Introduction
Regucalcin was first identifed in 1978 as a novel
calcium-binding protein that differs from calmodulin and other
calcium-related proteins (1–5).
The regucalcin gene (rgn) is localized on the X chromosome
(6,7), and it is identified in over 15
species consisting of the regucalcin family in vertebrate and
invertebrate species and is highly conserved in vertebrate species
(4,5). The organization of the rat
regucalcin gene consists of 7 exons and 6 introns and several
consensus regulatory elements exist upstream of the 5′-flanking
region (8). Activator protein 1
(AP-1), nuclear factor 1-A1 (NF1-A1), regucalcin gene promoter
region-related protein p117 (RGPR-p117), β-catenin and other
factors have been found to be transcription factors in the
enhancement of regucalcin gene promoter activity (5). The transcriptional activity of the
regucalcin gene is enhanced through intracellular signaling factors
that are mediated through the phosphorylation and dephosphorylation
of nuclear protein in vitro (5). As previously demonstrated,
regucalcin mRNA and protein are pronouncedly expressed in the liver
and kidney cortex of rats (9,10).
The mRNA expression of regucalcin in the liver and kidney cortex
has been shown to be stimulated by hormonal factors (including
calcium, calcitonin, parathyroid hormone, insulin, estrogen and
dexamethasone) (11).
Regucalcin, which plays a pivotal role as a
suppressor protein in cell signal transduction, plays a
multifunctional role in the regulation of various types of cells
and tissues (reviewed in refs. 12–14). Regucalcin translocates from the
cytoplasm to the nucleus in various cell types (15). Regucalcin has been shown to play a
role in maintaining intracellular calcium homeostasis, the
inhibition of various protein kinases and protein phosphatases in
the cytoplasm and nucleus, and nuclear DNA and RNA synthesis
(14,15). Nuclear regucalcin has also been
shown to regulate the gene expression of various proteins (15). Moreover, regucalcin suppresses
cell proliferation and apoptotic cell death that is mediated
through various signaling factors (16,17). Regucalcin has been suggested to
play a physiological role in maintaining cell homeostasis and
function as the regulatory protein of intracellular signaling
systems (13,14).
There is growing evidence that regucalcin plays a
pathophysiological role in metabolic disorder (16,18,19). In recent years, regucalcin has
been shown to be involved in carcinogenesis (reviewed in ref.
16). Regucalcin gene and protein
expression has been shown to be decreased in the liver tumor tissue
of rats in vivo (20). Of
note, regucalcin gene expression has been demonstrated to be
downregulated during the development of carcinogenesis in the
chemical-treated regenerating rat liver in vivo, suggesting
that regucalcin plays a suppressive role in carcinogenesis
(21). Moreover, regucalcin gene
and protein expression has been shown to be suppressed in cloned
rat hepatoma H4-II-E cells in vitro (22–25). The overexpression of endogenous
regucalcin has been found to suppress the enhancement of cell
proliferation with fetal bovine serum in H4-II-E cells in
vitro due to the inhibition of various protein kinases and
phosphatases, nuclear DNA synthesis and the mRNA expression of
oncogenes (22–25). Thus, regucalcin may play a
suppressive role in the development of carcinogenesis. However, the
findings of regucalcin gene expression in human normal and tumor
tissues require further confirmation.
The present study was undertaken in order to
determine whether the regucalcin gene is expressed in various human
normal and tumor tissues including liver, kidney, brain and lung
tissues. We found that the full-length regucalcin mRNA and its
alternatively spliced variants were expressed in various human
tissues.
Materials and methods
Materials
Regucalcin was isolated from rat liver as previously
described (1). A polyclonal
rabbit anti-regucalcin antibody was prepared as previously
described (10). Other antibodies
for western blot analysis were purchased from Santa Cruz
Biotechnology (Santa Cruz, CA, USA). Reagents for polymerase chain
reaction (PCR) were purchased from Invitrogen (Carlsbad, CA). All
other reagents were purchased from Sigma-Aldrich Chemical Corp.,
(St. Louis, MO, USA) unless otherwise indicated.
Human cDNA and protein samples
This study was approved by the Ethics Committee of
Meijo University (Nagoya, Japan). The human cDNA and protein
samples, which were prepared from normal (non-tumor) and tumor
tissues, were obtained from BioChain Institute, Inc. (Newark, CA,
USA). In the expression analysis of regucalcin mRNA in the human
normal and tumor tissues, cDNA prepared from the following tissues
was used: normal liver from a 64-year-old male; normal kidney from
a 46-year-old male; normal brain from a 26-year-old male; normal
lung from a 41-year-old female; liver hepatocellular carcinoma from
a 51-year-old male; kidney transitional cell carcinoma froma a
68-year-old male; brain malignant meningioma from a 7-year-old
male; and lung non-small cell carcinoma from a 46-year-old male. In
the expression analysis of regucalcin mRNA in the human brain, cDNA
prepared from the following tissues was used: amygdale from a
60-year-old female, cerebral cortex from a 28-year-old male,
cerebellum from a 27-year-old male, diencephalon from a 26-year-old
male, hippocampus from a 28-year-old male, medulla oblongata from a
26-year-old male, optic nerve from a 22-year-old male, pituitary
gland from a 68-year-old male and thalamus from a 22-year-old male.
In the expression analysis of regucalcin protein in normal human
liver and kidney, total protein lysates prepared from the following
tissues were used: normal liver from a 24-year-old male and normal
kidney from a 62-year-old male. In the expression analysis of
regucalcin protein in human liver and kidney tumor tissues, the
total protein lysates prepared from the following tissues were
used: liver hepatocellular carcinoma and its adjacent normal liver
tissue from a 39-year-old male; and kidney clear cell carcinoma and
its adjacent normal kidney tissue from a 64-year-old male. These
normal and tumor tissues used in the preparation of cDNA and
protein samples were estimated by clinical diagnosis.
Detection of alternatively spliced
variants
Total RNA in the non-tumor (normal) and tumor
tissues from human liver, kidney, brain and lung tissues (BioChain
Institute) was isolated using the guanidine thiocyanate technique.
Total RNA (11 μg) was primed by an oligo(dT) primer and reverse
transcribed using reverse transcriptase in a 40 μl final volume.
The reaction was terminated by heating at 65°C for 10 min. cDNA was
delivered in 1X RT buffer (50 mM Tris-Cl, pH 8.3, 75 mM KCl, 3 mM
MgCl2, 10 mM dithiothreitol). The estimated cDNA
concentration was approximately 2.5 ng/μl. cDNA (2.5–5 ng/μl) was
used as the template in polymerase chain reaction (PCR). The amount
of cDNA, which was normalized to the actin mRNA levels, was used
for PCR. PCR was performed using the following protocol: initial
denaturation at 95°C for 45 sec, followed by 30 cycles of 94°C for
45 sec, 56°C for 1 min, and 72°C for 1 min, with a final extension
at 72°C for 3 min. The primers used for the detection of human
regucalcin cDNA were 5′-ATGTCTTCCATTAAGATTGAGTGTGT-3′ for the sense
primer and 5′-TCATCCCGCATAGGAGTAGGG AGCA-3′ for the antisense
primer. The amplified PCR products were visualized on a 1.8%
agarose gel prepared with the addition of ethidium bromide. To
determine the DNA sequences of the PCR products, amplified DNA
fragments were cloned into the pGEM-T Easy cloning vector and then
sequenced with an automated fluorescence DNA sequencer (3130
Genetic Analyzer; Applied Biosystems, Foster City, CA, USA) to
confirm the identity of the cloned inserts. The house keeping gene,
β-actin, was also amplified to normalize the cDNA content of each
sample.
Western blot analysis
Total protein lysates, which were obtained from
normal and tumor tissues of human liver and kidney (BioChain
Institute), were used for western blot analysis. The total protein
lysates were subjected to 12% SDS-polyacrylamide gel
electrophoresis, and proteins on the gel were transferred to PVDF
membranes. The membranes were blocked in TBS-T (20 mm Tris, pH 7.5,
150 mm NaCl and 0.1% Tween-20) supplemented with 5% non-fat dry
milk for 1 h at room temperature. The first regucalcin antibody
that we previously produced against a purified rat regucalcin
protein (10) was diluted
(1:1,000) in Can get signal immunoreaction solution 1 (Toyobo,
Tokyo, Japan). The secondary antibody, an affinity purified
horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG
(Invitrogen Corp.) was diluted (1:1,000) in Can get signal
immunoreaction solution 2 (Toyobo). The membrane was probed with a
rabbit polyclonal regucalcin antibody overnight at 4°C, followed by
HRP-conjugated goat anti-rabbit IgG polyclonal antibodies for 1 h
at room temperature. Following immunoprobing with primary and
secondary antibodies, the blots were washed 3 times with TBS-T. The
blots were developed using ECL chemiluminescence reagent according
to the manufacturer’s instructions (Amersham Biosciences,
Pittsburgh, PA, USA). For reprobing, the membranes were incubated
in a stripping buffer (Pierce, Rockford, IL, USA) for 20 min and
were then reprobed with anti-β-actin antibody (Santa Cruz
Biotechnology) as a loading control.
Results
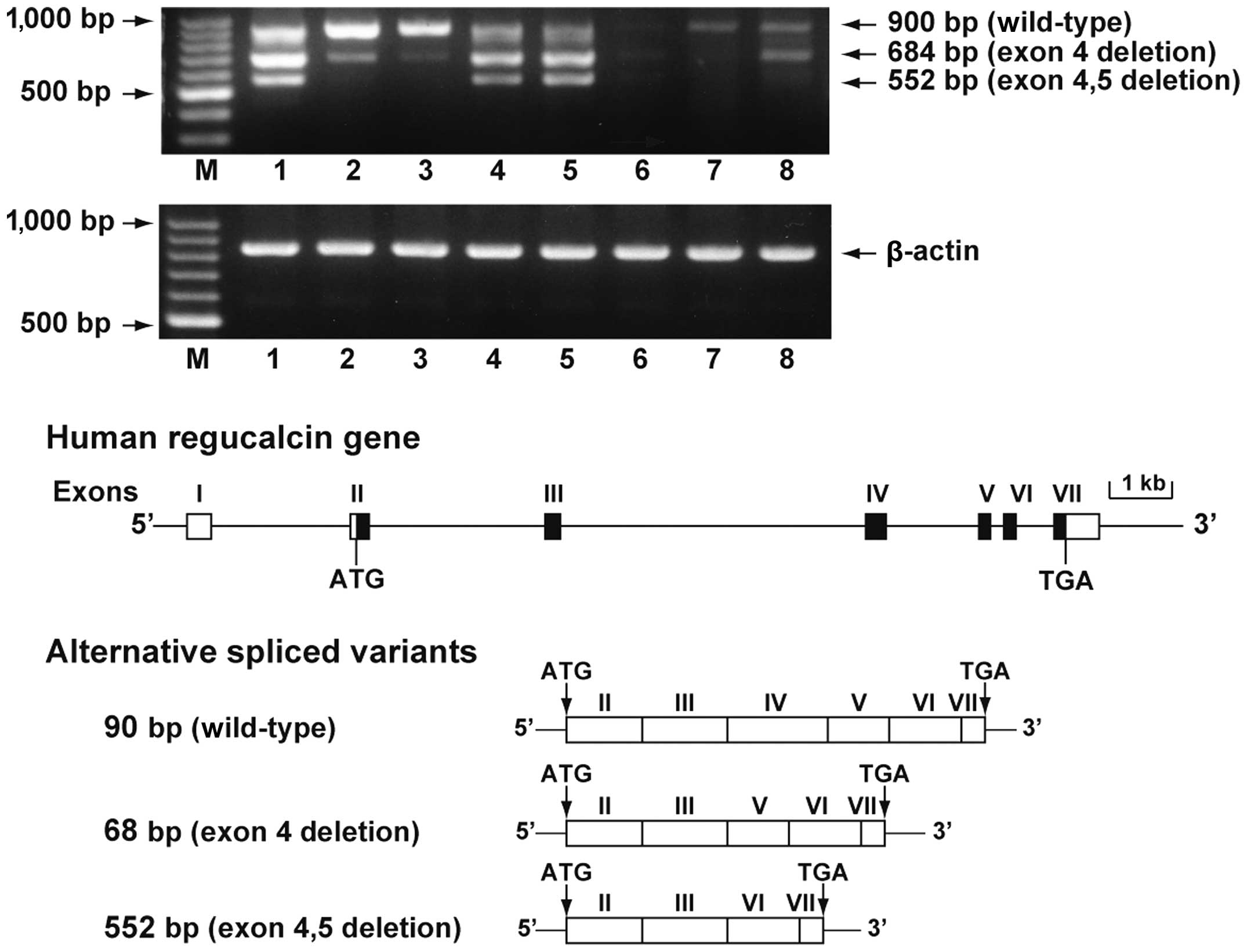
Regucalcin mRNA expression in the normal and tumor
tissues of human liver, kidney, brain and lung is shown in Fig. 1. The coding region of regucalcin
gene was amplified using cDNA from total RNA obtained from the
human tissues. Full-length regucalcin mRNA (900 bp) was expressed
in the normal tissues of the liver, kidneys, brain and lungs; its
expression was suppressed in tumor tissues, including liver
hepatocellular carcinoma, kidney transitional cell carcinoma, brain
malignant meningioma and lung non-small cell carcinoma with
clinical diagnosis. We determined the DNA sequence of 2 PCR
products derived from transcripts. The cloned PCR products consist
of 684-bp fragment with exon 4 deletion and a 552-bp fragment with
exon 4 and 5 deletions. These results indicated that 2
alternatively spliced variants were present in regucalcin mRNA. The
2 spliced transcripts encode the 2 predicted proteins of 227 amino
acids (~25 kDa) and 183 amino acids (~20 kDa). The 2 alternatively
spliced variants were detected in normal tissues, including the
liver, kidneys, brain and lungs. The expression levels of the 2
alternatively spliced variants were high in the normal liver and
lung tissue. Of note, the expression of the 2 alternatively spliced
variants appeared to be decreased in the tumor tissues of the
liver, kidneys, brain and lungs.
Regucalcin mRNA expression in the various brain
tissues obtained from healthy humans is shown in Fig. 2. Full-length regucalcin mRNA was
expressed in the amygdala, cerebral cortex, cerebellum,
diencephalon, hippocampus, medulla oblongata, optic nerve,
pituitary and thalamus. Alternatively the spliced variants were
also detected in the cerebral cortex and pituitary tissues. In
other brain tissues, the expression of these variants was
minimal.
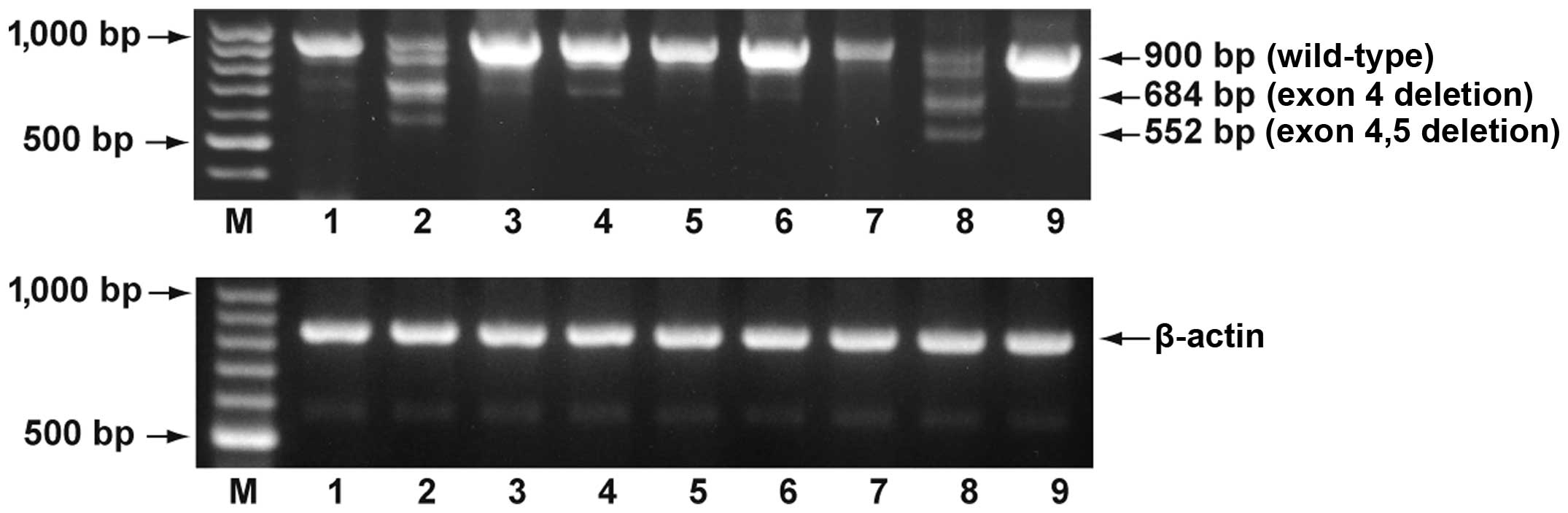 | Figure 2Expression of regucalcin mRNA in
various brain tissues. The coding region of the regucalcin gene was
amplified using cDNA for total RNA obtained from human normal brain
tissues with the method of PCR. The amplified PCR products were
visualized on a 1.8% agarose gel containing ethidium bromide. Data
are representative of one of 3 experiments. M, 100 base DNA-ladder;
lane 1, amygdala; lane 2, cerebral cortex; lane 3, cerebellum; lane
4, diencephalon; lane 5, hippocampus; lane 6, medulla oblongata;
lane 7, optic nerve; lane 8, pituitary; lane 9, thalamus. |
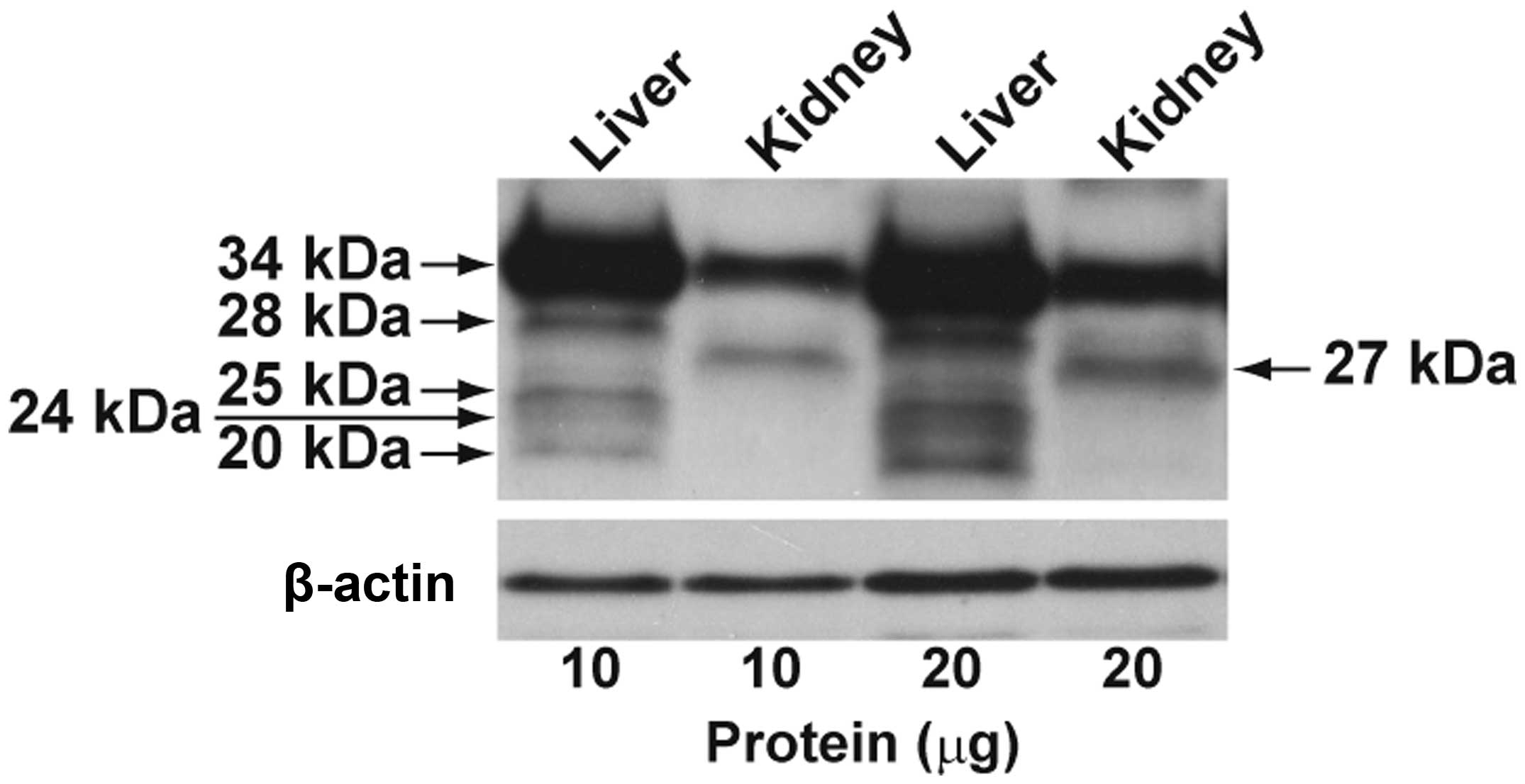
The expression of regucalcin protein in the liver
and kidney tissues obtained from healthy humans is shown in
Fig. 3. Full-length regucalcin
protein of 34 kDa was markedly detected in the normal liver and
kidney tissues. In addition, 4 weak bands with 28, 25, 24 and 20
kDa were observed in the liver tissues, and the band with 27 kDa
was found in the kidney tissue. The 2 alternatively spliced
transcripts indicated in Fig. 1
encoded 2 predicated proteins of approximately 25 and 20 kDa. The
liver protein bands of 25 and 20 kDa were associated with the
calculated molecular weights of the regucalcin splice variant
proteins. The expression levels of 2 splice variant proteins were
minimal as compared with those of the full-length protein.
Furthermore, the liver protein bands of 28 and 24 kDa and kidney
protein band of 27 kDa, which were not comparable to the predicted
molecular weights of the splice variant proteins, were detected at
minimal levels.
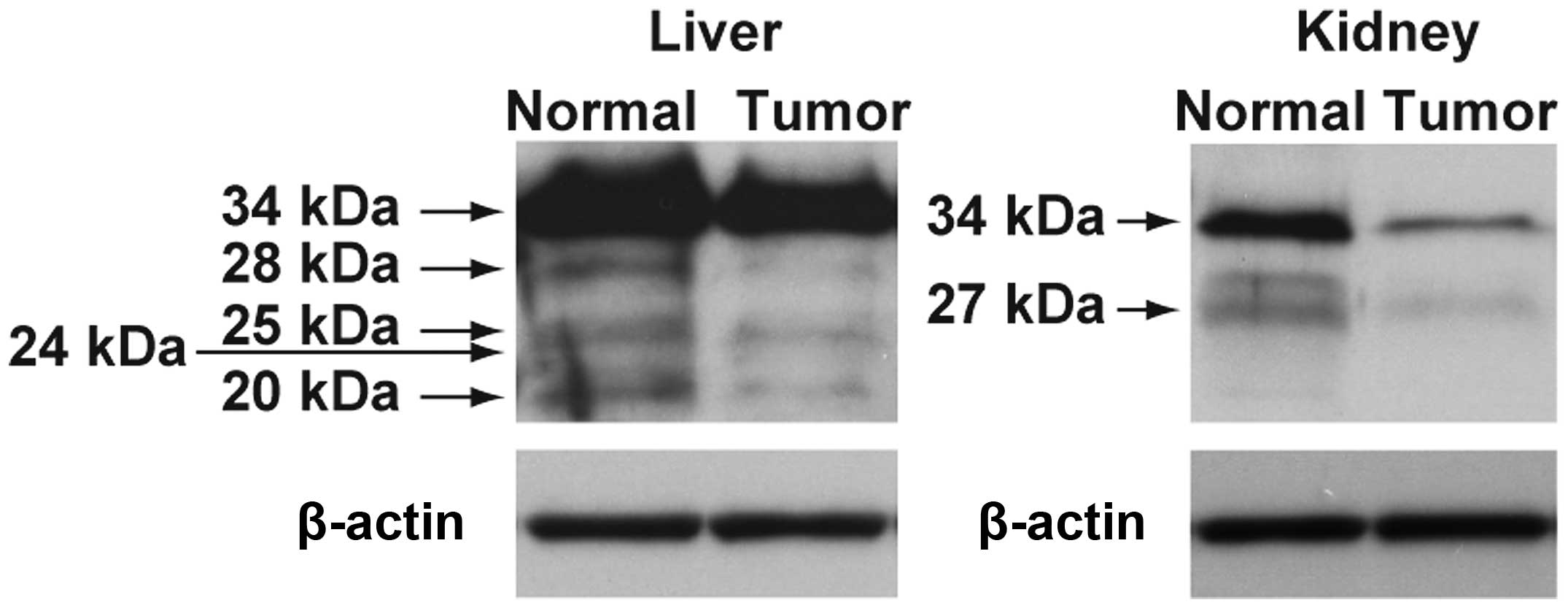
The expression of regucalcin protein in the normal
and tumor tissues of liver and kidney obtained from humans is shown
in Fig. 4. The expression of
regucalcin proteins with different molecular weight appeared to be
decreased in the tumor tissues of liver hepatocellular carcinoma
and kidney clear cell carcinoma as compared with those of the
normal tissues.
Alternatively spliced variants for regucalcin mRNA
and its related proteins (including 28, 25, 24 and 20 kDa) were not
observed in the liver and kidney of rats and mice (data not
shown).
Discussion
This study demonstrates that regucalcin mRNA
expression is suppressed in various human tumor tissues, including
hepatocellular carcinoma, kidney transitional cell carcinoma, brain
malignant meningioma and lung non-small cell carcinoma evaluated by
the clinical diagnosis of human subjects. Regucalcin protein was
clearly decreased in the liver and kidney tumor tissues, although
its levels were not determined in the brain and lung tumor tissues.
Regucalcin has also been reported to be underexpressed in human
hepatocellular carcinoma (26),
breast and prostate cancers (27).
It has been reported that several transcripts of
regucalcin mRNA are present in human breast (MCF-7) and prostate
(LNCap) cancer cell lines (27).
In this study, we found 3 transcripts for regucalcin mRNA in the
normal (non-tumor) and tumor tissues of human liver, kidney, brain
and lung. Moreover, we identified 2 alternatively spliced variants
lacking exon 4 or exons 4 plus 5 of the human regucalcin gene. The
identified alternatively spliced variants encoded 2 predicted
proteins of 227 amino acid residues (~25 kDa) and 183 amino acid
residues (~20 kDa). This observation on the alternative splicing
was in accordance with previous findings on non-neoplastic human
breast and prostate tissues and human breast (MCF-7) and prostate
(LNCap) cancer cells (27). We
also found that the expression of the alternatively spliced
regucalcin transcripts was decreased in the human tumor tissues.
Accordingly, it is interesting whether the alternative splicing
event of the regucalcin gene has a biological significance in the
human normal (non-tumor) and tumor tissues.
Two alternatively spliced regucalcin transcripts
[684 bp (exon 4 deletion) and 552 bp (exon 4 and 5 deletion)] were
not found in the various tissues (including liver, kidney cortex,
heart, brain and the others) of normal rat and mouse (data not
shown), in accordance with previous studies (5,9).
However, the alternatively spliced variants of the regucalcin gene
were found in various human tissues. This physiological
significance in the appearance of alternatively spliced variants
for the regucalcin gene expression in human tissues is unknown. It
is possible, however, that these variants play a role in the
physiological function of regucalcin in human tissues.
The characterization of regucalcin gene expression
in hepatoma cells is poorly understood. The mRNA expression,
post-translational processing and targeting of the gene may be
frequently altered in transformed cells. Regucalcin mRNA expression
in tumor tissues have been shown to be decreased as compared with
that in non-tumor tissues of chemically-fed rats, while regucalcin
mRNA expression has been observed in transplantable Morris hepatoma
cells (20). The sequencing of
cDNA cloning for regucalcin in human liver tissues and cloned human
hepatoma (HepG2) cells has been previously compared (28). We found that the human gene for
regucalcin gives rise to transcripts with different 5′-UTR
sequences in hepatoma cells; these genes contain identical coding
regions, differing only in their untranslated sequences. Moreover,
northern blot analysis using poly (A)+ RNA from human
liver and HepG2 cells revealed that regucalcin mRNA in the HepG2
cells was longer than that of the liver (28). These observations demonstrate the
existence of transcript heterogeneity of the human gene for
regucalcin in cancer cells. Regucalcin mRNA expression has been
shown to be decreased in HepG2 cells (30), and this reduction may be
implicated in the transcriptional alteration (30). From these findings, it can be
hypothesized that the suppressed regucalcin gene expression in
tumor tissues plays a role in transcriptional alteration.
We also examined the expression of regucalcin
protein in the liver and kidneys using normal human tissues.
Full-length regucalcin protein (34 kDa) was expressed in the human
normal liver and kidney tissues. The regucalcin splice variant
proteins (25 and 20 kDa) were found in the normal liver, but not in
the normal kidney. As regucalcin splice variant proteins were not
found in the normal kidney, their expression may be tissue
specific. It has been reported that the proteolytic processing of
full-length regucalcin protein (34 kDa) leads to the production of
2 molecular weight proteins of 28 and 24 kDa (29). In our study, the liver proteins of
28 or 24 kDa and the kidney protein of 27 kDa, which did not
correspond to the predicted molecular weight of the alternatively
spliced variant proteins, were detected at minimal levels. These
proteins were specifically observed in each tissue and appeared to
be produced by proteolysis. It is unknown whether these proteins
have a functional role.
Furthermore, we found that the expression of
full-length regucalcin protein is suppressed in human liver
hepatocellular carcinoma and kidney clear cell carcinoma.
Regucalcin mRNA expression and its protein levels have been shown
to be decreased in cloned rat hepatoma H4-II-E cells as compared to
those of normal rat liver (24,25). The overexpression of endogenous
regucalcin has been found to suppress the proliferation of cloned
rat hepatoma H4-II-E cells enhanced with serum stimulation
(17,24,25). Regucalcin has been demonstrated to
cause G1 and G2/M phase cell cycle arrest in H4-II-E cells
(25). In addition, regucalcin
has been shown to have suppressive effects on cell proliferation,
inducing G1 and G2/M phase cell cycle arrest in cloned normal rat
kidney proximal tubular epithelial NRK52E cells (31). The suppressive effects of
regucalcin on cell proliferation may be mediated through depression
in various Ca2+ signaling-dependent protein kinases,
protein phosphatases and PI3 kinase activities and the suppression
of c-myc, Ha-ras, c-jun and chk2 mRNA
expression or the enhancement of p53 and Rb mRNA
expression (24,25,32). Moreover, regucalcin has been shown
to suppress protein synthesis and nuclear DNA and RNA synthesis
(16,17,19). Suppressed regucalcin gene
expression may lead to the development of carcinogenesis (16,21).
In conclusion, our study demonstrates that the
full-length and alternatively spliced variants of regucalcin mRNA
were expressed in various normal human tissues, including the
liver, kidneys, brain and lungs, and that the expression of
full-length regucalcin protein was found in human normal liver and
kidney tissues. We also found that the spliced variant proteins of
regucalcin appeared in the human normal liver but not in the kidney
tissues.
References
|
1
|
Yamaguchi M and Yamamoto T: Purification
of calcium binding substance from soluble fraction of normal rat
liver. Chem Pharm Bull (Tokyo). 26:1915–1918. 1978. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yamaguchi M: A novel
Ca2+-binding protein regucalcin and calcium inhibition.
Regulatory role in liver cell function. Calcium Inhibition. Kohama
K: Japan Sci Soc Press, Tokyo and CRC Press; Boca Raton: pp. 19–41.
1992
|
|
3
|
Shimokawa N and Yamaguchi M: Molecular
cloning and sequencing of the cDNA coding for a calcium-binding
protein regucalcin from rat liver. FEBS Lett. 327:251–255. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Misawa H and Yamaguchi M: The gene of
Ca2+-binding protein regucalcin is highly conserved in
vertebrate species. Int J Mol Med. 6:191–196. 2000.
|
|
5
|
Yamaguchi M: The transcriptional
regulation of regucalcin gene expression. Mol Cell Biochem.
346:147–171. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shimokawa N, Matsuda Y and Yamaguchi M:
Genomic cloning and chromosomal assignment of rat regucalcin gene.
Mol Cell Biochem. 151:157–163. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Thiselton DL, McDowall J, Brandau O,
Ramser J, d’Esposito F, Bhattacharga SS, Ross MT, Hardcastle AJ and
Meindl M: An integrated, functionally annotated gene map of the
DXS8026-ELK1 internal on human Xp11.3-Xp11.23: Potential hotspot
for neurogenetic disorders. Genomics. 79:560–572. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yamaguchi M, Makino R and Shimokawa N: The
5′end seguences and exon organization in rat regucalcin gene. Mol
Cell Biochem. 165:145–150. 1996.
|
|
9
|
Shimokawa N and Yamaguchi M: Calcium
administration stimulates the expression of calcium-binding protein
regucalcin mRNA in rat liver. FEBS Lett. 305:151–154. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yamaguchi M and Isogai M: Tissue
concentration of calcium-binding protein regucalcin in rats by
enzyme-linked immunoadsorbent assay. Mol Cell Biochem. 122:65–68.
1993. View Article : Google Scholar
|
|
11
|
Yamaguchi M: Hormonal regulation of
regucalcin gene expression: Involvement in cell metabolism. Horm
Stud. 1:12013. View Article : Google Scholar
|
|
12
|
Yamaguchi M: Role of regucalcin in calcium
signaling. Life Sci. 66:1769–1780. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yamaguchi M: Role of regucalcin in
maintaining cell homeostasis and function. Int J Mol Med.
15:372–389. 2005.
|
|
14
|
Yamaguchi M: Regucalcin and cell
regulation: role as a suppressor in cell signaling. Mol Cell
Biochem. 353:101–137. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yamaguchi M: Role of regucalcin in cell
nuclear regulation: Involvement as a transcription factor. Cell
Tissue Res. 354:331–341. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yamaguchi M: Suppressive role of
regucalcin in liver cell proliferation: Involvement in
carcinogenesis. Cell Prolif. 46:243–253. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yamaguchi M: The anti-apoptotic effect of
regucalcin is mediated through multisignaling pathways. Apoptosis.
18:1145–1153. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yamaguchi M: Regucalcin and metabolic
disorder: osteoporosis and hyperlipidemia are induced in regucalcin
transgenic rats. Mol Cell Biochem. 327:53–63. 2010.PubMed/NCBI
|
|
19
|
Yamaguchi M and Murata T: Involvement of
regucalcin in lipid metabolism and diabetes. Metabolism.
62:1045–1051. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Makino R and Yamaguchi M: Expression of
calcium-binding protein regucalcin mRNA in hepatoma cells. Mol Cell
Biochem. 155:85–90. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Suzuki S, Asamoto M, Tsujimura K and
Shirai T: Specific differences in gene expression profile revealed
by cDNA microarray analysis of glutathione S-transferase placental
form (GST-P) immunohistochemically positive rat liver foci and
surrounding tissue. Carcinogenesis. 25:439–443. 2004. View Article : Google Scholar
|
|
22
|
Inagaki S and Yamaguchi M: Regulatory role
of endogenous regucalcin in the enhancement of nuclear
deoxyribonucleic acid synthesis with proliferation of cloned rat
hepatoma cells (H4-II-E). J Cell Biochem. 82:704–711. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Misawa H, Inagaki S and Yamaguchi M:
Suppression of cell proliferation and deoxyribonucleic acid
synthesis in cloned rat hepatoma H4-II-E cells overexpressing
regucalcin. J Cell Biochem. 84:143–149. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tsurusaki Y and Yamaguchi M:
Overexpression of regucalcin modulates tumor-related gene
expression in cloned rat hepatoma H4-II-E cells. J Cell Biochem.
90:619–626. 2003. View Article : Google Scholar
|
|
25
|
Yamaguchi M and Daimon Y: Overexpression
of regucalcin suppresses cell proliferation in cloned rat hepatoma
H4-II-E cells: Involvement of intracellular signaling factors and
cell cycle-related genes. J Cell Biochem. 95:1169–1177. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou SF, Mo FR, Bin YH, Hou GQ, Xie XX and
Luo GR: Serum immunoreactivity of SMP30 and its tissues expression
in hepatocellular carcinoma. Clin Biochem. 44:331–336. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Maia C, Santos C, Schmitt F and Socorro S:
Regucalcin is under-expressed in human breast and prostate cancers:
Effect of sex steroid hormones. J Cell Biochem. 107:667–676. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Misawa H and Yamaguchi M: Transcript
heterogeneity of the human gene for Ca2+-binding protein
regucalcin. Int J Mol Med. 5:283–287. 2000.PubMed/NCBI
|
|
29
|
Arun P, Aleti V, Parikh K, Manne V and
Chilukuri N: Senescence marker protein 30 (SMP30) expression in
eukaryotic cells: existence of multiple species and membrane
localization. PLoS One. 6:e165452011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Murata T, Synya N and Yamaguchi M:
Expression of calcium-binding protein regucalcin mRNA in the cloned
human hepatoma cells (HepG2): stimulation by insulin. Mol Cell
Biochem. 175:163–168. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nakagawa T, Sawada N and Yamaguchi M:
Overexpression of regucalcin suppresses cell proliferation of
cloned normal rat kidney proximal tubular epithelial NRK52E cells.
Int J Mol Med. 16:637–643. 2005.PubMed/NCBI
|
|
32
|
Tsurusaki Y and Yamaguchi M: Role of
regucalcin in liver nuclear function: Binding of regucalcin to
nuclear protein or DNA and modulation of tumor-related gene
expression. Int J Mol Med. 14:277–281. 2004.PubMed/NCBI
|