Introduction
Diabetic neuropathy (DN), a frequent chronic
complication of diabetes (type I and II) (in approximately 60 to
75% of cases) (1), is a major
cause of morbidity and mortality, potentially affecting the distal
sensory, motor and autonomic nerves (2). DN typically manifests as autonomic
dysfunction with postural hypotension, fainting, diarrhea and
peripheral neuropathy with the loss of the sensation of pain and
temperature followed by a pattern suggestive of a length-dependent
degeneration of nerve fibers (3).
The accurate and timely detection, characterization and
quantification of DN are critical for the identification of
patients at risk, the estimation of expected deterioration, the
monitoring of progression, the assessment of novel therapies, and,
ultimately, for the reduction of the incidence and cost of
treatment for this disease. Current treatments for DN provide
symptomatic relief rather than ameliorating disease progression
(4). A previous clinical study on
type II diabetes demonstrated that the insulin sensitizer
rosiglitazone (Rosi) (Avandia), a synthetic agonist of peroxisome
proliferator-activated receptor-γ (PPAR-γ) that is used to improve
insulin resistance, is a potential drug for the treatment of DN
(5). Another study however,
demonstrated the undesirable effects of Rosi on the cardiovascular
system. These findings suggested that treatment with Rosi may be
harmful and should be used with caution in cardiovascular patients
(6). Considering the fact that
the etiology and pathogenesis of DN are not entirely understood,
multicenter trials on the pathogenic mechanisms of DN are in
urgently required. Moreover, there is an urgent need for the
development of novel drugs with long-term effects on DN.
Streptozotocin (STZ), which selectively destroys
pancreatic β cells, rapidly induces diabetes in mammals in a model
of insulin-dependent diabetes (7). Gabapentin, pregabalin,
amitriptyline, mexiletine and morphine, but not diclofenac, inhibit
allodynia in a rodent model of STZ-induced DN, suggesting that the
STZ-induced model of DN is suitable for evaluating the clinical
potential of compounds for the treatment of painful DN (8).
In the present study, by microarray data analysis,
differentially expressed genes (DEGs) were screened between a
control (healthy) and a group of mice with STZ-induced diabetes, a
control and a group of mice treated with Rosi, as well as between a
group of mice with STZ-induced diabetes and mice with STZ-induced
diabetes treated with Rosi. Based on functional annotation
clustering analysis, processes significantly associated with this
disease were identified in order to further elucidate the
pathogenic mechanisms of this disese. Genes co-regulated by both
STZ and Rosi were identified and the protein structure of the
target genes was predicted. Proteins encoded by the identified
genes are potential targets in the treatment of DN.
Materials and methods
Data source
All the microarray data were downloaded from the
Gene Expression Omnibus database under the accession number
GSE11343 (9), which included data
on 4 control samples (GSM286159, GSM286160, GSM286163 and
GSM286165), 5 samples from mice with diabetes induced by
streptozotocin (STZ) (GSM286169, GSM286173, GSM286176, GSM286178
and GSM286430), 5 samples from normal (healthy) mice treated with
Rosi (GSM286431, GSM286432, GSM286433, GSM286434 and GSM286436) and
5 samples from mice with diabetes induced by STZ and treated with
Rosi (GSM286437, GSM286438, GSM286591, GSM286592 and GSM286593).
The data contained expression profiles of 45,101 probes in total.
The annotation platform was GPL1261 [Mouse430_2] Affymetrix Mouse
Genome 430 2.0 Array.
Identification of DEGs
Using an Affy pack with R programming language, 19
ChIP samples were pre-processed for statistical analysis. The
significant analysis of microarray (SAM) method (10) was applied in order to identify the
DEGs between the control and the group with STZ-induced diabetes,
the control and the Rosi-treated group, as well as between the
group with STZ-induced diabetes and the group with STZ-induced
diabetes treated with Rosi (STZ + Rosi group). DEGs with a fold
change ≥2 and Q-value <0.05 were selected.
Function analysis of DEGs
The screened DEGs may control and regulate the
development of DN. We wished to identify the DEGs affected by the
utilization of both STZ and Rosi, potential genes regulating the
development of DN, as well as the signaling pathways these genes
are involved in. Thus, Gene Ontology (GO) term and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway (11) analyses were performed using The
Database for Annotation, Visualization and Integrated Discovery
(DAVID) (12). For each GO term
(13), the P-value of function
clustering and the p-value following multiple detecting correction,
such as Benjamini correction or false discovery rate (FDR)
correction, were calculated in DAVID. Moreover, the regulatory
network of DEGs and the protein-protein interaction (PPI) network
of proteins encoded by the DEGs was searched using the BioCarta and
STRING online tools (14),
respectively.
Prediction of crystal structure of
proteins
We searched the Protein Data Bank (PDB) database and
universal protein resource (UniProt) database for the crystal
structure of proteins encoded by genes (15,16). If there was no information
available, the alignment of protein sequences was performed using
BLAST software, as previously described (17) and we searched for the structure of
homologous proteins in the PDB and UniProt databases.
Prediction of novel potential drug
targets
Rosi, belonging to the class of drugs known as
thiazolidinediones, targets the PPAR-γ protein (18), and as previously reported, its
application is limited by the side-effects of thiazolidinediones
(19). Therefore, we further
mined genes indirectly associated with PPAR-γ among potential
regulatory genes as novel drug targets in the treatment of DN. The
MOLCAD module in the SYBYL software (20) was used to predict the pockets of
proteins and AutoDock software was adopted for docking (21).
Results
Identification of DEGs
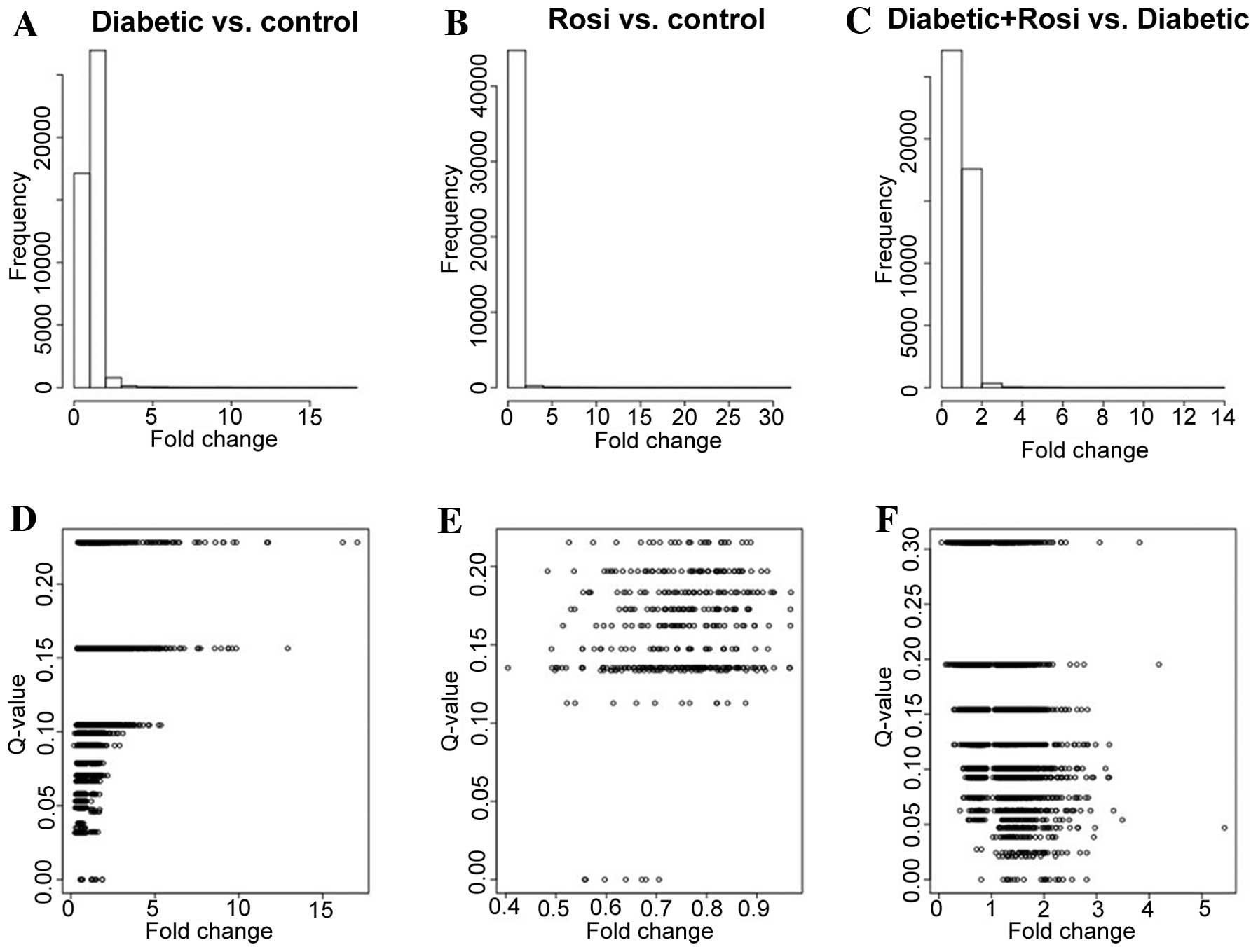
Compared with the control samples, there were 1,384
DEGs identified in the mice with STZ-induced diabetes with a fold
change ≥2, including 85 DEGs with a Q-value <0.05; 7 DEGs were
identified in the healthy mice treated with Rosi with a fold change
≥2, with no DEGs with a Q-value <0.05. There were 518 DEGs
identified between the mice in the STZ + Rosi group and the mice
with STZ-induced diabetes with a fold-change ≥2, including 41 DEGs
with a Q-value <0.05 (Fig.
1).
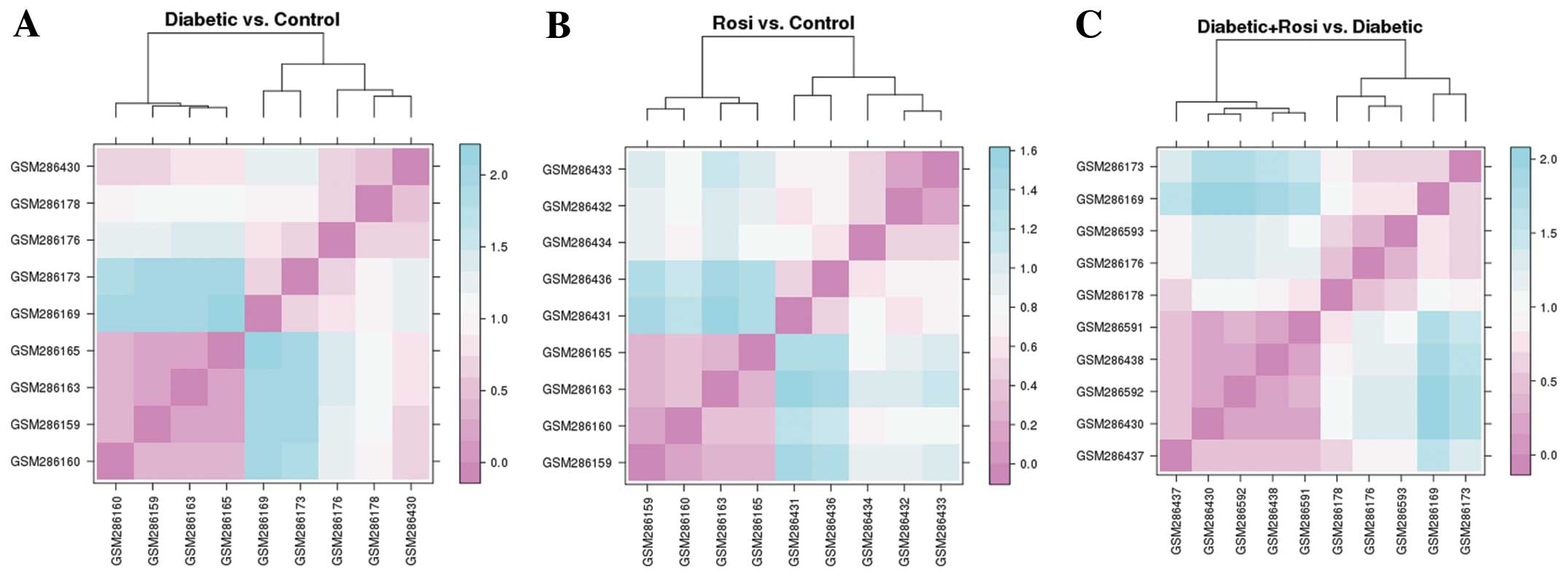
The samples from the mice with STZ-induced diabetes
and the healthy mice treated Rosi were effectively separated from
the control samples (Fig. 2A and
B), while the samples from the STZ + Rosi group could not be
separated from those of the mice with STZ-induced diabetes
completely (Fig. 2C). However,
the samples from the mice in the STZ + Rosi group clustered in the
first category (left), while the samples from the mice with
STZ-induced diabetes gathered in the second category (right).
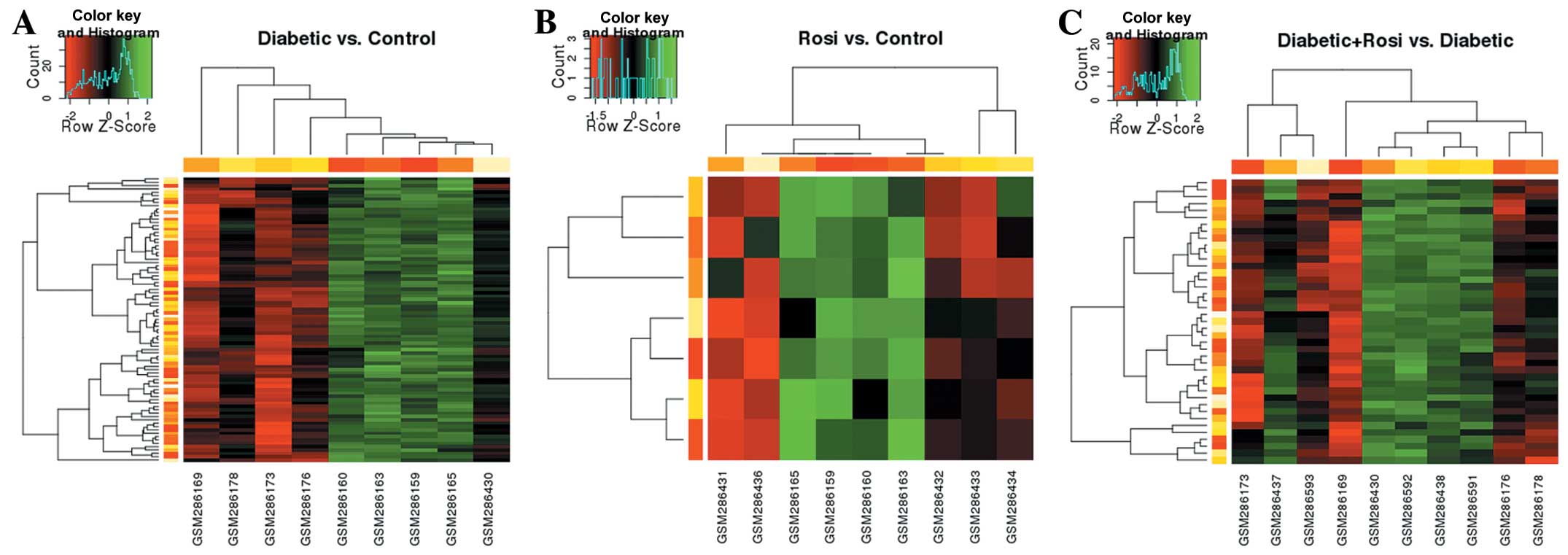
We further analyzed the DEGs in the samples
following drug administration. As shown in Fig. 3A and B, following the
administration of STZ or Rosi, the expression of all the DEGs was
generically downregulated compared with the control samples. As
shown in Fig. 3C, compared with
the samples from the mice with STZ-induced diabetes, the expression
of the DEGs in the samples from the STZ + Rosi group was mainly
upregulated.
Functional analysis of DEGs
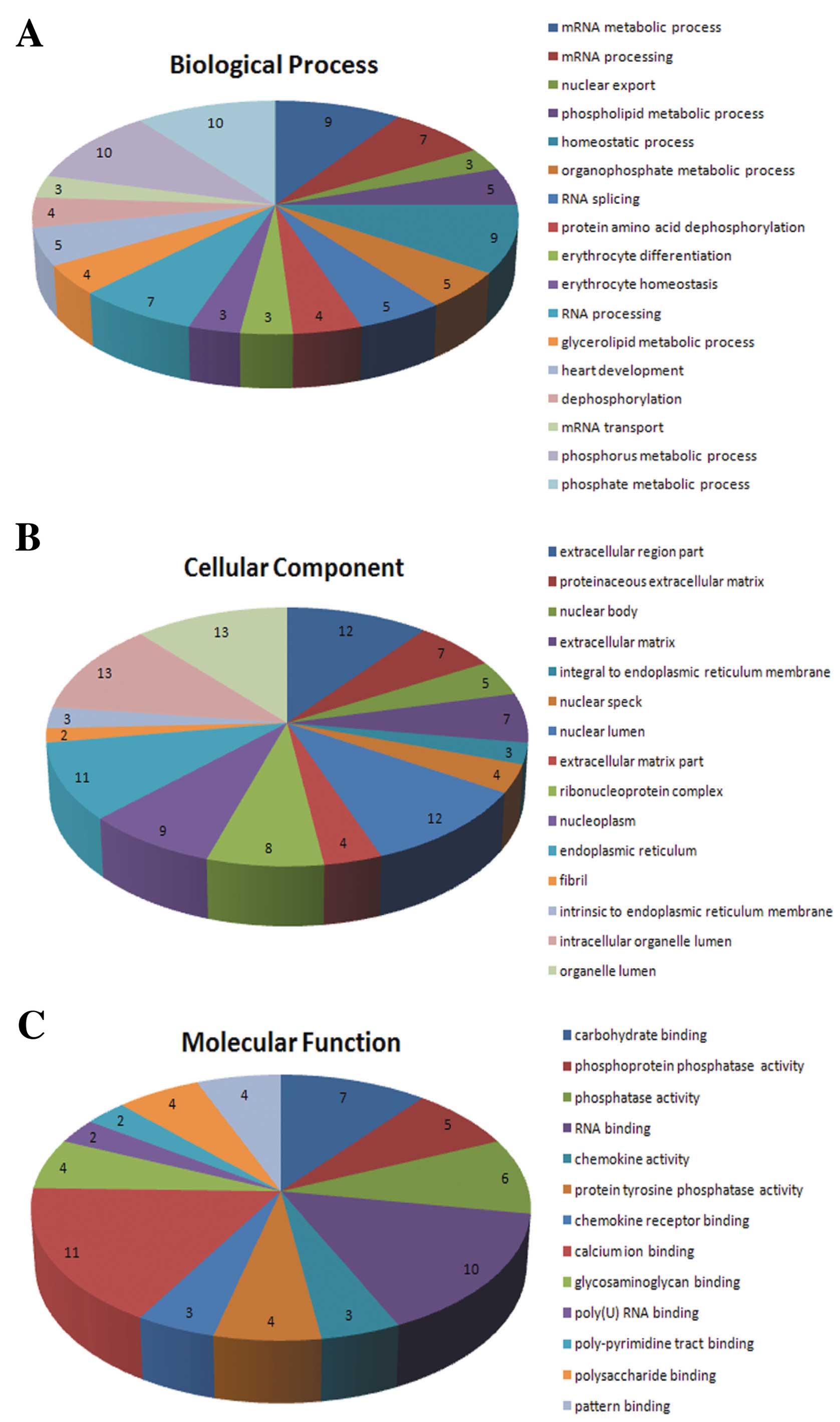
After combining all the DEGs (fold change ≥2,
Q-value <0.05), 123 DEGs were obtained. In the GO annotation
analysis (P<0.05) of these genes, there were 17 biological
process (BP) items, 15 cellular component (CC) items and 13
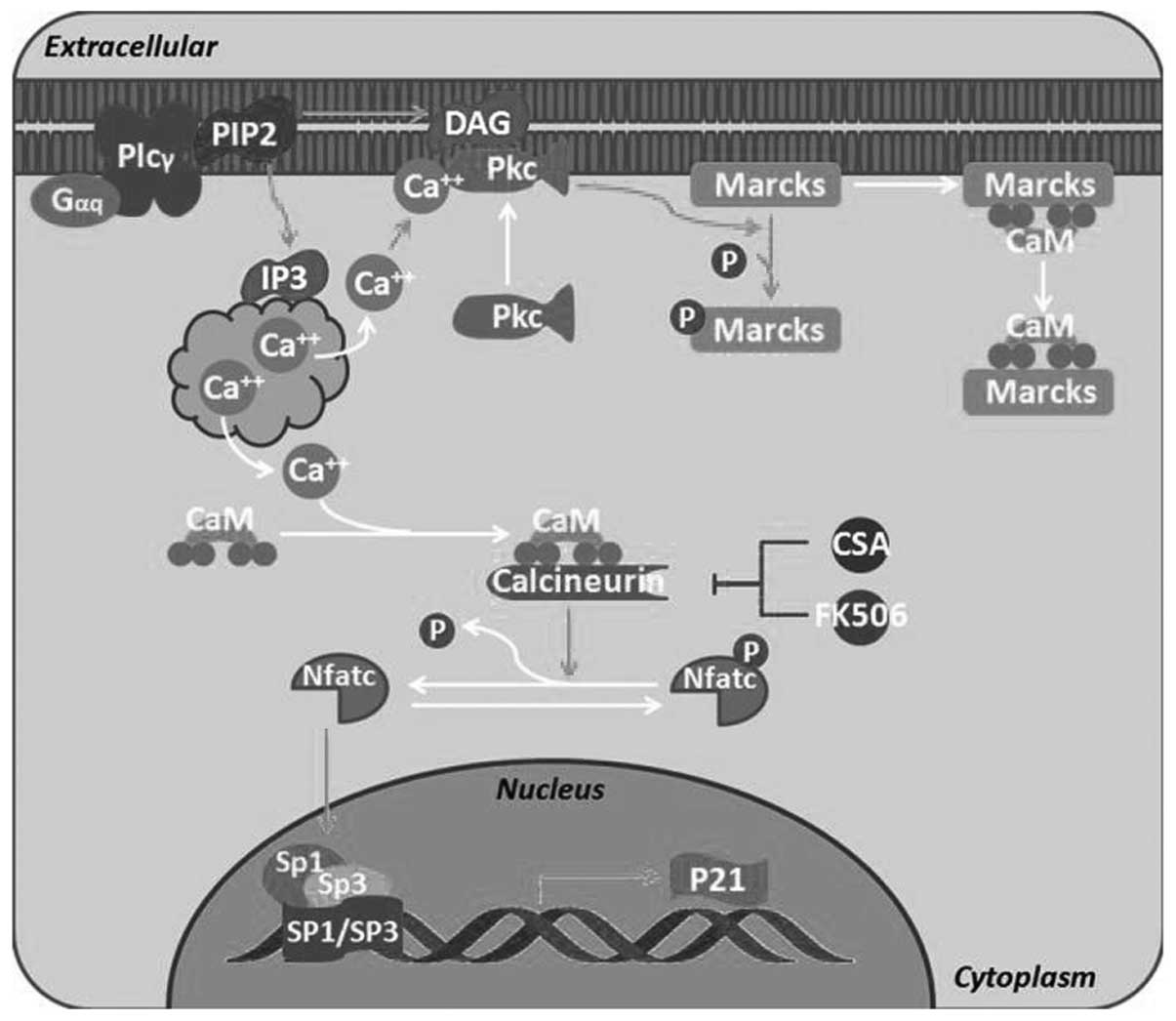
molecular function (MF) items identified (Fig. 4). The calmodulin nerve phosphatase
pathway (P=0.098) (Fig. 5) and
the chemokine signaling pathway (P=0.086) were identified as the
most significant signaling pathways in enrichment analysis.
Exploration of potential regulatory
genes
By analyzing the 85 DEGs identitied in the mice with
STZ-induced diabetes and the 41 DEGs identitied in the mice in the
STZ + Rosi group, 3 genes were found to be co-regulated by STZ and
Rosi; these were myristoylated alanine-rich protein kinase C
substrate (Marcks), GLI pathogenesis-related 2
(Glipr2) and centrosomal protein 170 kDa (Cep170). As
shown in Table I, these 3 genes
were all downregulated in the mice with STZ-induced diabetes and
upregulated in the mcie treated with Rosi. The recovery of gene
expression illustrated that treatment with Rosi resulted in the
upregulation of these genes. On the other hand, the downregulation
of these genes following the administration of STZ suggests that
the downregulation of these genes may contribute to the development
of DN.
 | Table IGenes co-regulated by STZ and
Rosi. |
Table I
Genes co-regulated by STZ and
Rosi.
| Gene name | STZ log2 (FC)a | Rosi log2
(FC)b | Functionc |
|---|
| Marcks | −1.238 | 1.154 | Marcks is the most
prominent cellular substrate for protein kinase C (PKC); binds to
calmodulin, actin and synapsin; inhibits F-actin cross-linking
activity |
| Glipr2 | −1.373 | 1.075 | Unannotated |
| Cep170 | −1.139 | 1.005 | Plays a role in
microtubule organization |
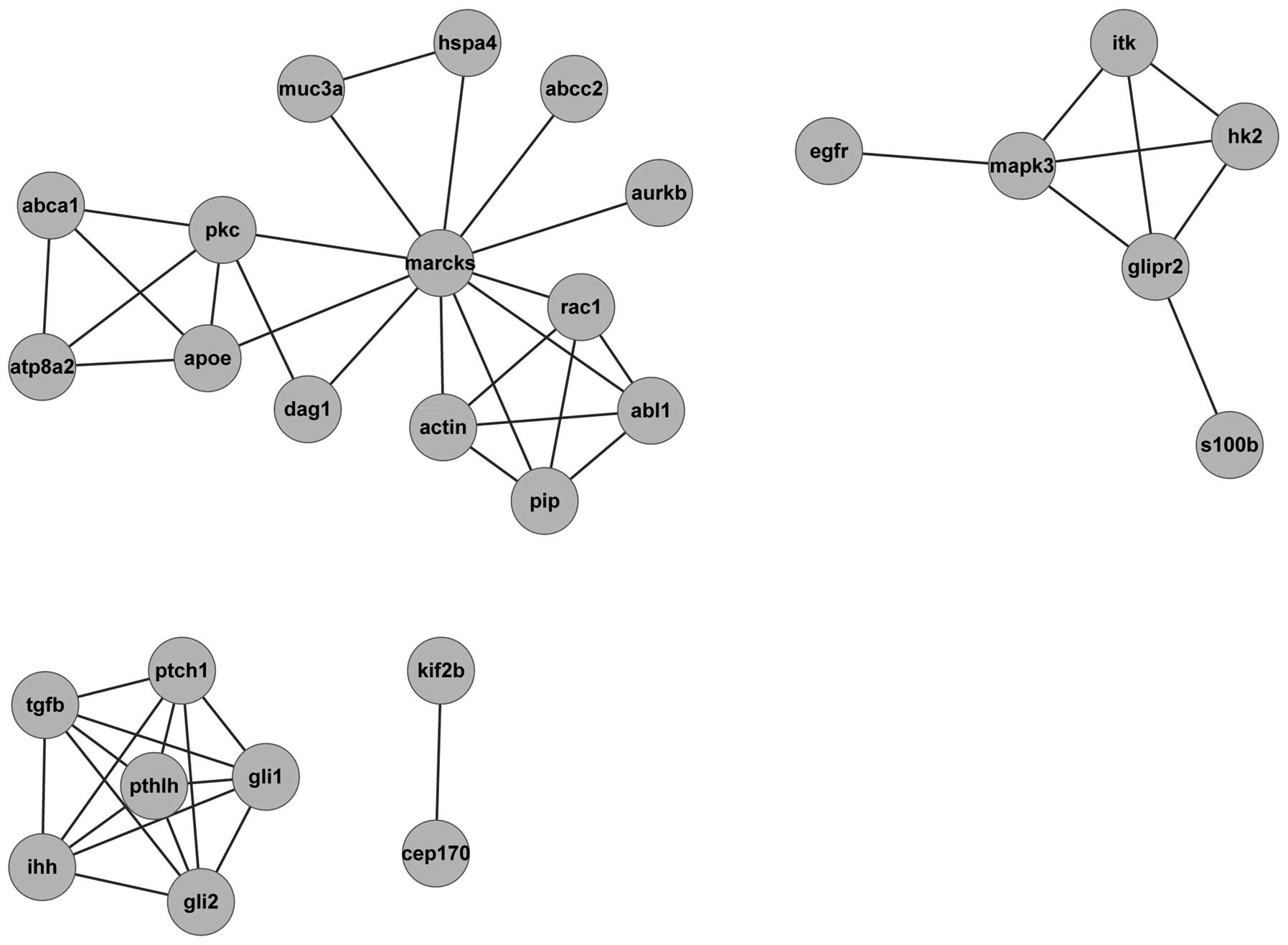
In addition, the PPI network of the proteins encoded
by these 3 genes was constructed using Cytoscape (Fig. 6). The protein regulating the
upstream and downstream domains may regulate the 3 genes
indirectly. For example, the actin protein connected with
Marcks was related to axon guidance, which may be involved
in the occurrence and development of DN.
Prediction of novel potential
targets
We searched the PDB crystal structure database in
order to verify whether the proteins encoded by Cep170,
Marcks and Glipr2 were analyzed. It was found that
Glipr2 was not analyzed in any type of crystal structure, while
Cep170 and Marcks were only analyzed in the human crystal
structure. The Cep170 and Marcks protein sequences from humans and
mice were found in the UniProt database. The identity of first
class sequences through BLAST comparison was 88 and 77%, with high
homology. Considering that it has been previously reported that
Rosi targets PPAR-γ (18), we
selected the human crystal structure of PPAR-γ for comparison
(identity was 62% compared with the mouse protein sequence).
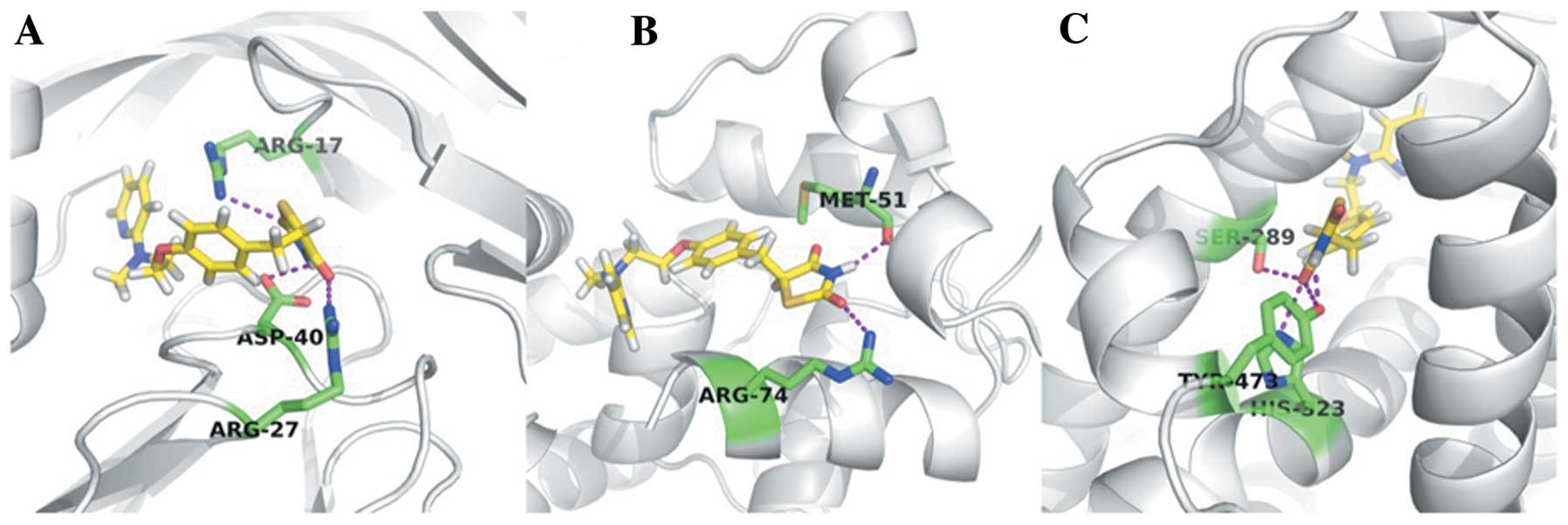
With the MOLCAD module using SYBYL software, the
binding pockets of the 3 proteins were predicted (Table II). Using AutoDock, STZ and Rosi
were docked to the crystal structures of Cep170 and Marcks,
respectively. While docking Rosi to the crystal structure of
PPAR-γ, a certain binding activity was found, as well as some key
hydrogen bond interactions (Table
II). The docking pattern of Rosi to the 3 proteins is shown in
Fig. 7, which illustrates that
Cep170 and Marcks may be the relevant targets of diabetes and DN,
although the binding capacity of the 2 proteins was weaker than
that of PPAR-γ.
 | Table IIInformation of screened protein
crystal structure and optimal combining capacity of butt joint. |
Table II
Information of screened protein
crystal structure and optimal combining capacity of butt joint.
| Gene name | PDB IDa | Resolution
(Å)b | No. of predicted
pocketsc | Ligandd | ePredicted optimal binding
energy/affinity |
|---|
| Cep170 | 4JON | 2.15 | 2 | STZ | −4.83 kcal/mol
(289.52 μM) |
| | | | Rosi | −6.70 kcal/mol (12.29
μM) |
| Marcks | 1IWQ | 2.00 | 2 | STZ | −4.67 kcal/mol
(378.72 μM) |
| | | | Rosi | −6.01 kcal/mol (39.01
μM) |
| PPAR-γ | 3V9V | 1.60 | 1 | Rosi | −8.00 kcal/mol
(1.37 nM) |
Discussion
DN is a common complication of type 1 and 2
diabetes, and affects approximately 20% of adult diabetic patients
(22). At present, the pathology
and pathogenesis of DN are not completely understood, and the
majority of researchers consider that it is caused by multiple
factors. In the present study, in GO annotation analysis of the
identified DEGs, there were 17 BP items, 15 CC items and 13 MF
items. The calmodulin nerve phosphatase pathways and chemokine
signaling pathways were the main enriched signaling pathways
identified. Additionally, in the study by Price et al
(23) on the dorsal root ganglia
of STZ-treated male Wistar rats, the authors examined diabetic
peripheral nerves and found that the GO categorizing on glucose
metabolism, oxidoreductase activity and manganese ion binding were
significantly enriched for the regulated genes. The results of the
current study are consistent with these findings. Price et
al (23) also reported other
enriched categories that we did not reproduce. This may either
reflect inherent differences in gene expression between mice and
rats, as well as between dorsal root ganglia and sciatic nerves;
alternatively, this difference may also be due to the more
stringent significance criteria in the current study. Therefore,
our study provides the basis for further research on the pathogenic
mechanisms of DN.
In this study, 3 DEGs (Cep170, Marcks
and Glipr2) were found to be co-regulated by both STZ and
Rosi. These 3 genes were downregulated in the mice with STZ-induced
diabetes and their expression began to increase following treatment
with Rosi. This result suggests that the downregulation of these
genes may be responsible for the development of DN. A total of 85
DEGs regulated by STZ and 41 differentially expressed genes
regulated by Rosi were verified, through the gene expression data
analysis of 19 samples from mice with STZ-induced diabetes and
those treated with Rosi, as well as from the mice in the STZ + Rosi
group. The expression of these genes was increased following
treatment with Rosi, which indicated that the proteins encoded by
these 3 genes were potential targets in the treatment of DN.
Guarguaglini et al (24)
demonstrated that Cep170 is expressed throughout the cell cycle. It
is associated with the centrosome in the interphase and with the
spindle apparatus during mitosis. The overexpression of Cep170 in
U2OS cells induced strong microtubule bundling, suggesting that
Cep170 was able to bind microtubules with high affinity, and that
Cep170 was involved in the regulation of microtubule dynamics
(24). The dynamic properties of
the cell cortex and its actin cytoskeleton determine important
aspects of cell behavior and are a major target of cell regulation.
The expression of Marcks has been found to be locally abundant and
to correlate with morpho-genic processes and cell motility, and the
protein accumulates at plasmalemmal rafts, where it is
co-distributed with PI(4,5) P(2)
and promotes its retention and clustering (25). DN is the most common cause of
end-stage renal disease (ESRD) among type 2 diabetes mellitus
patients (DM) in Malaysia (26).
Glipr2 has been identified as an upregulated gene in the
study by Lokman et al (27). Huang et al (28) found that the expression of Glipr2
was elevated in the kidney tissue samples of patients with DN.
However, further research on the function of Cep170 in DN or
other diabetic diseases were is required.
Rosi has been compared to metformin and glyburide as
monotherapy in patients with recently diagnosed type 2 diabetes in
A Diabetes Outcome Progression Trial (ADOPT) study. The results
revealed a significantly lower risk reduction of failing
monotherapy with Rosi compared to metformin and glyburide after 5
years of treatment (29).
In conclusion, in the present study, using a systems
biology approach, we found significant changes in gene expression
induced by treatment with STZ and Rosi. Furthermore, 3
differentially expressed genes were found to be potential novel
targets in the treatment of DN. In addition, STZ and Rosi were
tentatively docked to the binding pockets of these encoded
proteins; the combined capacity of STZ and Rosi was weaker than
that of PPAR-γ. If the molecular structure of Rosi could be
modified, or if new molecules were found to target only Cep170 or
Marcks rather than PPAR-γ, the side-effects of thiazolidinediones
could be avoided. In brief, Glipr2, Cep170 and
Marcks are novel potential targets in the treatment of
DN.
References
|
1
|
Workowski KA and Berman SM: Centers for
Disease Control and Prevention sexually transmitted diseases
treatment guidelines. Clin Infect Dis. 44(Suppl 3): S73–S76. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Skljarevski V and Malik RA: Clinical
diagnosis of diabetic neuropathy. Diabetic Neuropathy. Veves A and
Malik R: 2nd edition. Springer; New Jersey: pp. 275–292. 2007,
View Article : Google Scholar
|
|
3
|
Said G, Goulon-Goeau C, Slama G and
Tchobroutsky G: Severe early-onset polyneuropathy in
insulin-dependent diabetes mellitus. A clinical and pathological
study. N Engl J Med. 326:1257–1263. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Soumyanath A, Dimitrova D, Arnold G, et
al: P02.186. CAST (Centella asiatica selected triterpenes):
stability, safety, and effect on diabetic neuropathy (DN). BMC
Complement Altern Med. 12(Suppl 1): P2422012. View Article : Google Scholar :
|
|
5
|
Diep QN, El Mabrouk M, Cohn JS, et al:
Structure, endothelial function, cell growth, and inflammation in
blood vessels of angiotensin II-infused rats role of peroxisome
proliferator-activated receptor-gamma. Circulation. 105:2296–2302.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lygate CA, Hulbert K, Monfared M, Cole MA,
Clarke K and Neubauer S: The PPARγ-activator rosiglitazone does not
alter remodeling but increases mortality in rats post-myocardial
infarction. Cardiovasc Res. 58:632–637. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Szkudelski T: The mechanism of alloxan and
streptozotocin action in B cells of the rat pancreas. Physiol Res.
50:537–546. 2001.
|
|
8
|
Yamamoto H, Shimoshige Y, Yamaji T, Murai
N, Aoki T and Matsuoka N: Pharmacological characterization of
standard analgesics on mechanical allodynia in
streptozotocin-induced diabetic rats. Neuropharmacology.
57:403–408. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Edgar R, Domrachev M and Lash AE: Gene
Expression Omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar :
|
|
10
|
Tusher VG, Tibshirani R and Chu G:
Significance analysis of microarrays applied to the ionizing
radiation response. Proc Natl Acad Sci USA. 98:5116–5121. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kanehisa M: The KEGG database. Novartis
Found Symp. 247:91–103. 119–128. 244–252. 2002. View Article : Google Scholar
|
|
12
|
Dennis G Jr, Sherman BT, Hosack DA, et al:
DAVID: database for annotation, visualization, and integrated
discovery. Genome Biol. 4:P32003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nishimura D: BioCarta. Biotech Software
and Internet Report. 2:117–120. 2001. View Article : Google Scholar
|
|
14
|
Franceschini A, Szklarczyk D, Frankild S,
et al: STRING v9.1: protein-protein interaction networks, with
increased coverage and integration. Nucleic Acids Res.
41:D808–D815. 2013. View Article : Google Scholar :
|
|
15
|
Berman HM, Westbrook J, Feng Z, et al: The
Protein Data Bank. Nucleic Acids Res. 28:235–242. 2000. View Article : Google Scholar
|
|
16
|
Bairoch A, Apweiler R, Wu CH, et al: The
universal protein resource (UniProt). Nucleic Acids Res.
33:D154–D159. 2005. View Article : Google Scholar :
|
|
17
|
Altschul SF, Madden TL, Schäffer AA, et
al: Gapped BLAST and PSI-BLAST: a new generation of protein
database search programs. Nucleic Acids Res. 25:3389–3402. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yue TI TL, Chen J, Bao W, et al: In vivo
myocardial protection from ischemia/reperfusion injury by the
peroxisome proliferator-activated receptor-γ agonist rosiglitazone.
Circulation. 104:2588–2594. 2001. View Article : Google Scholar
|
|
19
|
Hussein Z, Wentworth JM, Nankervis AJ, et
al: Effectiveness and side effects of thiazolidinediones for type 2
diabetes: real-life experience from a tertiary hospital. Med J
Aust. 181:536–539. 2004.PubMed/NCBI
|
|
20
|
Vanopdenbosch N, Cramer R and Giarrusso
FF: Sybyl, the Integrated Molecular Modeling System. J Mol Graph.
3:110–111. 1985.
|
|
21
|
Morris GM, Huey R, Lindstrom W, et al:
AutoDock4 and AutoDockTools4: Automated docking with selective
receptor flexibility. J Comput Chem. 30:2785–2791. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
De Groot M, Anderson R, Freedland KE,
Clouse RE and Lustman PJ: Association of depression and diabetes
complications: a meta-analysis. Psychosom Med. 63:619–630. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Price SA, Zeef LaH, Wardleworth L, Hayes A
and Tomlinson DR: Identification of changes in gene expression in
dorsal root ganglia in diabetic neuropathy: correlation with
functional deficits. J Neuropathol Exp Neur. 65:722–732. 2006.
View Article : Google Scholar
|
|
24
|
Guarguaglini G, Duncan PI, Stierhof YD,
Holmström T, Duensing S and Nigg EA: The forkhead-associated domain
protein Cep170 interacts with Polo-like kinase 1 and serves as a
marker for mature centrioles. Mol Biol Cell. 16:1095–1107. 2005.
View Article : Google Scholar :
|
|
25
|
Laux T, Fukami K, Thelen M, Golub T, Frey
D and Caroni P: GAP43, MARCKS, and CAP23 modulate PI(4,5)P(2) at
plasmalemmal rafts, and regulate cell cortex actin dynamics through
a common mechanism. J Cell Biol. 149:1455–1472. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hooi LS, Wong HS and Morad Z: Prevention
of renal failure: The Malaysian experience. Kidney Int Suppl.
67:S70–S74. 2005. View Article : Google Scholar
|
|
27
|
Lokman FE, Seman NA, Al-Safi Ismail A, et
al: Gene expression profiling in ethnic Malays with type 2 diabetes
mellitus, with and without diabetic nephropathy. J Nephrol.
24:778–789. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang S, Liu F, Niu Q, et al: GLIPR-2
overexpression in HK-2 cells promotes cell EMT and migration
through ERK1/2 activation. PLoS One. 8:e585742013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Krentz A: Thiazolidinediones: effects on
the development and progression of type 2 diabetes and associated
vascular complications. Diabetes Metab Res Rev. 25:112–126. 2009.
View Article : Google Scholar : PubMed/NCBI
|