Introduction
Selenium deficiency is considered a causative factor
in various types of heart failure, including Keshan disease (KD),
which is prevalent in China and is also one of the most harmful
endemic diseases (1). Heart
failure patients with reduced left ventricular contractility or
systolic heart failure have lower blood selenium levels (2–4).
In a small clinical study, selenium improved left ventricular
function and quality of life in systolic heart failure (5). However, the selenium deficiency and
cardiac dysfunction link remains to be understood.
microRNAs (miRNAs or miRs) are a class of endogenous
non-coding small RNAs that are involved in the modulation of
numerous biological processes by base-pairing, usually imperfectly,
to the 3′ untranslated region of a target mRNA, leading to
post-transcriptional inhibition and occasionally mRNA cleavage
(6). miRNAs have been identified
in the majority of types of cells and tissues. The current estimate
is that 10–30% of genes may be regulated by miRNAs, particularly
the members of signal transduction networks. Additionally, there is
increasing evidence that miRNAs are involved in a numerous
biological processes, including cell proliferation,
differentiation, apoptosis and tumorigenesis (7). Aberrant and/or absent miRNA
expression is frequently associated with pathophysiological
disorders (8–10). However, miRNA expression changes
in the condition of selenium deficiency and whether selenium
deficiency is involved in cardiac dysfunction remains unclear.
Wnt/β-catenin signaling is subjected to multiple
levels of molecular control. The canonical Wnt/β-catenin signaling
pathway is initiated when Wnt ligands bind to its receptor(s),
Frizzled (11) and low-density
lipoprotein receptor-related protein-5 or -6 (LRP5 or LRP6)
(12). The stabilized β-catenin
accumulates in the cytoplasm and translocates into the cell nucleus
following activation, where it forms a β-catenin-lymphoid
enhancer-binding factor (LEF)/transcription factor (TCF)
transcriptional complex and induces transcription of downstream
genes implicated in carcinogenesis. In the absence of Wnt-ligand
stimulation, β-catenin is sequestered in the ‘destruction complex,’
which leads to β-catenin degradation by the ubiquitin-proteasome
mechanism and ultimately the inactivation of β-catenin signaling
(13–15). The secretion of antagonists of the
Wnt pathway, including Wnt inhibitory factor-1, secreted
Frizzled-related proteins and Dickkopf1 (16) also regulate the suppression of
β-catenin signaling. Wnt proteins belong to a large family of
cysteine-rich secreted glycoproteins, which are highly conserved
during evolution, and they activate a highly conserved
intracellular signaling cascade with a significant influence in
early embryogenesis and pattern regulations (17,18). These proteins have diverse
mediated effects, including proliferation, apoptosis, migration,
polarization, stem cell maintenance and differentiation (19). Wnt/β-catenin signaling plays
significant roles in animal development and tissue homeostasis, and
misregulation of this pathway has been associated in numerous
pathological states, such as cancer, heart disease and Alzheimer’s
(20).
Given the potential role of miRNAs, their expression
was profiled in selenium-deficient rats by microarray. The
differentially expressed miRNAs were selected and validated. Due to
the importance of Wnt/β-catenin signaling in cardiac function, it
was further investigated in an attempt to provide a novel insight
into the limited understanding of the biological process and
mechanism of cardiac dysfunction in selenium-deficient rats.
Materials and methods
Animals
A total of 60 male weaning Sprague-Dawley rats
(3-weeks old; specific-pathogen-free class; body weight, 75±10 g)
were raised under controlled-environmental conditions (12-h
light-dark cycle; temperature, 25±1°C; humidity, 65±4%). The
standard diet (containing 0.2 mg selenium/kg food) was produced by
the Animal Experimental Center of Xi’an Jiaotong University (Xi’an,
China) and the low-selenium diet (<0.02 mg selenium/kg food) was
produced by Trophic Animal Feed High-tech Co. (Jiangsu, China)
according to the AIN-93 M formula. The food was stored at 4°C and
fresh tap water was allocated continuously. The study was carried
out in strict accordance with the recommendations in the Guide for
the Care and Use of Laboratory Animals of the National Institutes
of Health. The protocol was approved by the Committee on the Ethics
of Animal Experiments of Xi’an Jiaotong University. All the
surgeries were performed under chloral hydrate anesthesia, and all
efforts were made to minimize suffering.
Groups and treatments
All the animals were randomly assigned into three
groups: Control (n=20), low selenium (LS) (n=20) and selenium
supplementation (SS) (n=20). In the control group, the animals were
fed with the standard diet for 14 weeks and were treated by
intraperitoneal injection of physiological saline every day for 21
days. In the LS group, the animals were fed with the low-selenium
diet for 14 weeks and were treated by intraperitoneal injection of
physiological saline every day for 21 days. In the SS group, the
animals were fed with the low-selenium diet for 14 weeks and were
treated by intraperitoneal injection of sodium selenite (0.05 mg/kg
bodyweight; Sigma-Aldrich, St. Louis, MO, USA) as selenium
supplementation every day for 21 days (21). All the animals were monitored
every two days by observing their mental status and activities. Two
animals succumbed as a result of Se deficiency. Following this,
these two animals were immediately quarantined and sent to the
Animal Center for unified treatment.
Selenium concentration detection
Samples of whole blood and hearts harvested from
rats were wet-washed by mixed acids (nitric acid and perchloric
acid) in borosilicate reaction tubes. Following removal of the
excess acids, the samples were titrated to 10 ml by ultra-pure
water (Aqulix 5 water purification system; Merck Millipore,
Billerica, MA, USA). The selenium concentration was subsequently
determined by the flameless atomic absorption spectrophotometry
method using a Z-5000 spectrophotometer (Hitachi, Ltd., Tokyo,
Japan) with a cathode lamp of Se (resonance line, 196.0 nm;
Photron, Victoria, Australia). Standard selenium solutions were
used to calibrate the results.
Glutathione peroxidase activity
assay
Cardiac glutathione peroxidase (GPx) activity in
heart extract was assessed by a spectrophotometry method using the
Glutathione Peroxidase Cellular Activity Assay kit (Sigma-Aldrich),
following the manufacturer’s instructions. The assay is based on
the reaction in which oxidation of glutathione (GSH) to oxidized
glutathione (GSSG) is catalyzed by GPx. As NADPH is consumed when
GSSG is recycled back to GSH, the decrease in NADPH absorbance at
340 nm (Tecan Sunrise Absorbance Reader, Tecan, Austria) can be
utilized to calculate the activity of GPx indirectly.
Plasma brain natriuretic peptide (BNP)
assay
Following femoral artery puncture, whole blood
samples were collected using EDTA-Na2 vacuum blood
collection tubes. The supernatant was collected after
centrifugation at 377 × g for 20 min. The samples were processed by
the Triage BNP assay (Biosite, San Diego, CA, USA) within 1 h after
collection at room temperature, according to the manufacturer’s
instructions.
Electrocardiography (ECG)
ECG in lead II was recorded (PowerLab 4/25;
ADInstrument, New South Wales, Australia) during the experiment
time. Prior to electrocardiography, all the rats were anesthetized
by intraperitoneal injection of chloral hydrate (10%, 0.03 ml/kg
bodyweight). The left upper limb, right upper limb and right lower
limb electrodes were placed for leads I. Subsequently, the ECG was
analyzed for changes in the ST segment, T wave, AV block and
arrhythmia (premature ventricular contraction, ventricular
tachycardia and fibrillation and atrial fibrillation). Cardiac
conduction of rats was evaluated by the number of ventricular
arrhythmic events (VAEs) within 15 min.
Echocardiography
Echocardiographic tests were performed according to
the instructions described in previous studies (22,23). Animals were anesthetized with
chloral hydrate (10%, 0.03 ml/kg bodyweight) intraperitoneally 10
min before imaging. In order to maintain an optimal image quality,
the hair of the anterior chest wall was removed by 7% sodium
sulfite solution and the rats were placed left laterally. Vivid 7
dimension (GE Healthcare, Pittsburgh, PA, USA) with a probe (12 L)
working at 10 MHz was utilized. The probe was placed parallel to
the left margin of the sternum and adjusting the image depth,
ranging 2.0–4.0 cm. Regurgitant jets were assessed by
two-dimensional color and continuous Doppler. M-mode tracings were
applied to determine the left ventricular end-systole diameter
(LVESD) and left ventricular end-diastole diameter (LVEDD).
Finally, the functional parameters, including left ventricular
fractional shortening (LVFS%), which is the percentage of blood
ejected by the left ventricle for each heart beat, and left
ventricular ejection fraction (LVEF%), which measures the change in
the left ventricular diameter from each diastole to systole were
calculated respectively: LVFS% = ((LVEDD − LVESD)/LVEDD)%; and
LVEF% = [(LVEDV − LVESV)/LVEDV]%.
Hemodynamic parameters
Hemodynamic determination was conducted according to
methods described in a previous study (24). The animals were anesthetized with
chloral hydrate (10%, 0.03 ml/kg bodyweight) intraperitoneally. A
catheter connected to the Powerlab 4/25 biological analysis system
was intubated from the right carotid artery into the left
ventricle. Left ventricular systolic pressure (LVSP), left
ventricular end-diastolic pressure (LVEDP), maximum rising rate of
left ventricular pressure (LVdp/dtmax) and maximum
dropping rate of left ventricular pressure (LVdp/dtmin)
were measured.
Histological examination
The animals were anesthetized with chloral hydrate
(10%, 0.03 ml/kg bodyweight) intraperitoneally. Subsequently, they
were sacrificed by removing the hearts following complete
anesthesia. The harvested hearts were rinsed in phosphate-buffered
saline, fixed in 4% paraformaldehyde for 24 h, embedded in paraffin
and cross-sectioned into 10-μm slices. The sections were stained
with hematoxylin and eosin (HE) for cells alignment according to
the general procedure. The morphological structures of the heart
were observed by light microscopy.
miRNA microarray analysis
Total RNA was isolated using TRIzol (Invitrogen) and
the miRNeasy mini kit (Qiagen) according to the manufacturer’s
instructions, which efficiently recovered all RNA species,
including miRNAs. RNA quality and quantity was measured by using
nanodrop spectrophotometer (ND-1000, Nanodrop Technologies,
Wilmington, DE, USA) and RNA integrity was determined by gel
electrophoresis. Following RNA isolation from the samples, the
miRCURY™ Hy3™/Hy5™ Power labeling kit (Exiqon, Vedbaek, Denmark)
was used according to the manufacturer’s guideline for miRNA
labeling. Each sample (1 μg) was 3′-end-labeled with the Hy3TM
fluorescent label, using T4 RNA ligase as follows: RNA in 2.0 μl
water was combined with 1.0 μl calf intestinal alkaline phosphatase
(CIP) buffer and CIP (Exiqon). The mixture was incubated for 30 min
at 37°C, and was terminated by incubation for 5 min at 95°C.
Subsequently, 3.0 μl labeling buffer, 1.5 μl fluorescent label
(Hy3TM), 2.0 μl dimethyl sulfoxide and 2.0 μl labeling enzyme were
added into the mixture. The labeling reaction was incubated for 1 h
at 16°C, and terminated by incubation for 15 min at 65°C.
Subsequent to stopping the labeling procedure, the Hy3TM-labeled
samples were hybridized on the miRCURYTM LNA Array (v.16.0)
(Exiqon) according to the array instructions. The total mixture (25
μl) from Hy3TM-labeled samples with 25 μl hybridization buffer were
first denatured for 2 min at 95°C, incubated on ice for 2 min and
hybridized to the microarray for 16–20 h at 56°C in a 12-Bay
Hybridization System (Hybridization System-Nimblegen Systems, Inc.,
Madison, WI, USA), which provides an active mixing action and
constant incubation temperature to improve hybridization uniformity
and to enhance the signal. Following hybridization, the slides were
obtained, washed several times using Wash buffer kit (Exiqon), and
dried by centrifugation for 5 min at 100 × g. Subsequently, the
slides were scanned using the Axon GenePix 4000B microarray scanner
(Axon Instruments, Foster City, CA). The scanned images were
imported into the GenePix Pro 6.0 software (Axon Instruments) for
grid alignment and data extraction. Replicated miRNAs were averaged
and miRNAs with intensities >50 in all the samples were chosen
for calculating normalization factor. Expressed data were
normalized using the Median normalization (25). Following normalization,
differentially expressed miRNAs were identified through fold change
filtering. Hierarchical clustering was performed using the MEV
software (Dana-Farber Cancer Institute, Boston, MA, USA).
Gene ontology (GO) analysis
GO analysis was applied in order to organize genes
into hierarchical categories and uncover the miR-gene regulatory
network on the basis of biological process and molecular function.
In detail, two-side Fisher’s exact test was used to classify the GO
category, and the false discovery rate (FDR) was calculated to
correct the P-value. Only GOs that had a P-value of <0.001 and
an FDR of <0.05 were chosen. Within the significant category,
the enrichment rare earth (Re) was:
Re=(nf/n)/(Nf)/N),
where nf is the number of flagged genes
within the particular category, n is the total number of
genes within the same category, Nf is the
number of flagged genes in the entire microarray and N is
the total number of genes in the microarray. Subsequently, the
apoptosis-related network of miRNA-mRNA interaction, representing
the critical miRNAs and their targets, was established according to
the degree of miRNA (26).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) of miRNA
Differentially expressed miRNAs of miR-374,
miR-16, miR-199a-5p, miR-195,
miR-30e*, miR-3571, miR-675 and
miR-450a* were selected for verification. Total
RNA was extracted from harvested heart using the MiniBEST Universal
RNA Extraction kit (Takara, Otsu, Japan). cDNA was synthesized from
total RNA with the SYBR PrimeScript™ miRNA RT-PCR kit (Takara). The
expression of miRNA was analyzed by qPCR using SYBR Premix Ex
TaqTMII (Takara). PCR was performed in triplicate for each sample.
The relative amount of miRNAs was normalized against U6
small nuclear RNA. Detection of mRNA was performed as described
previously (27). The sequences
of the primers are shown in Table
I.
 | Table IPrimers used in TaqMan RT-qPCR. |
Table I
Primers used in TaqMan RT-qPCR.
| Gene primer | Product size,
bp | Number gene primer
(5′→3′) |
|---|
| U6 | 62 | F:
5′-GCTTCGGCAGCACATATACTAAAAT-3′
R: 5′-CGCTTCACGAATTTGCGTGTCAT-3′ |
|
rno-miR-374 | 68 | F:
5′-CTCGGATGGATATAATACA-3′
R: 5′-CTCGGACAATAATAATACA-3′ |
|
rno-miR-16 | 62 | F:
5′-GGGGTAGCAGCACGTAAATA-3′
R: 5′-GTGCGTGTCGTGGAGTCG-3′ |
|
rno-miR-199a-5p | 94 | F:
5′-CTTCTGGAGATCCTGCTC-3′
R: 5′-TGCCCAGTCTAACCAATG-3′ |
|
rno-miR-195 | 78 | F:
5′-AACTCTCCTGGCTCTAGC-3′
R: 5′-GCCTGGAGCAGCACAG-3′ |
|
rno-miR-30e | 86 | F:
5′-GGGCAGTCTTTGCTACTG-3′
R: 5′-CCTGCCGCTGTAAACATC-3′ |
|
rno-miR-3571 | 96 | F:
5′-GGACATTACCTACCCAA-3′
R: 5′-TAGTGCCTACTCAGAGC-3′ |
|
rno-miR-675 | 70 | F:
5′-GGACTGGTGCGGAAAGG-3′
R: 5′-AGACCCAGGGACTGAGC-3′ |
|
rno-miR-450a | 70 | F:
5′-AGAGATGCGGAGCTGTT-3′
R: 5′-TATGCAAAATGTTCCCAAT-3′ |
Western blotting
Frozen cardiac tissue was homogenized in
radioimmunoprecipitation assay lysis buffer system (Santa Cruz
Biotechnology, Inc., Dallax, TX, USA) with phenylmethylsulfonyl
fluoride (Santa Cruz Biotechnology, Inc.). All the procedures
followed manufacturer’s instructions. The sample protein
concentration was detected by using bicinchoninic acid protein
assay kit (Santa Cruz Biotechnology, Inc.). The sample protein was
boiled in 1x SDS-PAGE loading buffer, separated by electrophoresis
in 10% SDS-polyacrylamide gel and subsequently transferred to a
polyvinylidene fluoride membrane. Antibodies against Wnt (ab15251)
and β-catenin (ab6302) (Abcam, Cambridge, MA, USA) were applied to
incubate the bolts at 4°C overnight. Tris-buffered saline
(containing 0.02% Tween 20) was used to wash the membranes, which
were subsequently incubated with goat polyclonal secondary antibody
to rabbit immunoglobulin G conjugated to horseradish peroxidase
(Abcam). The membranes were developed using Super Signal West Pico
chemiluminescence reagent (Thermo Scientific, Waltham, MA, USA) and
were visualized on X-ray films.
Statistical analysis
All the results were expressed as mean ± standard
deviation. Statistical analysis was performed with one-way analysis
of variance for multiple comparisons. P<0.05 was considered to
indicate a statistically significant difference.
Results
Se concentration in blood samples
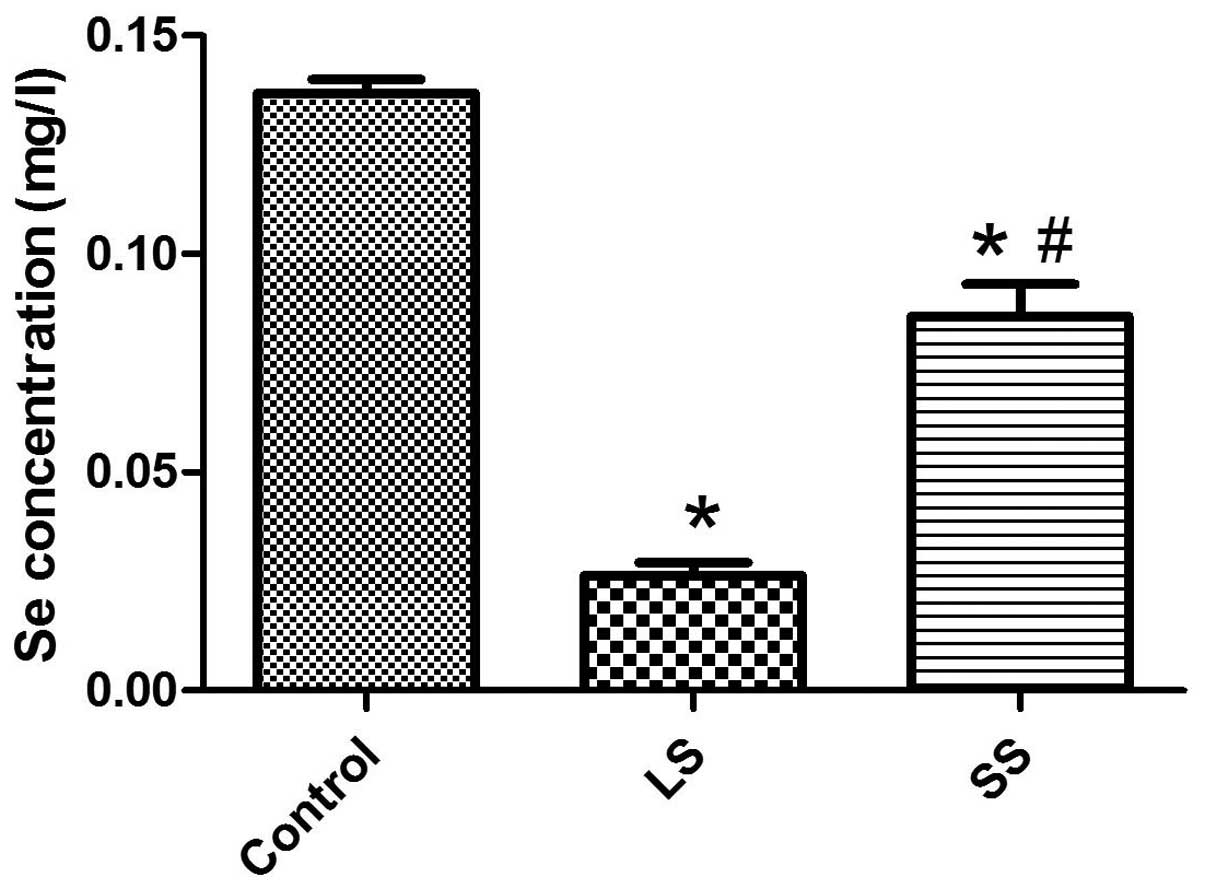
The Se concentration in blood is shown in Fig. 1. Significant changes of Se
concentration were observed in the LS and SS groups. Compared to
the control group, the Se concentration in blood decreased
significantly in the LS and SS groups (P<0.05). Se
intraperitoneal injection was proved to increase the Se
concentration in blood in the SS group compared to the LS group
(P<0.05).
Effects of Se deficiency and Se
supplementation on GPx activity
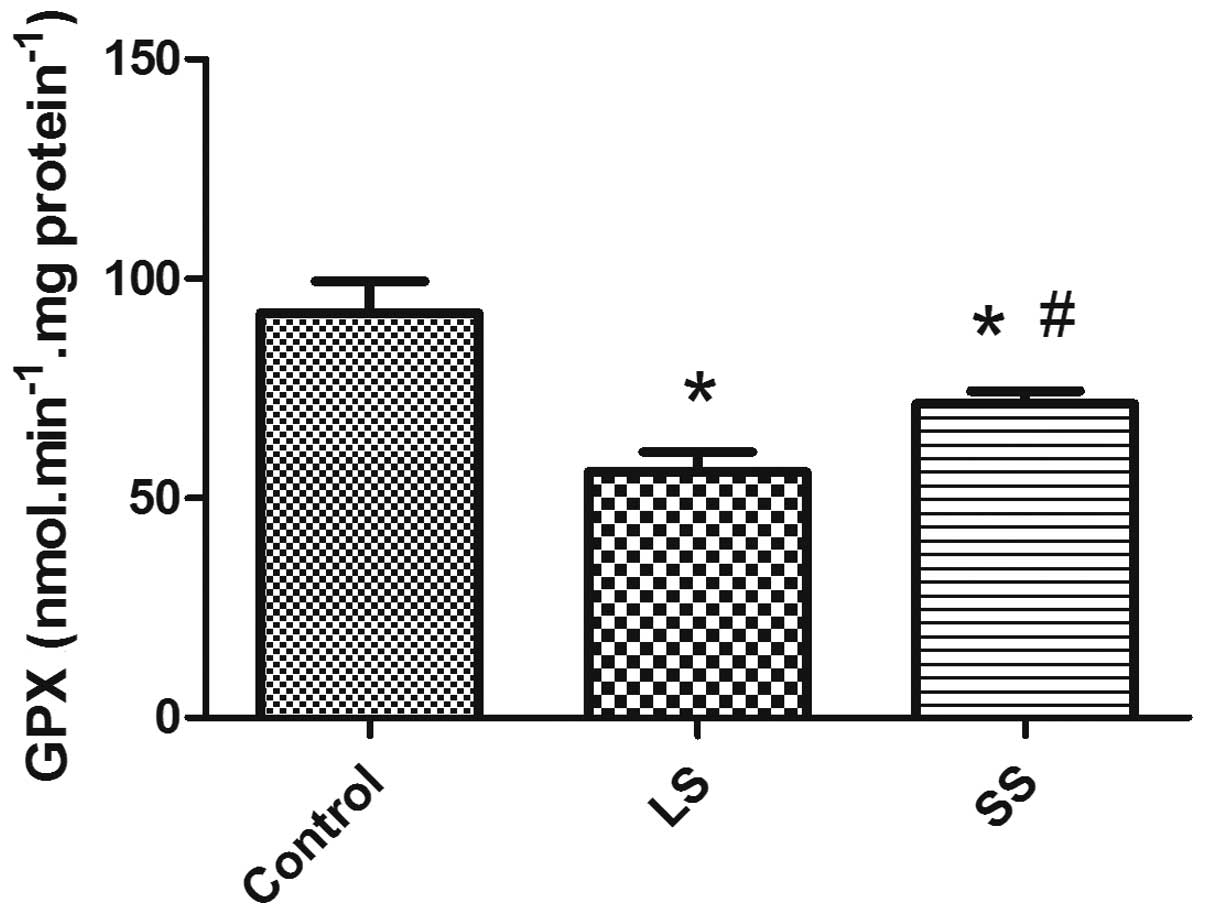
As shown in Fig.
2, the activity of GPx in the LS and SS groups decreased
significantly compared to the control group (P<0.05). The
activity of GPx was observed to reduce more significantly in the LS
group. However, an evident recovery of GPx activity in the SS group
was confirmed by the GPx activity assay when compared to the LS
group (P<0.05).
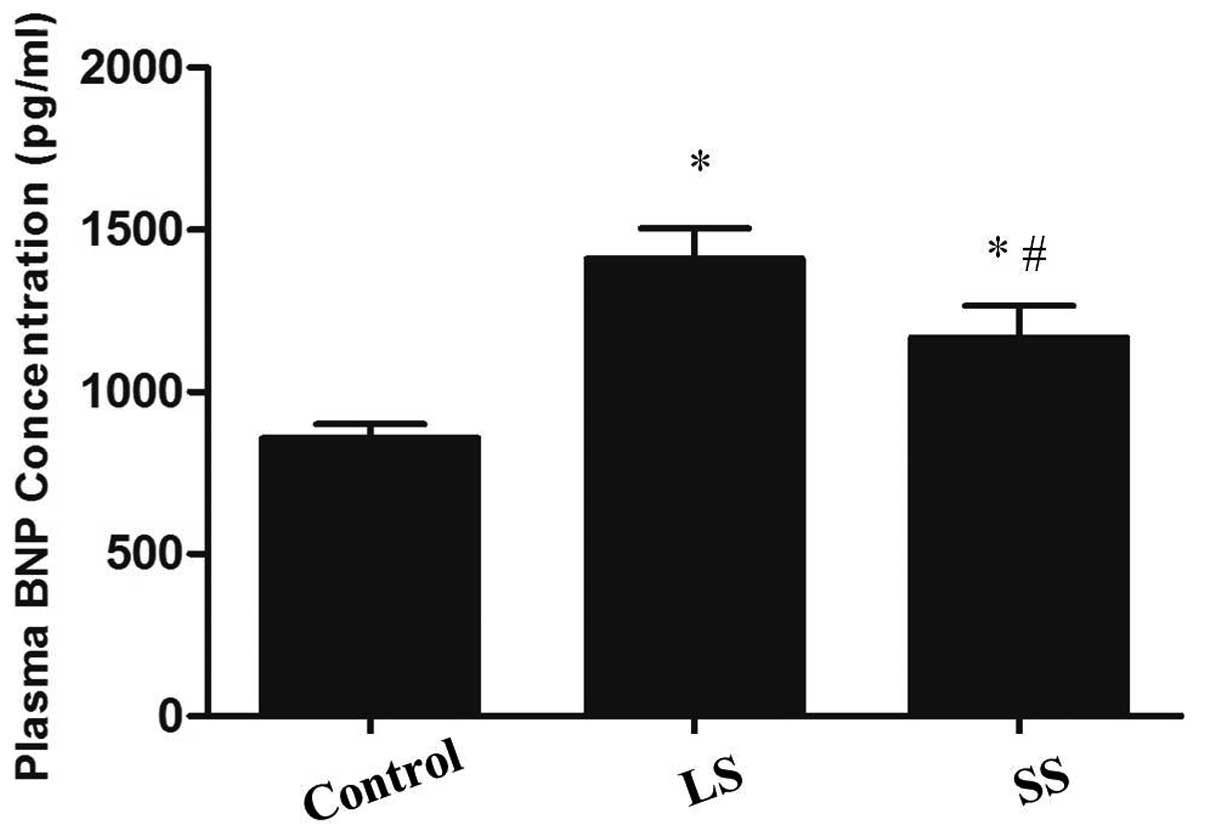
Plasma BNP
The Triage BNP assay showed that the plasma BNP
level increased significantly in the LS and SS groups compared to
the control group (P<0.05). A clearer increase of plasma BNP was
found in the LS group. Following Se supplementation by
intra-peritoneal injections, a reduction in the plasma BNP level in
the SS group was identified compared to the LS group (P<0.05)
(Fig. 3).
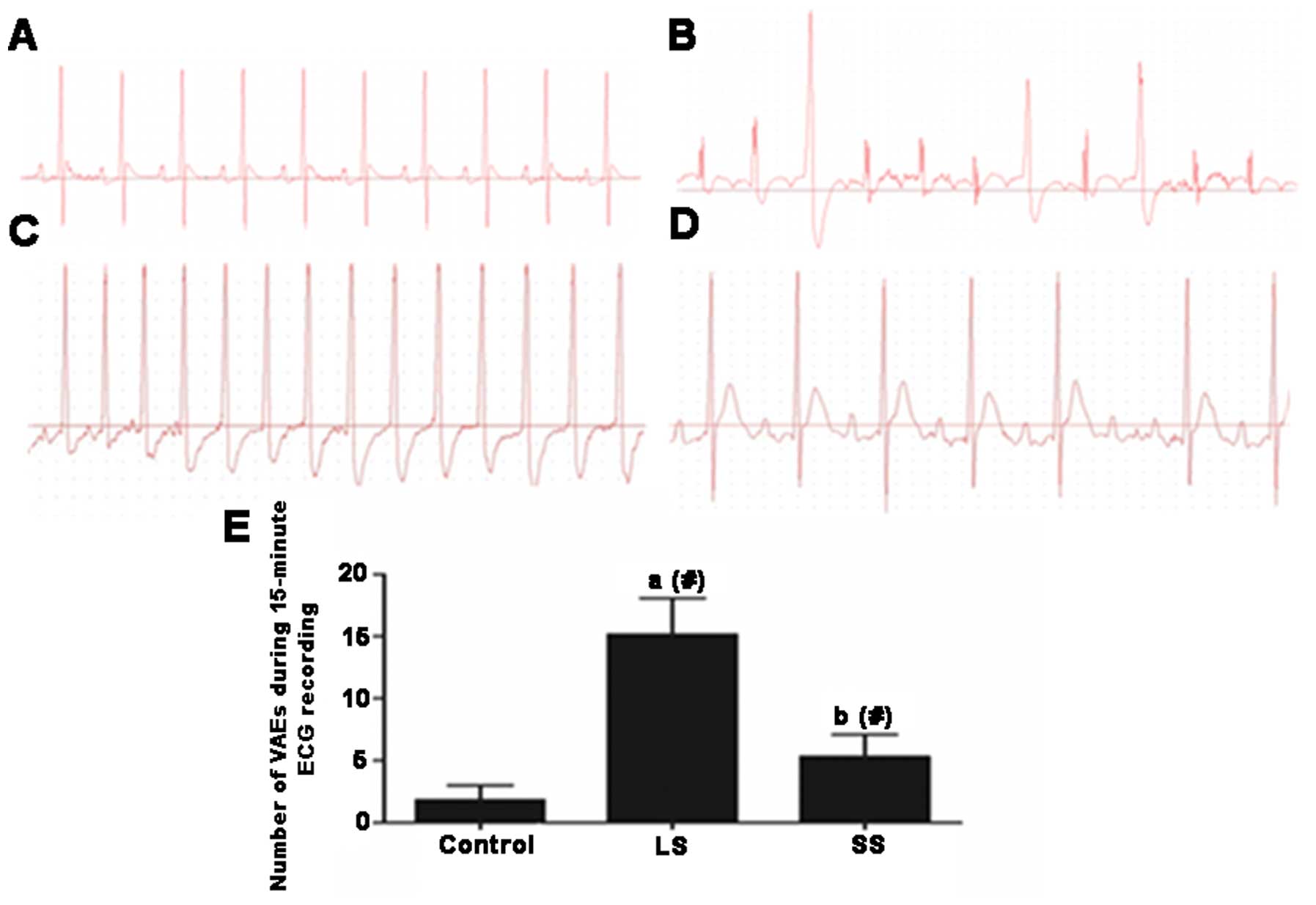
ECG features
ECG of rats in the control group was normal
(Fig. 4A). However, ECG of rats
in the LS group showed significant differences. In the LS group, a
few rats exhibited normal ECG, but the majority had abnormal ECG,
which manifested as premature ventricular contraction and
paroxysmal supraventricular tachycardia (Fig. 4B and C). The majority of ECG in
the SS group were normal, but there remained a few abnormal ECG
(Fig. 4D). VAEs in the LS group
were higher than those of the control group (P<0.05), but
following Se supplementation the VAEs significantly decreased
compared to the LS group (P<0.05) (Fig. 4E).
Echocardiographic detection
The echocardiographic parameters, including LVEDD,
LVESD, LVEF and LVFS, were detected and calculated in the present
study as shown in Table II. An
overall increase of LVEDD and LVESD accompanied by an overall
decrease of LVEF and LVFS were found in the LS and SS groups.
However, there was a significant increase of LVEDD and LVESD and a
significant decrease of LVEF and LVFS in the LS group compared to
the SS group. A significant recovery of cardiac function marked by
echocardiographic parameters was found in the SS group compared to
the LS group, which was evidenced by restoration of LVEDD, LVESD,
LVEF and LVFS following Se supplementation.
 | Table IIEchocardiographic parameters of the
rats in the different groups. |
Table II
Echocardiographic parameters of the
rats in the different groups.
| Group | No. | LVEDD, mm | LVESD, mm | LVEF, % | LVFS, % |
|---|
| Control | 10 | 4.14±0.38 | 1.13±0.77 | 85.60±11.02 | 75.64±8.30 |
| LS | 10 |
5.29±0.26a* |
2.77±0.35a,c |
68.93±10.92a,c |
50.44±5.73a,d |
| SS | 10 |
4.92±0.79b* |
2.05±0.23b,c |
79.20±9.41b,c |
97.73±6.28b,d |
Hemodynamic detection
The hemodynamic parameters, including LVEDP, LVSP,
LVdp/dtmax and LVdp/dtmin, were detected and
calculated in the study as shown in Table III. An overall increase of LVEDP
and LVSP accompanied by an overall decrease of
LVdp/dtmax and LVdp/dtmin were found in the
LS and SS groups. However, there was a significant increase of
LVEDP and LVSP and decrease of LVdp/dtmax and
LVdp/dtmin in the LS group compared to the SS group. A
significant recovery of cardiac function marked by hemodynamic
parameters was found in the SS group compared to the LS group,
which was evidenced by restoration of LVEDP, LVSP,
LVdp/dtmax and LVdp/dtmin following Se
supplementation.
 | Table IIIHemodynamic parameters of the rats in
the different groups. |
Table III
Hemodynamic parameters of the rats in
the different groups.
| Group | No. | LVEDP, mmHg | LVSP, mmHg |
LVdp/dtmax, mmHg/sec |
LVdp/dtmin, mmHg/sec |
|---|
| Control | 7 | 47.62±6.41 | 185.32±13.28 | 3794.55±127.47 | 2887.65±154.13 |
| LS | 8 |
67.81±5.50a* |
157.69±14.24a,c |
2640.31±144.20a,c |
2216.28±138.69a,c |
| SS | 8 |
53.29±5.13b** |
162.30±14.68b,c |
2948.19±152.35b,c |
2677.00±142.98b,c |
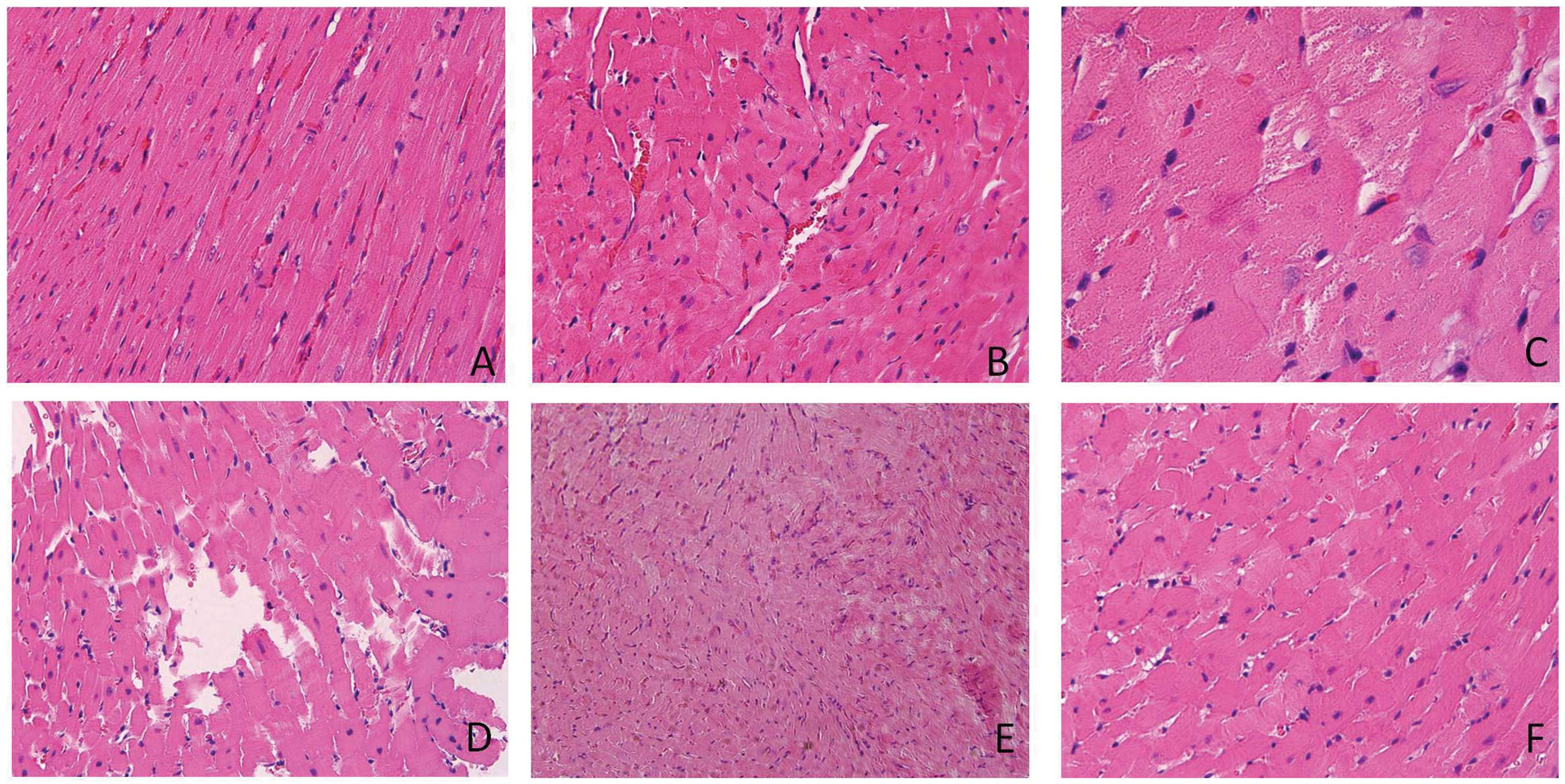
Histological characterization of the
heart
HE staining showed that the structure of myocardial
tissue in the control group was clear, including well-connected
intercalated discs and normally arranged filaments (Fig. 5A). In the LS group, the muscle
fibers of the rats were swelling and disorganized (Fig. 5B). Myocardial necrosis, nuclear
condensation and partially intermittent muscle fibers were observed
in the LS group (Fig. 5C and D).
In addition, the small myocardial necrosis focus accompanied by
inflammatory infiltration can be observed in certain areas
(Fig. 5E). Following Se
supplementation, the myocardial fibers were mildly swelling
compared to the control group, but their arrangements were orderly
(Fig. 5F).
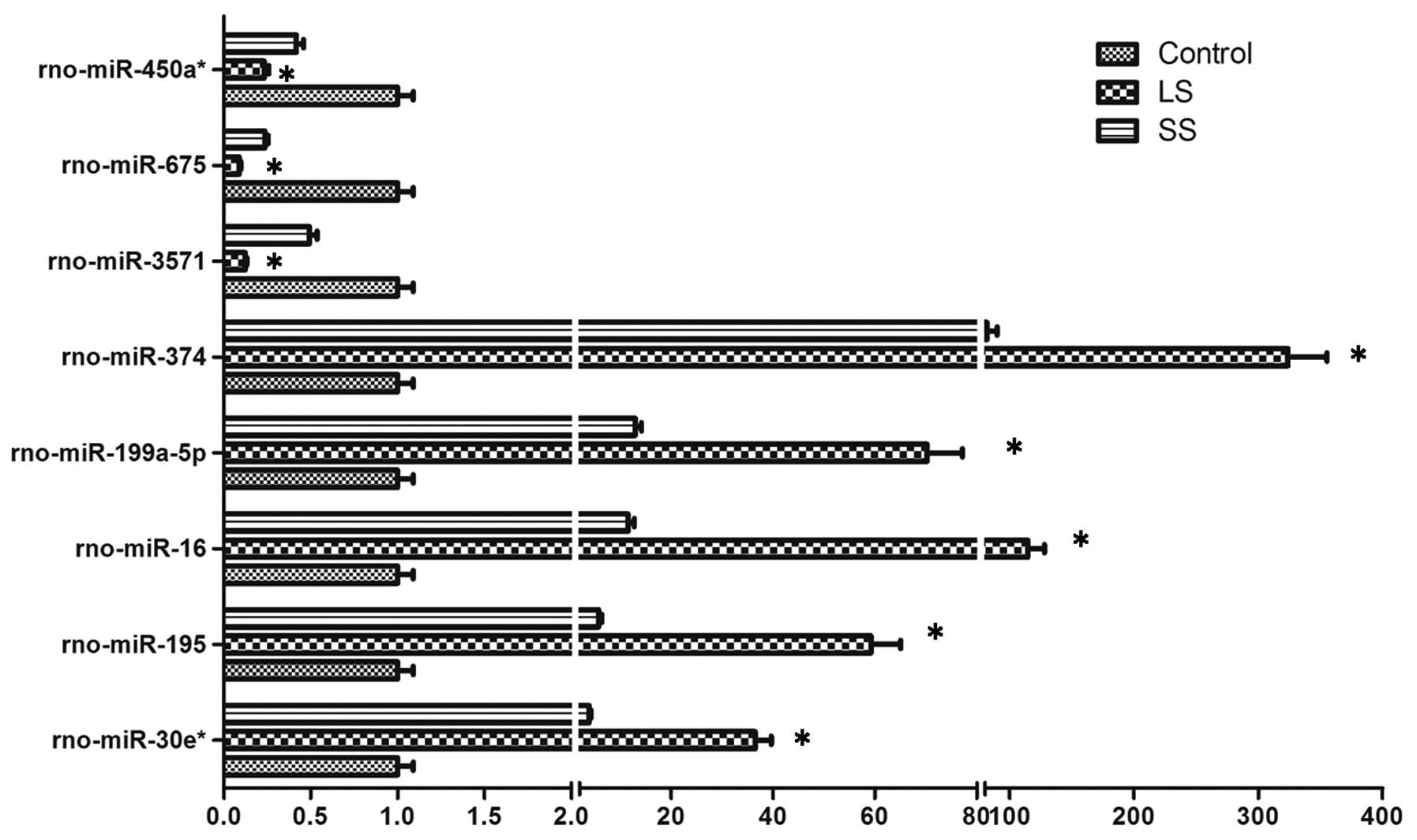
miRNA expression profiles
Using the miRCURY LNA Array platform, the miRNAs
expression profiles were assessed in Se-deficient rats. The
expression profiles of 116 miRNAs were regulated between the
control, LS and SS groups, and the samples were separated into
biologically interpretable groups. Among these, 5 miRNAs were
identified to be upregulated >5-fold in the LS group compared to
the SS group, whereas 3 miRNAs were less than the threshold level
(3-fold) set during the progression of Se deficiency, but following
selenium supplementation these miRNAs were >1.5-fold compared to
Se deficiency. These 8 miRNAs were validated to be significantly
different (P<0.05). As shown in Fig. 6, the levels of miR-374,
miR-16, miR-199a-5p, miR-195 and
miR-30e* were upregulated in rats with selenium
deficiency, while miR-3571, miR-675 and
miR-450a* showed an opposite expression pattern,
in agreement with the results of microarray hybridization.
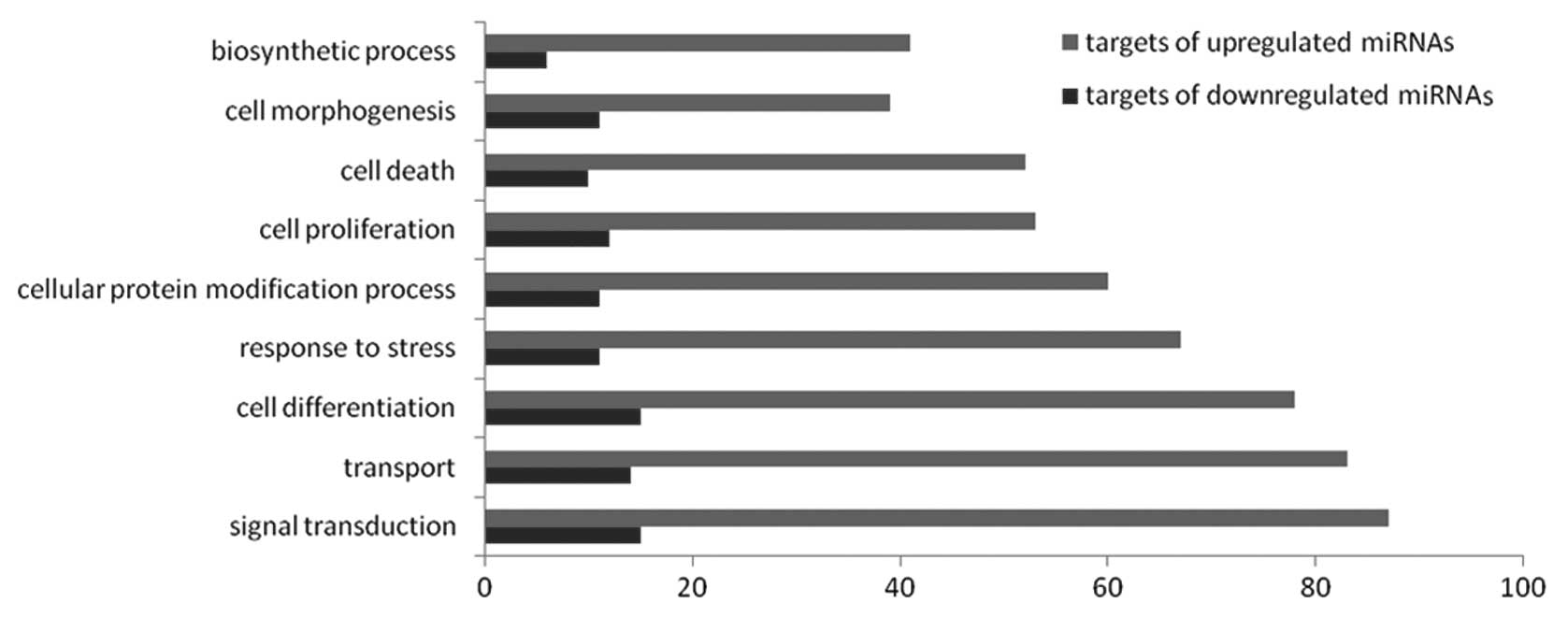
Microarray-based GO analysis
According to the threshold of GOs that were
significantly regulated by miRNAs, the P-value and FDR was
<0.001 and <0.05, respectively. The high-enrichment GOs
targeted by upregulated miRNAs included signal transduction,
transport, cell differentiation and response to stress. By
contrast, the significant GOs corresponding to downregulated miRNAs
appeared to include signal transduction, cell differentiation,
transport and cell proliferation. Among these, the
maximum-enriched-GO associated with signal transduction, together
with the numerous miRNAs that interacted with signal
transduction-related genes, suggested them to have an important
role in the activation of selenium deficiency (Fig. 7).
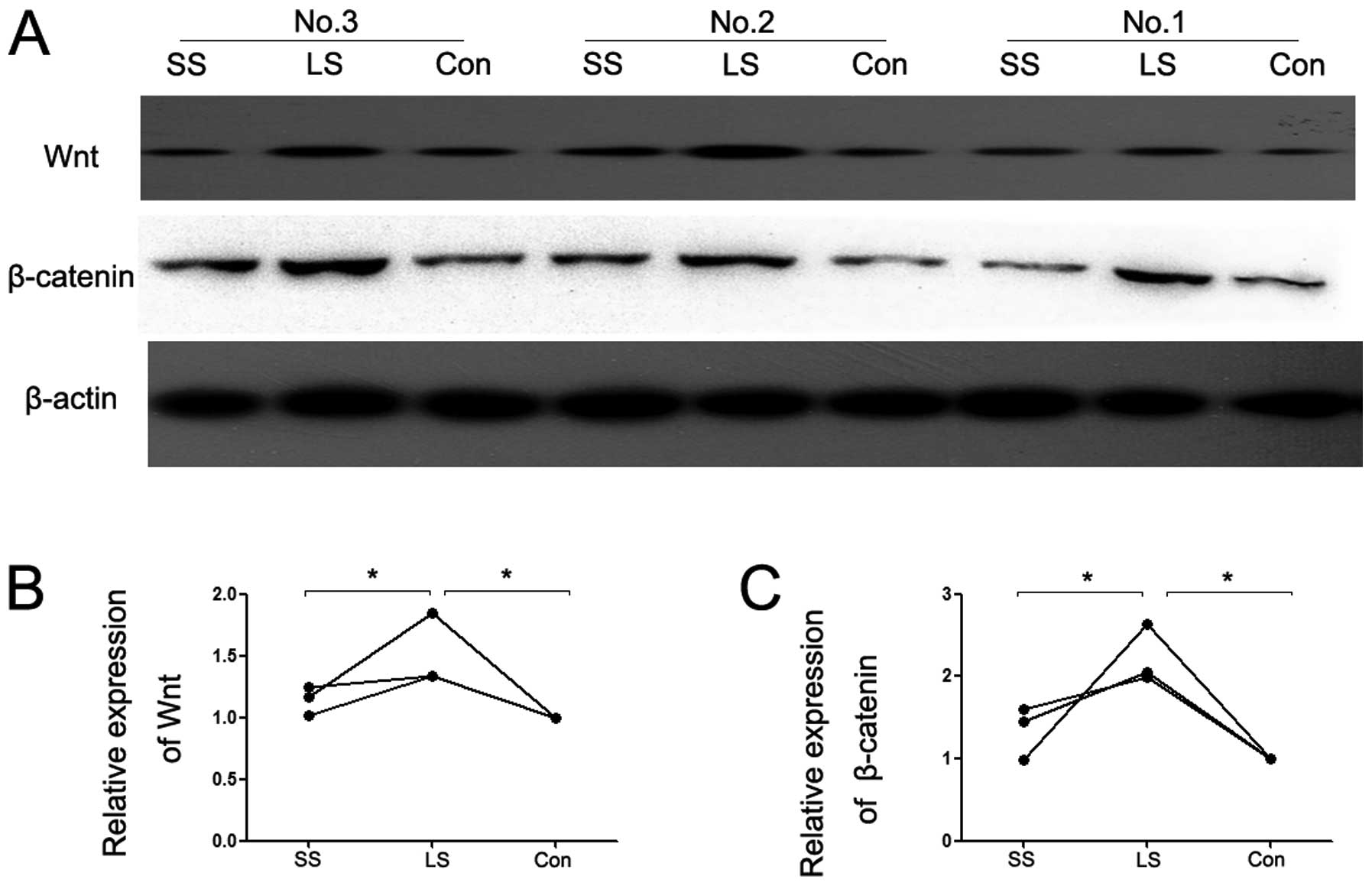
Expression of Wnt/β-catenin
Western blotting was employed to detect the changes
of Wnt and β-catenin expression in the present study. As shown in
Fig. 8, the protein expression of
Wnt and β-catenin in cardiac tissue were highest in the LS group
compared to the control group. Increasing expression of Wnt and
β-catenin in the LS group was decreased by Se supplementation.
However, it appeared that Se supplementation reversed the
increasing expression of Wnt and β-catenin in the LS group.
Discussion
In China, KD is the most detrimental and widely
distributed endemic cardiomyopathy. In an observational
epidemiological study and from population-based intervention trial
results, it was concluded that Se deficiency is the critical
etiological factor for KD (28–31). An early population-based clinical
study showed that a low selenium level was associated with
cardiovascular outcomes (32). In
a recent prospective cohort study of patients with coronary artery
disease, the individual baseline serum selenium concentration was
inversely associated with mortality of acute coronary syndrome. Low
serum selenium was considered independent of classic risk factors,
including biomarkers of necrosis and LVEF, of acute coronary
syndrome (33). In an animal
study, Se deficiency was associated with evident worsening of
cardiac functional parameters and cardiac hypertrophy, associated
with a marked decrease of GPx activity in cardiac tissue (34). In the present study, the
deterioration of cardiac function in rats receiving a low-Se diet
by plasma BNP and echocardiographic determinations was determined.
The results demonstrated that the serum BNP level, LVEDD and LVESD
increased, and LVEF and LVFS decreased in low-Se-diet treated rats.
These results were in accordance with those in the above
studies.
Increasing evidence has confirmed miRNA as one of
the most significant and critical factors controlling gene
expression. Thus far, miRNAs have been noted to play an extremely
important role in cardiac development, homeostasis and pathobiology
(35,36). Although miRNAs function as
endogenous intracellular regulators of mRNA translation, they can
be detected in circulating blood or its components in a markedly
stable form and, therefore, should be considered as disease
biomarkers, for instance, in cardiovascular diseases (37,38). A series of significant advances
have been found in the research of cardiac hypertrophy (39), congenital heart disease (40), heart failure (41,42), arrhythmia (43), arterial atherosclerosis (44) and hypertension (45), and miRNAs have become a current
interest for international cardiovascular research. However, no
research has been reported regarding miRNAs and KD thus far.
Therefore, the miRNAs expression profile in rats with Se deficiency
was evaluated. In comparison to those of Se supplementation and
normal Se supply, the microarray revealed a set of differentially
expressed miRNAs, 5 significantly upregulated and 3 significantly
downregulated, in the rats with Se deficiency. Evaluation of
miR-374, miR-16, miR-199a-5p, miR-195,
miR-30e*, miR-3571, miR-675 and
miR-450a* further validated the reliability of
micoarray hybridization.
Molecular mechanisms underlying cardiac dysfunction
caused by Se deficiency should be intensively studied. Wnt
signaling is required for various features of cardiac and vascular
development, such as myocardial specification, cardiac
morphogenesis and cardiac valve formation, as well as endothelial
and vascular smooth muscle cell proliferation (46). Defective Wnt signaling can lead to
various cardiac and vascular abnormalities. Wnt signaling activity
is quite low under normal conditions in the adult heart and blood
vessels. However, during the pathological cardiac remodeling
induced by pressure overload, in injured arteries and following
myocardial infarction, this pathway is reactivated. Genetically
modified animal models have shown that inhibition of Wnt signaling
results in increased angiogenesis, improved infarct healing and an
attenuated hypertrophic response of the heart, indicating that
pharmacological inhibition of Wnt signaling provided a novel
therapeutic strategy to prevent excessive cardiac and vascular
remodeling (47). The β-catenin
gene is a critical component of the Wnt signaling, where it
transmits extracellular Wnt signals to the nucleus. The level of
β-catenin in the cytoplasm and nucleus is tightly regulated. In the
absence of the Wnt signal, cytoplasmic β-catenin is maintained at a
low level through ubiquitin-mediated proteasomal degradation, which
is controlled by the destruction complex consisting of glycogen
synthase kinase 3b/adenomatous polyposis coli/Axin. Upon receiving
Wnt signals, cell degradation of β-catenin by the destruction
complex and ubiquitin-proteasome complex is inhibited, resulting in
accumulation of β-catenin in the cytoplasm and translocation into
the nucleus. β-catenin heterodimerizes with members of the TCF/LEF
transcription factors to activate transcription of the target genes
in the nucleus. The nuclear translocation of β-catenin is involved
in epithelial-mesenchymal transformation, which is an initial and
critical step in fibrosing processes, including pulmonary fibrosis
and myocardial infarction repair (48,49). Zelarayán et al (50) found that β-catenin depletion was
beneficial in postinfarction LV remodeling partly through enhanced
differentiation of α-MHCpos cardiac resident progenitor
cells. Downregulation of β-catenin improved cardiac function
following experimental infarction. The study indicated that
endogenous cardiac regeneration contributes to LV remodeling
following chronic ischemia through differentiation of resident
precursor cells, amplified by β-catenin downregulation (50). In the present study, selenium
deficiency was demonstrated to result in cardiac dysfunction by
increasing the expression of Wnt and β-catenin, however, following
selenium supplementation cardiac function partially recovery and
Wnt and β-catenin expression decreased compared to the LS group.
Therefore, the Wnt/β-catenin signaling pathway may be involved in
the process of cardiac function deterioration.
In conclusion, Se deficiency is associated with
eight miRNAs: miR-374, miR-16, miR-199a-5p,
miR-195, miR-30e*, miR-3571,
miR-675 and miR-450a*. Their wide range of
actions may regulate cardiac function. The expression of
miR-374 was the highest, which may be significant in rats
with selenium deficiency. The possible mechanism of cardiac
dysfunction was associated with the Wnt/β-catenin signaling
pathway. These findings not only highlight the incidence of cardiac
dysfunction caused by Se deficiency, but also facilitate access to
a novel therapeutic strategy from a molecular level against cardiac
dysfunction and offer insight into its progression.
Acknowledgements
The present study was supported by the Natural
Science Foundation of China (NSFC) (grant no. 30972557). The
company Genminix provided technical assistance.
References
|
1
|
Ge K, Xue A, Bai J and Wang S: Keshan
disease-an endemic cardiomyopathy in China. Virchows Arch A Pathol
Anat Histopathol. 401:1–15. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Oster O, Prellwitz W, Kasper W and
Meinertz T: Congestive cardiomyopathy and the selenium content of
serum. Clin Chim Acta. 128:125–132. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
de Lorgeril M, Salen P, Accominotti M,
Cadau M, Steghens JP, Boucher F and de Leiris J: Dietary and blood
antioxidants in patients with chronic heart failure. Insights into
the potential importance of selenium in heart failure. Eur J Heart
Fail. 3:661–669. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Arroyo M, Laguardia SP, Bhattacharya SP,
Nelson MD, Johnson PL, Carbone LD, et al: Micronutrients in
African-Americans with decompensated and compensated heart failure.
Transl Res. 148:301–308. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Witte KK, Nikitin NP, Parker AC, von
Haehling S, Volk HD, Anker SD, et al: The effect of micronutrient
supplementation on quality-of-life and left ventricular function in
elderly patients with chronic heart failure. Eur Heart J.
26:2238–2244. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Djuranovic S, Nahvi A and Green R: A
parsimonious model for gene regulation by miRNAs. Science.
331:550–553. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Viader A, Chang LW, Fahrner T, Nagarajan R
and Milbrandt J: MicroRNAs modulate Schwann cell response to nerve
injury by reinforcing transcriptional silencing of
dedifferentiation-related genes. J Neurosci. 31:17358–17369. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Roldo C, Missiaglia E, Hagan JP, Falconi
M, Capelli P, Bersani S, et al: MicroRNA expression abnormalities
in pancreatic endocrine and acinar tumors are associated with
distinctive pathologic features and clinical behavior. J Clin
Oncol. 24:4677–4684. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhao JJ, Hua YJ, Sun DG, Meng XX, Xiao HS
and Ma X: Genome-wide microRNA profiling in human fetal nervous
tissues by oligonucleotide microarray. Childs Nerv Syst.
22:1419–1425. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bhanot P, Brink M, Samos CH, Hsieh JC,
Wang Y, Macke JP, et al: A new member of the frizzled family from
Drosophila functions as a Wingless receptor. Nature. 382:225–230.
1996. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tamai K, Semenov M, Kato Y, Spokony R, Liu
C, Katsuyama Y, et al: LDL-receptor-related proteins in Wnt signal
transduction. Nature. 407:530–535. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kishida S, Yamamoto H, Ikeda S, Kishida M,
Sakamoto I, Koyama S and Kikuchi A: Axin, a negative regulator of
the wnt signaling pathway, directly interacts with adenomatous
polyposis coli and regulates the stabilization of beta-catenin. J
Biol Chem. 273:10823–10826. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sakanaka C, Weiss JB and Williams LT:
Bridging of beta-catenin and glycogen synthase kinase-3beta by axin
and inhibition of beta-catenin-mediated transcription. Proc Natl
Acad Sci USA. 95:3020–3023. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Behrens J, Jerchow BA, Wurtele M, Grimm J,
Asbrand C, Wirtz R, et al: Functional interaction of an axin
homolog, conductin, with beta-catenin, APC, and GSK3beta. Science.
280:596–599. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kawano Y and Kypta R: Secreted antagonists
of the Wnt signalling pathway. J Cell Sci. 116:2627–2634. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cadigan KM: Wnt-beta-catenin signaling.
Curr Biol. 18:R943–R947. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rao TP and Kuhl M: An updated overview on
Wnt signaling pathways: a prelude for more. Circ Res.
106:1798–1806. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Klaus A and Birchmeier W: Wnt signalling
and its impact on development and cancer. Nat Rev Cancer.
8:387–398. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Katoh M and Katoh M: Wnt signaling pathway
and stem cell signaling network. Clin Cancer Res. 13:4042–4045.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dursun N, Taskin E, Yerer AM and Sahin L:
Selenium-mediated cardioprotection against adriamycin-induced
mitochondrial damage. Drug Chem Toxicol. 34:199–207. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gu L, Pandey V, Geenen DL, Chowdhury SA
and Piano MR: Cigarette smoke-induced left ventricular remodelling
is associated with activation of mitogen-activated protein kinases.
Eur J Heart Fail. 10:1057–1064. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ma J, Qian J, Ge J, Zeng X, Sun A, Chang
S, et al: Changes in left ventricular ejection fraction and
coronary flow reserve after coronary microembolization. Arch Med
Sci. 8:63–69. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jiang K, Shui Q, Xia Z and Yu Z: Changes
in the gene and protein expression of K (ATP) channel subunits in
the hippocampus of rats subjected to picrotoxin-induced kindling.
Brain Res Mol Brain Res. 128:83–89. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang L, Yang M, Marks P, White LM, Hurtig
M, Mi QS, Divine G and Gibson G: Serum non-coding RNAs as
biomarkers for osteoarthritis progression after ACL injury.
Osteoarthritis Cartilage. 20:1631–1637. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guo CJ, Pan Q, Li DG, Sun H and Liu BW:
miR-15b and miR-16 are implicated in activation of the rat hepatic
stellate cell: An essential role for apoptosis. J Hepatol.
50:766–778. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guan H, Song L, Cai J, Huang Y, Wu J, Yuan
J, et al: Sphingosine kinase 1 regulates the Akt/FOXO3a/Bim pathway
and contributes to apoptosis resistance in glioma cells. PLoS One.
6:e199462011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen J: An original discovery: selenium
deficiency and Keshan disease (an endemic heart disease). Asia
Pacific J Clin Nutr. 21:320–326. 2012.
|
|
29
|
Xu GL: The effectiveness of sodium
Selenite on prevention of acute attacks ofKeshan diseases. Chin Med
J. 92:471–476. 1979.
|
|
30
|
Chen X, Yang G, Chen J, Chen X, Wen Z and
Ge K: Studies on the relationships of selenium and Keshan disease.
Biol Trace Elem Res. 2:91–107. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li Q, Liu M, Hou J, Jiang C, Li S and Wang
T: The prevalence of Keshan disease in China. Int J Cardiol.
168:1121–1126. 2013. View Article : Google Scholar
|
|
32
|
Salonen JT, Alfthan G, Huttunen JK,
Pikkarainen J and Puska P: Association between cardiovascular death
and myocardial infarction and serum selenium in a matched-pair
longitudinal study. Lancet. 2:175–179. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lubos E, Sinning CR, Schnabel RB, Wild PS,
Zeller T, Rupprecht HJ, et al: Serum selenium and prognosis in
cardiovascular disease: results from the AtheroGene study.
Atherosclerosis. 209:271–277. 2010. View Article : Google Scholar :
|
|
34
|
Lymbury RS, Marino MJ and Perkins AV:
Effect of dietary selenium on the progression of heart failure in
the ageing spontaneously hypertensive rat. Mol Nutr Food Res.
54:1436–1444. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Latronico MV and Condorelli G: MicroRNAs
and cardiac pathology. Nat Rev Cardiol. 6:419–429. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Porrello ER: microRNAs in cardiac
development and regeneration. Clin Sci (Lond). 125:151–166. 2013.
View Article : Google Scholar
|
|
37
|
Van Aelst LN and Heymans S: MicroRNAs as
biomarkers for ischemic heart disease. J Cardiovasc Transl Res.
6:458–470. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Nabialek E, Wanha W, Kula D, Jadczyk T,
Krajewska M, Kowalowka A, et al: Circulating microRNAs (miR-423-5p,
miR-208a and miR-1) in acute myocardial infarction and stable
coronary heart disease. Minerva Cardioangiol. 61:627–637.
2013.PubMed/NCBI
|
|
39
|
Sayed D, Hong C, Chen IY, Lypowy J and
Abdellatif M: MicroRNAs play an essential role in the development
of cardiac hypertrophy. Circ Res. 100:416–424. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xing HJ, Li YJ, Ma QM, Wang AM, Wang JL,
Sun M, et al: Identification of microRNAs present in congenital
heart disease associated copy number variants. Eur Rev Med
Pharmacol Sci. 17:2114–2120. 2013.PubMed/NCBI
|
|
41
|
Melman YF, Shah R and Das S: MicroRNAs in
heart failure: is the picture becoming less miRky? Circ Heart Fail.
7:203–214. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Oliveira-Carvalho V, Da SM, Guimaraes GV,
Bacal F and Bocchi EA: MicroRNAs: new players in heart failure. Mol
Biol Rep. 40:2663–2670. 2013. View Article : Google Scholar
|
|
43
|
Kim GH: MicroRNA regulation of cardiac
conduction and arrhythmias. Transl Res. 161:381–392. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Madrigal-Matute J, Rotllan N, Aranda JF
and Fernández-Hernando C: MicroRNAs and atherosclerosis. Curr
Atheroscler Rep. 15:3222013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kontaraki JE, Marketou ME, Zacharis EA,
Parthenakis FI and Vardas PE: Differential expression of vascular
smooth muscle-modulating microRNAs in human peripheral blood
mononuclear cells: novel targets in essential hypertension. J Hum
Hypertens. 28:5410–516. 2014. View Article : Google Scholar
|
|
46
|
Gessert S and Kühl M: The multiple phases
and faces of wnt signaling during cardiac differentiation and
development. Circ Res. 107:186–199. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
van de Schans VA, Smits JF and
Blankesteijn WM: The Wnt/frizzled pathway in cardiovascular
development and disease: friend or foe? Eur J Pharmacol.
585:338–345. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Blankesteijn WM, van Gijn ME,
Essers-Janssen YP, Daemen MJ and Smits JF: Beta-catenin, an inducer
of uncontrolled cell proliferation and migration in malignancies,
is localized in the cytoplasm of vascular endothelium during
neovascularization after myocardial infarction. Am J Pathol.
157:877–883. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chilosi M, Poletti V, Zamò A, Lestani M,
Montagna L, Piccoli P, et al: Aberrant Wnt/beta-catenin pathway
activation in idiopathic pulmonary fibrosis. Am J Pathol.
162:1495–1502. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zelarayán LC, Noack C, Sekkali B, Kmecova
J, Gehrke C, Renger A, et al: Beta-Catenin downregulation
attenuates ischemic cardiac remodeling through enhanced resident
precursor cell differentiation. Proc Natl Acad Sci USA.
105:19762–19767. 2008. View Article : Google Scholar : PubMed/NCBI
|