.H773_V774insPH | 1 | R |
| |
c.2322_2323insCCACGT |
p.V774_C775insPR | 1 | R |
| 21 | Missense | c.2483T>A |
p.L828* | 1 | U |
| | c.2506C>A | p.R836S | 1 | U |
| | c.2573T>G | p.L858R | 46 | S |
| | c.2582T>A | p.L861Q | 3 | S |
| | c.2591C>A | p.A864E | 1 | U |
| | c.2597A>T | p.E866V | 1 | U |
| | c.2602G>A | p.E868K | 1 | U |
| | c.2497T>G &
c.2504 A>T | p.H838V and
p.H835L | 3 | S |
| Doublets | |
c.2235_2249del15 | p.E746_A750del | 3 | S |
| | c.2573T>G | p.L858R | | |
| |
c.2235_2249del15 | p.E746_A750del | 1 | R |
| | c.2615G>T | p.T790M | | |
Five TKI-resistant mutations, including p.T790M,
p.D770_N771insG, p.H773_V774insPH, p.V774_C775insPR, and double
mutations in exons 20 and 21 (p.T790M and p.L858R) were detected.
In addition, 13 EGFR mutations, which did not correlate with
TKI response, are listed in Table
III. These were L718* and p.E709_T710del in exon 18,
point mutations (p.G729E, p.A743T, p.L747P and p.S752F) in exon 19,
point mutations (p.L777Q and p.L782N) in exon 20, and point
mutations (p.L828*, p.R836S, p.A864E, p.E866V and
p.E868K) in exon 21.
Correlation between EGFR mutation status
and smoking history
Among the 210 male patients, 162 were current
smokers, 27 were non-smokers, and 21 had an uknown smoking history
(Tables I and IV). All 146 female patients were
non-smokers. Overall, the rate of EGFR mutations was
significantly decreased in the smokers (38.27%, 62/162) compared
with the non-smokers (50.87%, 88/173, P=0.0141). The mutation rate
was significantly higher in the female non-smokers than in the male
smokers (53.42%, 78/146 vs. 38.27%, 62/162, P=0.0085). Among the
non-smokers, the EGFR mutation rates were comparable in the
male and female patients (53.42%, 78/146 vs. 37.04%, 10/27,
P=0.1439). Among the male patients, no statistically significant
difference was observed in the non-smokers, smokers and patients
with an unknown smoking history (37.04%, 38.27% and 33.33%;
Table IV).
 | Table IVMutation rates between smokers and
non-smokers. |
Table IV
Mutation rates between smokers and
non-smokers.
| All
n (%) | Wild-type
n (%) | Mutation
n (%) | P-value |
|---|
| Non-smoker | 173 (48.60) | 88 (56.05) | 85 (42.71) | 0.0141a |
| Males | 27 (7.59) | 17 (8.54) | 10 (6.37) | |
| Females | 146 (41.01) | 68 (34.17) | 78 (49.68) | |
| Smoker | 162 (45.51) | 62 (39.49) | 100 (50.25) | |
| Males | 162 (45.51) | 100 (50.25) | 62 (39.49) | |
| Females | 0 | 0 | 0 | |
| Unknown | 21 (5.89) | 7 (4.46) | 14 (7.04) | |
| Males | 21 (5.89) | 14 (7.04) | 7 (4.46) | |
| Females | 0 | 0 | 0 | |
Correlation between EGFR mutations and
histological parameters
The overall rate of activating EGFR mutations
was 39.04%, with 50.87% (88/173) in the adenocarcinoma and 25.00%
(14/56) in the squamous cell carcinoma samples. There was a
significant difference in the EGFR mutation rate between
adenocarcinoma and squamous cell carcinoma (P=0.0004). The
prevalence of activating EGFR mutation rates in other
subtypes of NSCLC are summarized in Table V. It should be noted that the
activating EGFR mutations often occurred in NSCLC with the
adenosquamous carcinoma (50.00%, 4/8) and alveolar cell carcinoma
subtypes (83.33%, 5/6). In addition, 25.84% (23/89) of EGFR
mutations were detected in poorly differentiated NSCLC. Although
the difference was not statistically significant, the activating
EGFR mutations occurred more frequently in females compared
to males with the adenocarcinoma subtype (56.18% and 50/89 vs.
45.24% and 38/84, P=0.1722; Table
VI).
 | Table VAssociation of activating mutation
rates with histological subtypes of NSCLC. |
Table V
Association of activating mutation
rates with histological subtypes of NSCLC.
| Histological
subtype | Total | Frequency (%) | Mutation rate
(%) | Mutation |
|---|
| Adenocarcinoma | 173 | 48.60 | 88 | 50.87 |
| Squamous cell | 56 | 15.73 | 14 | 25.00 |
| Large cell | 11 | 3.09 | 3 | 27.27 |
| Adenosquamous | 8 | 2.25 | 4 | 50.00 |
| Alveolar cell | 6 | 1.69 | 5 | 83.33 |
| Adenocarcinoma and
alveolar cell | 5 | 1.40 | 1 | 20.00 |
| Squamous and large
cell | 2 | 0.56 | 1 | 50.00 |
| Neuroendocrine | 2 | 0.56 | 0 | 0 |
| Adenocarcinoma and
large cell | 2 | 0.56 | 0 | 0 |
| Sarcomatodes | 1 | 0.28 | 0 | 0 |
| Adenoid cystic | 1 | 0.28 | 0 | 0 |
| Poorly
differentiated | 89 | 25.00 | 23 | 25.84 |
| Total | 356 | 100.00 | 139 | 39.04 |
 | Table VIComparison of the activating mutation
rate between males and females with adenocarcinoma. |
Table VI
Comparison of the activating mutation
rate between males and females with adenocarcinoma.
| Adenocarcinoma | All
n (%) | Wild-type
n (%) | Mutation
n (%) | P-value |
|---|
| Males | 84 (58.99) | 44 (55.00) | 38 (50.32) | 0.1722 |
| Females | 89 (41.01) | 36 (45.00) | 50 (49.68) | |
Between the smoking and non-smoking male patients
with the adenocarcinoma subtype, the rate of activating EGFR
mutations was similar (48.44 vs. 50.00%, P=0.5814). Of note, the
activating EGFR mutations occurred more frequently in
females compared to males with the squamous cell carcinoma subtype
(40.00 vs. 23.68%, P=0.4253; Table
VII).
 | Table VIIComparison of the activating mutation
rate between males and females, and smokers and non-smokers. |
Table VII
Comparison of the activating mutation
rate between males and females, and smokers and non-smokers.
| Males | |
|---|
|
| |
|---|
| Smokers | Non-smokers | Unknown | Females |
|---|
| Adenocarcinoma |
| Total | 64 | 15 | 5 | 89 |
| Mutation | 31 | 6 | 1 | 50 |
| Mutation rate
(%) | 48.44 | 50.00 | 20.00 | 56.18 |
| Squamous cell
carcinoma |
| Total | 38 | 3 | 5 | 10 |
| Mutation | 9 | 0 | 1 | 4 |
| Mutation rate
(%) | 23.68 | 0 | 20.00 | 40.00 |
Correlation between EGFR mutation status
and sample type
Among all the samples analyzed, 112 (31.46%) were
bronchoscopic biopsies, 224 (62.93%) were surgical resections and
20 (5.61%) were pleural effusion cytological samples. The pleural
effusion cytological samples were collected from patients diagnosed
with NSCLC and confirmed by a pathologist to contain tumor cells.
The corresponding activating EGFR mutation rates were 45.00,
30.36 and 42.86%, respectively (Table VIII). It is important to note
that the rate of EGFR mutations identified from the
bronchoscopic biopsies was lower than that from the surgical
resections or pleural effusion, and was also lower than the overall
rate. A statistically significant difference in the EGFR
mutation rate in the bronchoscopic biopsies and surgical resections
was observed (P=0.0425).
 | Table VIIIComparison of the activating mutation
rate among different sample types. |
Table VIII
Comparison of the activating mutation
rate among different sample types.
| All
n (%) | Wild-type
n (%) | Mutation
n (%) | Mutation rate
(%) | P-value |
|---|
| Pleural
effusion | 20 (5.61) | 11 (5.53) | 9 (6.47) | 45.00 | 0.8158 |
| Bronchoscopic
biopsies | 112 (31.46) | 71 (35.17) | 34 (24.46) | 30.36 | 0.0425a |
| Surgical
resections | 224 (62.93) | 117 (59.30) | 96 (69.07) | 42.86 | 0.0618 |
Detection of EGFR mutations by different
methods
A total of 86 samples was successfully analyzed by
PCR amplification followed by direct sequencing. Direct sequencing
identified EGFR mutations in 34 (39.53%) samples. The
EGFR mutation rates in pleural effusion, bronchoscopic
biopsies and surgical resections were 14.29 (1/7), 30.00 (3/10) and
43.47% (30/69), respectively. Although there was no statistically
significant difference, EGFR mutations were more frequently
identified in the surgical resections than the other two types of
samples by direct sequencing.
A total of 120 samples was analyzed by ARMS assay
and 43 (35.83%) were identified as EGFR mutation-positive.
The EGFR mutation rates detected by ARMS were 55.56 (5/9),
30.30 (20/66) and 40.00% (18/45) for the individual sample types. A
total of 150 samples was analyzed by HRM assay followed by
sequencing verification. HRM combined with sequencing verification
detected EGFR mutations in 62 (41.33%) of the analyzed
samples. The EGFR mutation rates were 75.00 (3/4), 30.56
(11/36) and 43.64% (48/110) for the three sample types (Table IX). Taken together, for the
tissue samples, such as bronchoscopic biopsy and surgical
resection, the rate of EGFR mutations detected by any of the
three methods was comparable. For detecting mutations in pleural
effusion, ARMS and HRM assays were possibly superior to direct
sequencing. Nevertheless, it should be noted that no statistically
significant difference between direct sequencing and ARMS assay
(P=0.1451) or direct sequencing and HRM assay (P=0.0879) with
respect to the mutation frequency was obtained due to the limited
sample numbers. Regardless of the method used, the EGFR
mutation rate detected in the bronchoscopic biopsies was the
lowest.
 | Table IXComparison of the activating mutation
rate detected by different methods in different sample types. |
Table IX
Comparison of the activating mutation
rate detected by different methods in different sample types.
| All | Pleural fluid | Bronchoscopic
biopsies | Surgical
resections |
|---|
|
|
|
|
|---|
| Method | Total | Rate (%) | Total | Mutation | Rate (%) | Total | Mutation | Rate (%) | Total | Mutation | Rate (%) |
|---|
| Sequencing | 86 | 39.53 | 7 | 1 | 14.29 | 10 | 3 | 30.00 | 69 | 30 | 43.47 |
| ARMS | 120 | 35.83 | 9 | 5 | 55.56 | 66 | 20 | 30.30 | 45 | 18 | 40.00 |
| HRM and Se | 150 | 41.33 | 4 | 3 | 75.00 | 36 | 11 | 30.56 | 110 | 48 | 43.64 |
Method correlation with sequencing
The EGFR mutation status of 114 samples
detected by ARMS assay using the AmoyDx™EGFR 29 Mutations Detection
kit (Amoy Diagnostics Co.) was tested again by direct sequencing.
We were unable to perform additional sequencing for six samples due
to insufficient gDNA. Forty EGFR mutation-positive and 44
EGFR mutation-negative samples reached a consensus in the
two methods (Table X). The
concordance rate between the two methods was 73.68%.
 | Table XComparison of the results of the
EGFR mutation analysis between ARMS and direct
sequencing. |
Table X
Comparison of the results of the
EGFR mutation analysis between ARMS and direct
sequencing.
| Mutation
status | Sequencing | Total |
|---|
|
|---|
| + | − |
|---|
| ARMS |
| + | 40 | 15 | 55 |
| − | 15 | 44 | 59 |
| Total | 55 | 59 | 114 |
Fifteen EGFR mutation-positive samples
detected by ARMS assay were found to be EGFR
mutation-negative by sequencing. It is important to note that
another 15 EGFR mutation-negative samples detected by ARMS
assay were found to be EGFR mutation-positive by sequencing.
Direct sequencing identified 11 rare mutations that were not
designed to be detected by ARMS assay, including p.E709_T710del,
p.G729E, p.G729V, p.L747P, p.A864E and p.E866V. A discrepancy was
observed in another four samples between the two methods. The
sensitivity of ARMS assay was 72.73% and the specificity
74.58%.
A total of 150 samples was tested by HRM assay for
the detection of mutations in EGFR exons 18–21. HRM assay
detected more positive samples than sequencing and detected 97
samples as positive for mutations. Among these, 68 samples were
confirmed as positive by sequencing and 29 samples were not
confirmed, which were possibly false positives. Most of these are
likely to be true false positives due to degraded DNA extracted
from FFPE specimens. Fifty samples were detected as negatives by
both methods used. Three samples that were detected as EGFR
mutation-negative by HRM were detected as EGFR
mutation-positive by sequencing (Table XI). The sensitivity and
specificity for the samples that were suspected of having mutations
by HRM assay was 95.77 and 63.29% when compared to sequencing, with
an accuracy rate of 78.67%. For each patient, EGFR mutation
testing was carried out on the same gDNA, avoiding the
inconsistency potentially resulting from intra-tumor heterogeneity.
Collectively, the discordance was 26.32% between direct sequencing
and ARMS and was 21.33% between direct sequencing and HRM.
 | Table XIComparison of the results of the
EGFR mutation analysis between HRM and sequencing. |
Table XI
Comparison of the results of the
EGFR mutation analysis between HRM and sequencing.
| Mutation
status | Sequencing | Total |
|---|
|
|---|
| + | − |
|---|
| HRM |
| + | 68 | 29 | 97 |
| − | 3 | 50 | 53 |
| Total | 71 | 79 | 150 |
Sensitivity testing by direct sequencing,
ARMS and HRM
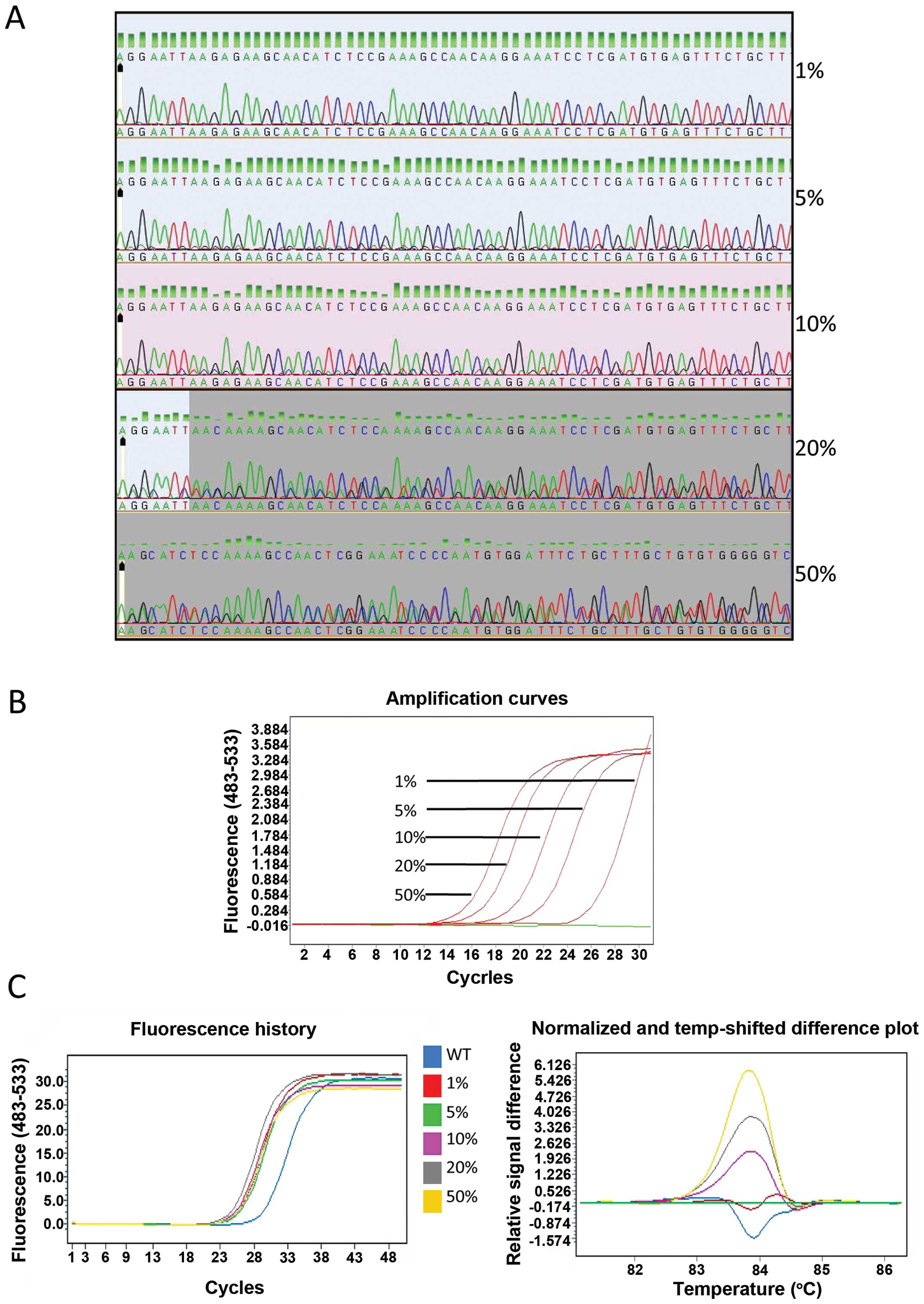
The gDNA of the PC-9 cells was serially diluted into
A549 gDNA at ratios of 100, 40, 20, 10 and 2% to yield mutant
allele frequencies of 50, 20, 10, 5 and 1%. The relative
sensitivity of direct sequencing, ARMS and HRM was evaluated using
the diluted DNA. The mutation was detectable (at a low peak) by
direct sequencing when the mutant frequency was higher than 10%.
However, when the mutation frequency was at 5%, it was only
distinguishable from the background. When the mutant frequency was
below 5%, the mutation was not detectable (Fig. 2A). ARMS assay positively detected
the deletions in EGFR exon 19 in the sample containing down
to 1% mutant allele frequency (Fig.
2B). Using HRM, the melting curve from 1% mutant template
sufficiently differed from wild-type template (Fig. 2C), and this distinct melting
profile was consistently observed across all other templates
measured (5, 10, 20 and 50%). Thus, the sensitivity of direct
sequencing, ARMS and HRM was found to be 10, 1 and 1%,
respectively.
Discussion
Based on the observations from IPASS and other
studies (14,15), the ASCO provisional clinical
opinion (PCO) states that ‘patients with advanced NSCLC who are
being considered for first-line therapy with an EGFR-TKI should
have their tumor tested for EGFR mutations to determine
which is an appropriate therapy: an EGFR-TKI or chemotherapy’
(7,16). Therefore, evaluating the
EGFR mutation status is a matter of urgency in clinical
practice, particularly in patients with adenocarcinoma.
As a matter of fact, the selection of patients for
EGFR-TKI therapy based on mutation analysis is not an absolute
warranty for good response and approximately 20–30% of patients
harboring activating EGFR mutations do not benefit from TKI
treatment (17). The presence of
TKI-resistant or increased copy number (amplification) of the
MET oncogene contributes to resistance to TKI. Furthermore,
the low abundance of EGFR mutations affects the response to
TKI (18). False positives due to
the methodology used for mutation detection should not be
neglected, particularly when an extremely sensitive test is
performed. On the other hand, a proper interpretation of negative
results requires a thorough understanding of the technical
limitations of the assay and the type of specimen used for mutation
detection. One of the possible reasons that patients without
activating EGFR mutations respond to an EGFR-TKI is the
false negatives (13). For
example, as previously demonstrated, five out of 50 patients with
advanced NSCLC had discrepancies in the results of mutant-enriched
PCR, peptide nucleic acid-locked nucleic acid (PNA-LNA) PCR and PCR
clamp. All five patients were false-negative as they responded to
gefitinib (19).
In the present study, the prevalence of EGFR
mutations was higher in females than in males and the frequency of
activating EGFR mutations (39.04%) was similar to that
described in earlier studies conducted on patients with advanced
NSCLC in an East Asian group (20). Two types of mutations, the short
in-frame deletions in exon 19 (particularly p.E746_A750del) and the
point mutation in exon 21 (c.2573T>G, p.L858R) comprised up to
90% of mutations. This is also in line with what has been
previously described (3,4,21,22). Low-frequency mutations in exon 18
and exon 21, such as p.G719X and p.L861Q, were also found. Of note,
three cases carrying the complex mutations of p.L833V and p.H835L
in exon 21 were detected in this study. The occurrence frequency of
these types of mutation (2.16%, 3/139) was equivalent to that of
p.L861Q. A good response to EGFR-TKI therapy has been reported in
one patient harboring the p.L833V and p.H835L mutations (23); therefore, this specific type of
mutation was considered one of the activating EGFR
mutations. The correlation of p.L833V and p.H835L mutations with
TKI response requires further clinical investigation. In addition,
13 mutations that were not associated with TKI response were
detected in treatment-free samples.
All the females with advanced NSCLC in this study
were non-smokers and the EGFR mutation rate was
significantly higher in the non-smokers than in the smokers, which
is in line with previous studies reporting that EGFR
mutations were more frequently detected in patients without a
smoking history (5,15). A recent meta-analysis demonstrated
that non-smokers with NSCLC show a statistically significant
increase in the prevalence of somatic EGFR mutations
(20). The activating EGFR
mutations were more often detected in patients with adenocarcinoma
than in patients with squamous cell carcinoma. Among the
adenocarcinoma patients, the prevalence of activating EGFR
mutations was modestly higher in females than in males; however,
this difference was not statistically significant. Importantly,
among the male patients with adenocarcinoma, no difference was
observed in the mutation rate between smokers and non-smokers.
Almost all the male patients diagnosed with squamous cell carcinoma
were smokers, and the frequency of activating EGFR mutations
was much higher in the females (non-smokers) than in the male
smokers. Only three males with squamous cell carcinoma were
non-smokers and were unable to be analyzed statistically. Thus, we
suggest that adenocarcinoma, particularly in females, is a valuable
predictive factor for the occurrence of EGFR mutations. A
smoking history largely affected the EGFR mutation
occurrence in patients with squamous cell carcinoma rather than
adenocarcinoma.
In order to carry out a routine EGFR mutation
screening in clinical practice, good quality DNA in sufficient
quantity, including tumor content (particularly for cytological
material), and the most reliable method in terms of sensitivity and
specificity are an absolute requirement. Resected tumor tissues are
preferred, but they are not always available. Small biopsy samples
and cytological material, including that obtained from pleural
effusion, are increasingly used in clinical practice. In the
present study, three types of samples, i.e., surgical resections,
bronchoscopic biopsies and pleural effusion, were tested for
EGFR mutations. We found that the lowest frequency of
activating EGFR mutations was observed in the bronchoscopic
biopsies (30.36%). In clinical practice, it is not possible to
obtain both bronchoscopic biopsies and surgical sections from the
same patient. In the present study, the bronchoscopic biopsies are
obtained from advanced NSCLC patients with unresectable tumors,
while the surgical sections were obtained from NSCLC patients, at
early clinical stage, who had received surgical therapy. However,
the clinical features, including pathological type and gender (data
not shown), that significantly affected the EGFR mutation
frequency in the sample sets from the bronchoscopic biopsies and
surgical sections were comparable. The mutation rate in pleural
effusion (45.00%) was similar to that in surgical resections
(42.86%; Table VIII). Our
observations are in accordance with those from previous studies on
the detection of EGFR mutations in cytological samples
(5,24). Five methods, including
PCR-Invader, PNA-LNA PCR clamp, direct sequencing, cycleave PCR and
ARMS, show a comparable performance in the assessment of tissue and
cytology samples. Cytology-derived DNA is a suitable alternative to
FFPE samples and very useful when FFPE samples are unavailable for
molecular analysis (5,24).
Multiple sensitive techniques are employed as
alternatives to direct sequencing. Using cell lines with
heterozygous EGFR mutations, we found that the sensitivities
of direct sequencing, ARMS and HRM in our experimental setting were
10, 1 and 1%, respectively. The overall mutation rate detected by
ARMS assay was the lowest (35.83%), and the mutation rate detected
by direct sequencing (39.53%) was similar to that detected by HRM
assay (41.33%; Table IX). The
clinical characteristics of the samples tested by the three
methods, which significantly affected the EGFR mutation
frequency, including pathological type and gender (data not shown),
were comparable. In the 224 surgical resections, the difference
observed in the mutation rates (43.47, 40.00 and 43.64%) detected
by the three methods (sequencing, ARMS, and HRM and sequencing
together, respectively) was not considerable. In the 112
bronchoscopic biopsies, the mutation rates detected by the three
methods (30.00, 30.30 and 30.56%) were almost the same and clearly
lower than those detected in surgical resections. In the 20 pleural
effusion samples, the mutation rate detected by direct sequencing
was the lowest (14.29%), and the mutation rates detected by ARMS
(55.56%) and HRM assay (75.00%) were even higher than the average
rate of all samples (39.04%) (Table
IX). Collectively, sensitive methods, i.e., ARMS and HRM, were
not superior to direct sequencing in surgical resections and
bronchoscopic biopsies in terms of mutation detection frequency in
this study. The possible reasons for this were the quantity control
of tumor content (>20%) in FFPE sections by H&E staining and
improved sensitivity (10%) of direct sequencing by optimizing all
reaction conditions. It is also important to note that the
prevalence of EGFR mutations detected in bronchoscopic
biopsies using any of the three methods was the lowest.
Bronchoscopic biopsies usually contain smaller amounts of tissue
than surgical resections due to the limited tissue size; they also
provide relatively inadequate information a molecular evaluation
due to tumor heterogeneity. The amount of DNA extracted from small
biopsy specimens varies significantly, depending on the size of the
material, the tumor viability, etc. The minimum amount of DNA
extracted from FFPE samples for direct sequencing, ARMS and HRM
assay are 300, 100 and 150 ng, respectively. Therefore, ARMS assay
is preferred when the sample DNA is extremely low (4,13).
In addition, it was necessary to detect EGFR
mutations in pleural effusion, using a sensitive technique, such as
ARMS or HRM. The findings of the present study were consistent with
those of several other studies. ARMS assay is more sensitive in
detecting EGFR mutations than direct sequencing in
cytological samples from transbronchial needle aspirates or pleural
effusion (25,26). Other methods, including
pyrosequencing and HRM, have been reported (27,28). A recent review evaluating 33
studies using cytological samples for EGFR mutation testing
suggested that the use of sensitive methods is warranted when
cytological samples with low-tumor content are used (5).
In the present study, EGFR mutations were
identified in the same gDNA using two methods with low and high
sensitivity concomitantly. The concordance rate between direct
sequencing and ARMS assay was 73.68%. The discordance found in the
mutation status in direct sequencing and ARMS may be explained by
the different degree of of sensitivity, particularly for
identifying a low abundance of mutations. A total of 11 (out of 15)
mutations detected by direct sequencing were not detected by ARMS,
as the ARMS assay used was not designed to detect these rare
EGFR mutations. Clinical data on less common mutations are
being increasingly gathered. However, further research on the
analysis of predictable outcomes on TKI response is required
(23,29). An analysis of another four
mutations, which were detected only via direct sequencing, was not
carried out using ARMS due to insufficient materials. The
observations of the present study were in accordance with those of
other studies that compare direct sequencing with ARMS. Compared
with direct sequencing, 10–20% of mutations are missed by ARMS, but
20% of mutations detected by ARMS at low levels are missed by
direct sequencing (8,30,31). In another study, 32% of tumors
carrying activating EGFR mutations detected by direct
sequencing are missed by the commercially available ARMS kit. The
percentage of missed mutations is too high to recommend the use of
ARMS for diagnostic application (32).
The sensitivity of HRM in the present study was
95.77%, which is similar to that found in other studies (10,33,34). The concordance rate between HRM
and sequencing was 78.67%. The difference in sensitivity was one of
the reasons for the discrepancy. In addition, any DNA alteration
due to the interference of single nucleotide polymorphism (SNP) or
formalin fixation may produce an abnormal melting curve (35). The high rate of false positives in
FFPE samples indicates that an additional sequencing should be
performed.
Given the respective limitations of the currently
available testing methodologies, several laboratories tend to use a
combination of methodologies (5,30).
In a previous study, we also proposed a sequential detection
workflow using ARMS assay and/or direct sequencing (13). Thus, each method compensates for
the disadvantages of the other and reduces the frequency of false
negatives.
In conclusion, we recommend that the choice of
method should be made based on the sample type. An analysis of
samples obtained at the diagnostic stage, e.g., bronchoscopic
biopsies, should be performed using the ARMS assay for the
detection of mutations due to the limited amount of DNA extracted
from small biopsy specimens. A sensitive method, such as ARMS, is
necessary when mutations in cytological samples, such as those
obtained from pleural effusion, need to be detected. The choice of
method used for mutation detection in samples from surgical
resections is largely based on the expertise of the laboratory, but
direct sequencing is highly recommended. However, the low detection
rate of EGFR mutations by direct sequencing is possibly due
to limited sensitivity. The absence of EGFR mutations,
determined by methods that detect known mutations, such as ARMS,
cannot be the exclusion criterion for EGFR-TKI usage. Therefore, we
suggest performing two methods (direct sequencing and a sensitive
method) sequentially in clinical practice, due to the presence of
non-neglected discordance between any method from its own benefits
and drawbacks. In the future, we may be able to benefit from the
incorporation of next-generation sequencing into daily clinical
practice.
Acknowledgements
The present study was supported by the Jiangsu
Provincial Natural Science Foundation of China (grant no.
BK2011306) and the National Natural Science Foundation of China
(grant no. 81172433).
References
|
1
|
Lynch TJ, Bell DW, Sordella R, et al:
Activating mutations in the epidermal growth factor receptor
underlying responsiveness of non-small-cell lung cancer to
gefitinib. N Engl J Med. 350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Paez JG, Jänne PA, Lee JC, et al: EGFR
mutations in lung cancer: correlation with clinical response to
gefitinib therapy. Science. 304:1497–1500. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sharma SV, Bell DW, Settleman J and Haber
DA: Epidermal growth factor receptor mutations in lung cancer. Nat
Rev Cancer. 7:169–181. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lopez-Rios F, Angulo B, Gomez B, et al:
Comparison of molecular testing methods for the detection of EGFR
mutations in formalin-fixed paraffin-embedded tissue specimens of
non-small cell lung cancer. J Clin Pathol. 66:381–385. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ellison G, Zhu G, Moulis A, Dearden S,
Speake G and McCormack R: EGFR mutation testing in lung cancer: a
review of available methods and their use for analysis of tumour
tissue and cytology samples. J Clin Pathol. 66:79–89. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pirker R, Herth FJ, Kerr KM, et al:
Consensus for EGFR mutation testing in non-small cell lung cancer:
results from a European workshop. J Thorac Oncol. 5:1706–1713.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Beasley MB and Milton DT: ASCO provisional
clinical opinion: epidermal growth factor receptor mutation testing
in practice. J Oncol Pract. 7:202–204. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ellison G, Donald E, McWalter G, et al: A
comparison of ARMS and DNA sequencing for mutation analysis in
clinical biopsy samples. J Exp Clin Cancer Res. 29:1322010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wittwer CT: High-resolution DNA melting
analysis: advancements and limitations. Hum Mutat. 30:857–859.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Do H, Krypuy M, Mitchell PL, Fox SB and
Dobrovic A: High resolution melting analysis for rapid and
sensitive EGFR and KRAS mutation detection in formalin fixed
paraffin embedded biopsies. BMC Cancer. 8:1422008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Krypuy M, Newnham GM, Thomas DM, Conron M
and Dobrovic A: High resolution melting analysis for the rapid and
sensitive detection of mutations in clinical samples: KRAS codon 12
and 13 mutations in non-small cell lung cancer. BMC Cancer.
6:2952006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fukuoka M, Wu YL, Thongprasert S, et al:
Biomarker analyses and final overall survival results from a phase
III, randomized, open-label, first-line study of gefitinib versus
carboplatin/paclitaxel in clinically selected patients with
advanced non-small-cell lung cancer in Asia (IPASS). J Clin Oncol.
29:2866–2874. 2011. View Article : Google Scholar
|
|
13
|
Zhuang Y, Xu J, Ma H, et al: A sequential
method of epidermal growth factor receptor mutation detection
reduces false negatives: a new case with doublet mutations of L833V
and H835L in China. Clin Lung Cancer. 14:295–300. 2013. View Article : Google Scholar
|
|
14
|
Mok TS, Wu YL, Thongprasert S, et al:
Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N
Engl J Med. 361:947–957. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Douillard JY, Shepherd FA, Hirsh V, et al:
Molecular predictors of outcome with gefitinib and docetaxel in
previously treated non-small-cell lung cancer: data from the
randomized phase III INTEREST trial. J Clin Oncol. 28:744–752.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Keedy VL, Temin S, Somerfield MR, et al:
American society of clinical oncology provisional clinical opinion:
epidermal growth factor receptor (EGFR) mutation testing for
patients with advanced non-small-cell lung cancer considering
first-line EGFR tyrosine kinase inhibitor therapy. J Clin Oncol.
29:2121–2127. 2011. View Article : Google Scholar
|
|
17
|
Gazdar AF: Tyrosine kinase inhibitors and
epidermal growth factor receptor (EGFR) mutations in non-small cell
lung cancer: to test or not to test? Medicine (Baltimore).
90:168–170. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou Q, Zhang XC, Chen ZH, et al: Relative
abundance of EGFR mutations predicts benefit from gefitinib
treatment for advanced non-small-cell lung cancer. J Clin Oncol.
29:3316–3321. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ikeda T, Nakamura Y, Yamaguchi H, et al:
Direct comparison of 3 PCR methods in detecting EGFR mutations in
patients with advanced non-small-cell lung cancer. Clin Lung
Cancer. 13:369–74. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ren JH, He WS, Yan GL, Jin M, Yang KY and
Wu G: EGFR mutations in non-small-cell lung cancer among smokers
and non-smokers: a meta-analysis. Environ Mol Mutagen. 53:78–82.
2012. View Article : Google Scholar
|
|
21
|
Angulo B, Conde E, Suarez-Gauthier A, et
al: A comparison of EGFR mutation testing methods in lung
carcinoma: direct sequencing, real-time PCR and
immunohistochemistry. PLoS One. 7:e438422012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liang Z, Zhang J, Zeng X, Gao J, Wu S and
Liu T: Relationship between EGFR expression, copy number and
mutation in lung adenocarcinomas. BMC Cancer. 10:3762010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang TY, Tsai CR, Chen KC, Hsu KH, Lee HM
and Chang GC: Good response to gefitinib in a lung adenocarcinoma
harboring a heterozygous complex mutation of L833V and H835L in
epidermal growth factor receptor gene. J Clin Oncol. 29:e468–e469.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Goto K, Satouchi M, Ishii G, et al: An
evaluation study of EGFR mutation tests utilized for non-small-cell
lung cancer in the diagnostic setting. Ann Oncol. 23:2914–2919.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Horiike A, Kimura H, Nishio K, et al:
Detection of epidermal growth factor receptor mutation in
transbronchial needle aspirates of non-small cell lung cancer.
Chest. 131:1628–1634. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kimura H, Fujiwara Y, Sone T, et al: High
sensitivity detection of epidermal growth factor receptor mutations
in the pleural effusion of non-small cell lung cancer patients.
Cancer Sci. 97:642–648. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim HJ, Oh SY, Kim WS, et al: Clinical
investigation of EGFR mutation detection by pyrosequencing in lung
cancer patients. Oncol Lett. 5:271–276. 2013.PubMed/NCBI
|
|
28
|
Fassina A, Gazziero A, Zardo D, Corradin
M, Aldighieri E and Rossi GP: Detection of EGFR and KRAS mutations
on trans-thoracic needle aspiration of lung nodules by high
resolution melting analysis. J Clin Pathol. 62:1096–1102. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
He M, Capelletti M, Nafa K, et al: EGFR
exon 19 insertions: a new family of sensitizing EGFR mutations in
lung adenocarcinoma. Clin Cancer Res. 18:1790–1797. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Leary AF, Castro DG, Nicholson AG, et al:
Establishing an EGFR mutation screening service for non-small cell
lung cancer-sample quality criteria and candidate histological
predictors. Eur J Cancer. 48:61–67. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu Y, Liu B, Li XY, et al: A comparison
of ARMS and direct sequencing for EGFR mutation analysis and
tyrosine kinase inhibitors treatment prediction in body fluid
samples of non-smal-cell lung cancer patients. J Exp Clin Cancer
Res. 30:1112011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Penzel R, Sers C, Chen Y, et al: EGFR
mutation detection in NSCLC-assessment of diagnostic application
and recommendations of the German panel for mutation testing in
NSCLC. Virchows Arch. 458:95–98. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Takano T, Ohe Y, Tsuta K, et al: Epidermal
growth factor receptor mutation detection using high-resolution
melting analysis predicts outcomes in patients with advanced
non-small cell lung cancer treated with gefitinib. Clin Cancer Res.
13:5385–5390. 2007. View Article : Google Scholar
|
|
34
|
Gonzalez-Bosquet J, Calcei J, Wei JS, et
al: Detection of somatic mutations by high-resolution DNA melting
(HRM) analysis in multiple cancers. PLoS One. 6:e145222011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Franklin WA, Haney J, Sugita M, Bemis L,
Jimeno A and Messersmith WA: KRAS mutation: comparison of testing
methods and tissue sampling techniques in colon cancer. J Mol
Diagn. 12:43–50. 2010. View Article : Google Scholar : PubMed/NCBI
|