Introduction
The malignant transformation of normal cells may be
caused by aberrant gene expression, which disrupts the regulation
of cell proliferation, apoptosis, senescence and DNA repair. The
Bcl-2 antagonist of cell death (BAD)-mediated apoptotic pathway has
been demonstrated to play an important role in carcinogenesis
(1) and chemoresponse (2). Evidence suggests that the expression
levels of the BAD-mediated apoptotic pathway and BAD protein
influence endometrial and ovarian cancer cell resistance to
chemotherapy (3,4).
The BAD protein regulates apoptosis by binding to
anti-apoptotic members of the same family (5). The BAD protein itself is also
regulated by multiple kinases and phosphatases (5–7).
When non-phosphorylated, Bad selectively dimerizes with Bcl-xL and
Bcl-2, displacing Bax, which is then free to initiate mitochondrial
membrane permeability, which leads to apoptosis (8). When phosphorylated, BAD is unable to
heterodimerize with Bcl-2 or Bcl-xL and is sequestered into the
cytosol by 14-3-3 protein (9).
The phosphorylation of 3 serine residues (Ser-112, -136 and -155)
influences the activity of the BAD protein. BAD at Ser-112 is
phosphorylated by ribosomal protein S6 kinase alpha-1 (RPS6KA1/RSK)
and cAMP-dependent protein kinase [also known as protein kinase A
(PKA)]; BAD at Ser-136 is phosphorylated by protein kinase B
(PKB/Akt) (10) and BAD at
Ser-155 is preferentially phosphorylated by PKA (5,11,12). Conversely, the activity of a
series of phosphatases, including protein phosphatase(PP)1, PP2A
and PP2C (PPM1A), as well as calcineurin, has been shown to exert
pro-apoptotic effects through the de-phosphorylation of BAD
(13).
We and others have demonstrated that the expression
of the BAD-mediated apoptotic pathway and the phosphorylation
status of the BAD protein influence the chemosensitivity of cancer
cells, including ovarian and endometrial cancer cells (3,4,6,14).
Furthermore, we have previously demonstrated that ovarian cancer
samples from patients categorized as incomplete responders to
primary platinum therapy have a decreased expression of the BAD
protein Ser-155 phosphatase, PP2C, compared to samples from
patients categorized as complete responders (14). In this study, we investigated the
influence of the BAD pathway and the expression of PP2C in the
development of cancer. It has previously been suggested that the
phosphorylation of BAD at Ser-155 may convey an oncogenic potential
(15). In the present study, we
demonstrate that the expression of the BAD-mediated apoptotic
pathway is associated with the development of a variety of human
cancers and that phosphorylated BAD (pBAD) isoforms are
overrepresented in cancer cells when compared to immortalized
normal cells from the same tissues. Furthermore, using ovarian
cancer as a model, we demonstrate the expression of PP2C to be
decreased in ovarian cancer when compared to normal ovarian
epithelial samples and that depletion of PP2C in immortalized
normal ovarian cells, as well as cancer cells provides a growth
advantage. Our findings suggest that the BAD-mediated apoptotic
pathway influences the development of human cancer and that the
expression of PP2C may be an important mediator of oncogenic
potential.
Materials and methods
Cell lines
The cell lines, CRL-1831 (normal colon epithelial
cells), HCT-15 (colorectal adenocarcinoma cells), MCF-7 (breast
cancer cells), MCF-10A (mammary epithelial cells), MDA-MB-231
(human breast basal epithelial cells), HEC-1A (endometrial
adenocarcinoma cells) and IVAN (immortalized normal ovarian surface
epithelial cells), were obtained from the American Type Culture
Collection (ATCC; Manassas, VA, USA). OVCAR4 cells were a kind gift
from Dr Patricia Kruk, University of South Florida. A2780CP ovarian
cancer cells were obtained from the European Collection of Cell
Cultures (Salisbury, UK). The cells were cultured at 37°C, 5%
CO2 in RPMI-1640 or Dulbecco’s modified Eagle’s medium
(DMEM) containing 10% fetal bovine serum, 0.01% non-essential amino
acids, 1% sodium pyruvate, and 1% penicillin and streptomycin.
Mycoplasma testing was performed every 6 months in accordance with
the manufacturer’s instructions (Lonza, Rockland, ME, USA).
Development and evaluation of a
BAD-mediated apoptotic pathway gene expression signature
Principal component analysis (PCA) was used to
derive a BAD-mediated apoptotic pathway gene expression signature
with a corresponding ‘pathway score’ to represent an overall gene
expression level for the BAD-mediated apoptotic pathway genes (or
subsets thereof for datasets generated by the U133A or U95A
Affymetrix platforms, as previously described) (4,16).
In brief, PCA was applied to each dataset to reduce the data
dimension into a small set of uncorrelated principal components,
which were generated based on their ability to account for the
systematic variation in the data. In the PCA model, the X matrix
(gene expression values) can be described as follows: X =
t1*p1′ +
t2*p2′ +
t3*p3′ + … +
tA*pA′ + E (where ti
represents scores, pi represents loading and E
represents the residual matrix). The scores, ti, show
how similar samples are to each other, and the loading,
pi, explains which variables (genes) are important for
the principal component i. The first principal component
(PC1), which accounts for the largest variability in the data, was
used as the BAD-mediated apoptotic pathway PCA score to represent
the overall expression profile for the BAD-mediated apoptotic
pathway. It is known that directional signs of PCA scores are
recognized to be arbitrary and can vary between software and
algorithm used to calculate the PCA model (17). However, this does not affect the
interpretation of the PCA model and can be easily solved by
multiplying both scores and loadings by −1, a 180° rotation.
Reflecting this, and for the sake of consistency, in our analyses,
the PCA model was rotated so a high score corresponded to a normal
sample or to an increased survival time.
To evaluate the influence of the BAD-mediated
apoptotic pathway on the development and progression of cancer, the
BAD-mediated apoptotic pathway PCA score was tested in a series of
6 publically available Affymetrix, U133 Plus GeneChip mRNA
expression array datasets from a total of 427 patient samples as
follows: i) ovarian samples [28 normal samples, 78 cancer samples,
Moffitt Cancer Center (MCC)]; ii) breast samples (143 normal
samples, 42 cancer samples, GSE10780); iii) breast samples (8
atypical ductal hyperplasia samples, 23 ductal carcinoma in
situ samples and 30 invasive ductal carcinoma samples, MCC);
iv) colon samples (10 normal samples, 12 cancer samples, GSE4107);
v) endometrial samples (9 normal samples, 4 endometrial hyperplasia
samples and 20 endometrial cancer samples, MCC); and vi)
endometrial samples (10 normal samples, 10 ovarian endometriosis
samples, GSE7305). Gene expression data from the normal,
pre-invasive and invasive cancer samples for each tissue type were
subjected to background correction and normalization using the MAS5
algorithm (Affymetrix Expression Console). The BAD-mediated
apoptotic pathway score was evaluated between the normal and
cancer, normal and hyperplasia, and carcinoma in situ and
cancer samples, where available.
Patients
Patient samples and molecular and clinical data were
accessed from the MCC Total Cancer Care (TCC®)
repository. Patients enrolled in the TCC Protocol had provided
written informed consent, and sample collection was approved by the
University of South Florida Internal Review Board.
For the ovarian cancer samples, inclusion criteria
included age >18 years, ovarian, fallopian, or primary
peritoneal cancer, stage III and IV, serous, clear cell,
endometrioid, or mixed histology not containing mucinous type.
Retrieved clinical data included age at diagnosis, FIGO stage,
tumor grade, histological type, surgical cytoreductive status,
response to chemotherapy and survival (Table I). Of the total number of patients
(n=67), 62 had ovarian, 4 had primary peritoneal and 1 had
fallopian tube cancer, with 64 patients undergoing primary
debulking (43 optimal, 17 sub-optimal, based on FIGO criteria of
residual disease <1 cm, and 4 unknown). The remaining 3 patients
received neoadjuvant chemotherapy followed by optimal interval
debulking. Of the 9 patients who failed to complete at least 2
cycles of adjuvant chemotherapy, 2 were due to patient refusal, 3
due to death in the post-operative period, 1 patient died during
the second cycle, 1 had borderline pathology, 1 stopped due to
intolerance and 1 had the disease confined to the ovary (although
she was incompletely staged). The 38 patients having a complete
response included 3 patients who received neoadjuvant chemotherapy.
Complete response was defined by normalized CA-125 post-treatment,
disappearance of measurable disease on a CT scan, or negative
second-look surgery. Incomplete response (in 17 of our patients)
was defined as partial response (decrease in tumor size on a CT
scan or decrease but not normalization of CA-125), stable disease
(by CT), progression during treatment, or positive second-look
surgery. The demographic and baseline data for the patients are
listed in Table I.
 | Table IPatient demographics. |
Table I
Patient demographics.
| Characteristics | n | % |
|---|
| Age at diagnosis
(mean, 63 years) |
| <45 years | 3 | 4 |
| 45–65 years | 28 | 42 |
| >65 years | 3 | 48 |
| Unknown | 4 | 6 |
| FIGO stage |
| 3 | 53 | 79 |
| 4 | 11 | 16 |
| Other | 3 | 4 |
| Tumor grade |
| Low | 7 | 10 |
| Moderate | 9 | 13 |
| High | 51 | 76 |
| BRCA-positive | 2 | 3 |
| Debulking status
(n=64) |
| Optimal | 43 | 67 |
| Sub-optimal | 17 | 27 |
| Unknown | 4 | 6 |
| Response to
chemotherapy |
| CR | 38 | 57 |
| IR | 17 | 25 |
| Received <2
cycles | 9 | 13 |
| Unknown | 3 | 4 |
| Histology |
| Serous | 52 | 78 |
| Clear cell | 2 | 3 |
| Endometrioid | 1 | 1 |
| Mixed | 7 | 10 |
|
Undifferentiated | 3 | 4 |
| Other | 2 | 3 |
For the normal ovarian surface epithelial (NOSE)
samples, the surface epithelium was carefully scraped from a series
(n=9) of normal ovaries that had been resected from patients
undergoing surgery for non-ovarian pathology.
Sample preparation
Macro-dissection was employed to ensure >80%
tumor content. Total RNA extraction was performed using the RNeasy
mini kit (Qiagen Inc., Hilden, Germany) per manufacturer’s
instructions. Fifty nanograms of total RNA were converted into cDNA
using TaqMan High Capacity RNA to cDNA master mix (Applied
Biosystems/Life Technologies, Grand Island, NY, USA). The following
parameters were used for reverse transcription: 30-min hold at
16°C, 30-min hold at 42°C and 5-min hold at 85°C.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The NOSE (n=9) and ovarian cancer (n=67) samples
were subjected to quantitative (real-time) PCR (RT-qPCR), performed
using the relative standard curve method for quantification of
relative PP2C expression (StepOne Plus™ real-time PCR system,
TaqMan™ small RNA assay and TaqMan Universal PCR master mix;
Applied Biosystems/Life Technologies). Glyceraldehyde 3-phosphate
dehydrogenase (GAPDH) was used as an endogenous control to
normalize the RT-qPCR data. A total of 2 μl of cDNA was used
for each RT-qPCR reaction. The following parameters were used for
thermocycling: 10 min hold at 95°C, followed by 40 cycles of 15 sec
at 95°C, and 60 sec at 60°C. Analysis was performed using StepOne
software (version 2.1). The expression level of PP2C in an ovarian
cancer cell line, SKOV6 (kind gift from Dr Susan Murphy, Duke
University), was used as the reference point (relative expression
level of 1), to which all patient samples were compared.
Transfection and assessment of cell
proliferation
Transfection was performed through electroporation
using the Amaxa Nucleofactor II™ (Lonza). Cells (4×106)
were transfected with either PP2C siRNA or a non-targeting negative
control siRNA (Applied Biosystems/Life Technologies), with a final
siRNA concentration of 1 μM. Following transfection, the
cells were seeded in 96-well optical plates. Subsequent
proliferation was measured via MTS assay using CellTiter 96 Aqueous
One Solution (Promega, Madison, WI, USA) at 24-h intervals.
Baseline proliferation at 24 h post-transfection was used to
normalize subsequent assays. The proliferation of the cells
transfected with PP2C siRNA was expressed as a percentage of the
proliferation of the cells transfected with non-targeting negative
control siRNA.
Immunofluorescence microscopy
Immortalized normal and invasive cancer cell lines
of different cancer types, such as ovarian, colon, breast and
endometrial cancer, were cultured using standard techniques. Twenty
thousand cells were plated in each well of standard 12-well plates
for 24 h. The cells were then fixed with a solution of 95% ethanol
and 5% acetic acid for 1 min. The cells were washed 3 times with
phosphate-buffered saline (PBS) and then incubated with 2% bovine
serum albumin (BSA) in PBS. Primary antibody was added to a
blocking serum consisting of 2% BSA in PBS for 24 h. After 5
washes, the cells were incubated with fluorescent-labeled secondary
antibody in blocking serum for 1 h. The wells were then
counterstained and mounted with Prolong Gold containing
4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen/Life Technologies,
Grand Island, NY, USA). Fluorescence images were obtained using the
AxioCam MRm CCD camera and AxioVision (version 4.7). Exposure times
were identical for each antibody across the cell line pairs. The
intensity of fluorescence for each image was determined using
Definiens Developer XD 1.5 software. An algorithm was developed to
extract the fluorescence intensity per cell. The immunofluorescence
intensity of pBAD was expressed as a proportion of the intensity of
total BAD for n=50 cells of each cell line. The immortalized normal
cell line was then set as the reference to which its paired cancer
cell line was compared. Primary antibodies included rabbit
anti-phospho-BAD[Ser-112] (#A0029) and rabbit
anti-phospho-BAD[Ser-136] (#A01156) were acquired from Genscript
(Piscataway, NJ, USA). Rabbit anti-phospho-BAD[Ser-155] (#9297) and
rabbit anti-pan-BAD (#9292) were acquired from Cell Signaling
Technologies (Danvers, ME, USA). Secondary antibody included goat
anti-rabbit AlexaFluor 546 (#A11010) was obtained from
Invitrogen/Life Technologies.
Western blot analysis
The cells were harvested in medium and washed with
cold PBS containing 1X phosphatase inhibitor cocktail
(Sigma-Aldrich, St. Louis, MO, USA). Lysates were prepared with
sodium dodecyl sulfate (SDS) lysis buffer (2% SDS, 10% glycerol,
0.06 M Tris; pH 6.8) and evaluated for protein concentration using
the bicinchoninic acid method (Pierce, Rockford, IL, USA). Proteins
(75 μg) were separated on the same day as collection time on
12–15% SDS-polyacrylamide gel (PAGE) gels and transferred onto
polyvinylidene fluoride membranes. The membranes were blocked with
5% non-fat milk in Tris-buffered saline containing 0.05% Tween-20
(TBST) and incubated with primary antibody in 5% non-fat milk in
TBST overnight at 4°C. The membranes were washed 3 times for 5 min
with TBST and incubated with the appropriate secondary antibody in
5% non-fat milk in TBST for 60 min at room temperature. The
membranes were washed 4 times for 5 min with TBST prior to antibody
binding visualization using SuperSignal West Pico chemiluminescence
solution (Pierce) on autoradiography film (Midwest Scientific, St.
Louis, MO, USA). Primary antibodies included rabbit
anti-phospho-BAD[Ser-155] (#9297; Cell Signaling Technologies),
mouse anti-PP2C (#Sc-56956; Santa Cruz Biotech, Santa Cruz, CA,
USA), and mouse anti-GAPDH (#MAB374; Millipore, Temecula, CA, USA).
Secondary antibodies included donkey anti-rabbit-HRP (#NA9340V) and
sheep anti-mouse-HRP (#NA931V) from GE Healthcare, Pittsburgh, PA,
USA.
Statistical analysis
Differences in BAD pathway expression as defined by
the PC1 score as well as differences in percentage cell growth were
evaluated by the Student’s t-test. A p-value ≤0.05 was considered
to indicate a statistically significant difference. Spearman’s
correlation was used to evaluate differences in BAD pathway
expression between normal, hyperplasia and cancer samples. A
p-value ≤0.05 indicated a significant difference.
Results
BAD-mediated apoptotic pathway expression
is associated with the development of cancer
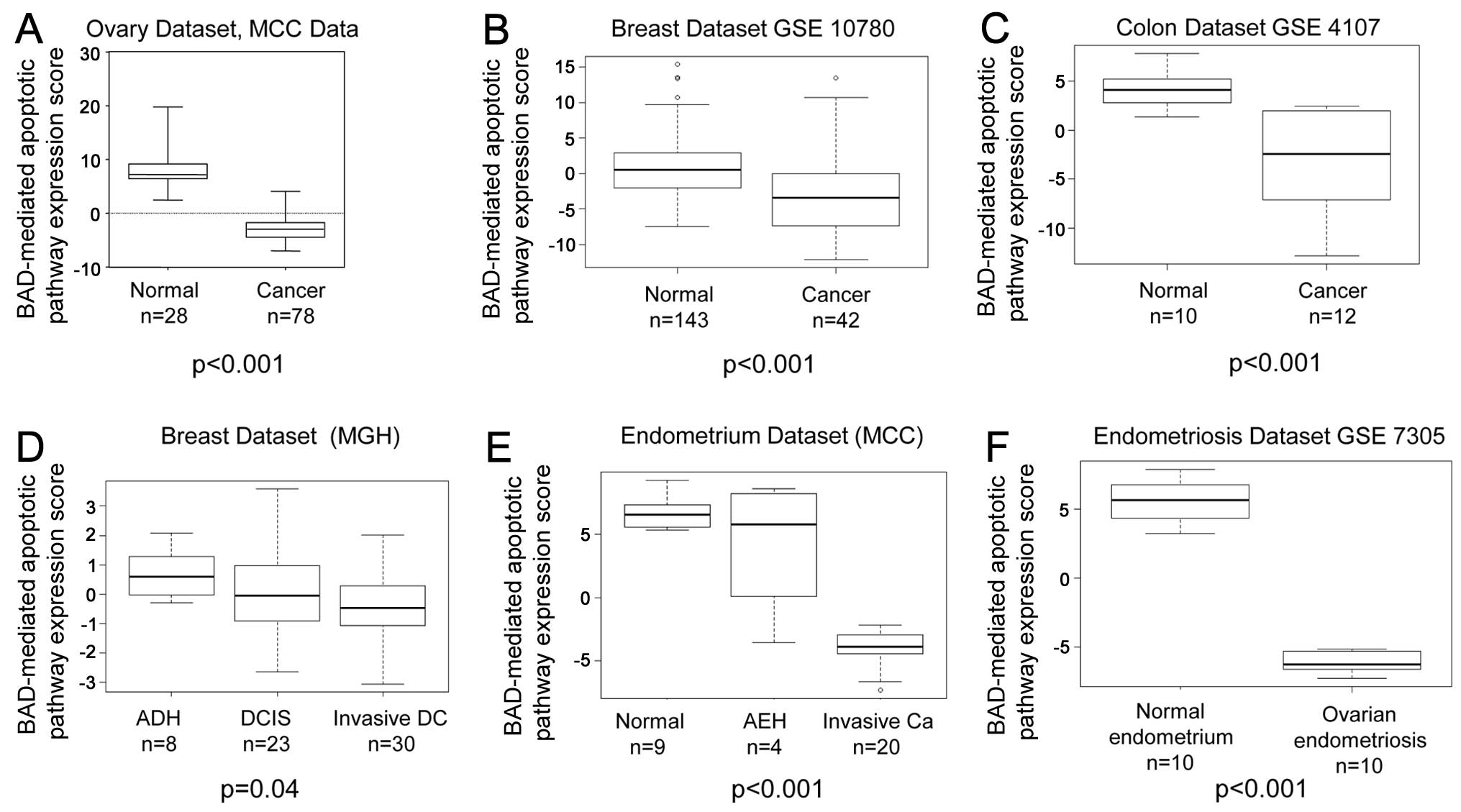
In each tissue type examined, the BAD-mediated
apoptotic pathway PCA score was higher in the normal tissue than in
the corresponding invasive carcinoma samples (Fig. 1). The normal ovary samples (n=28)
had a mean BAD-mediated apoptotic pathway expression score of
8.1968, whereas the ovarian cancer samples (n=78) had a mean score
of −2.9424 (P<0.001) (Fig.
1A). The normal breast samples (n=143) had a mean BAD-mediated
apoptotic pathway expression score of 2.721 vs. a score of −0.799
for the breast cancer samples (n=42; P<0.001) (Fig. 1B). The normal colon samples (n=10)
had a mean BAD-mediated apoptotic pathway expression score of 4.049
vs. a value of −3.374 for the colon cancer samples (n=12;
P<0.001) (Fig. 1C). When the
BAD-mediated apoptotic pathway expression results were compared
among the various stages of cancer progression, the expression
score was higher in the atypical ductal hyperplasia breast tissue
samples (mean expression score, 0.687, n=8) than in the ductal
carcinoma in situ samples (mean expression score, 0.046,
n=23) and higher in ductal carcinoma in situ than in
invasive ductal carcinoma (mean expression score, −0.298, n=30,
Spearman’s correlation estimate, −0.264, P=0.04) (Fig. 1D). BAD-mediated apoptotic pathway
expression was higher in the normal endometrial samples (mean
expression score, 6.745, n=9) than in the hyperplastic tissue
samples (mean expression score, 4.161, n=4) and higher in the
hyperplastic tissue samples than in the carcinoma samples (mean
expression score, −3.867, n=20, Spearman correlation estimate,
−0.795, P<0.001) (Fig. 1E).
The mean BAD-mediated apoptotic pathway expression score of the
normal endometrium samples (n=10) was 5.614 vs. −5.614 in the
ovarian endometriosis samples (n=10, P<0.001) (Fig. 1F).
BAD phosphorylation status and cancer
development
The post-translational modification of BAD
represents a key control point in the determination of cell
survival vs. apoptosis (5,19).
Previously, we demonstrated an inverse correlation between the
phosphorylation status of the BAD protein and BAD-mediated
apoptotic pathway PCA score (4).
In light of this and the identified differences in BAD-mediated
apoptotic pathway expression between normal and cancer tissues, we
evaluated differences in BAD phosphorylation between normal and
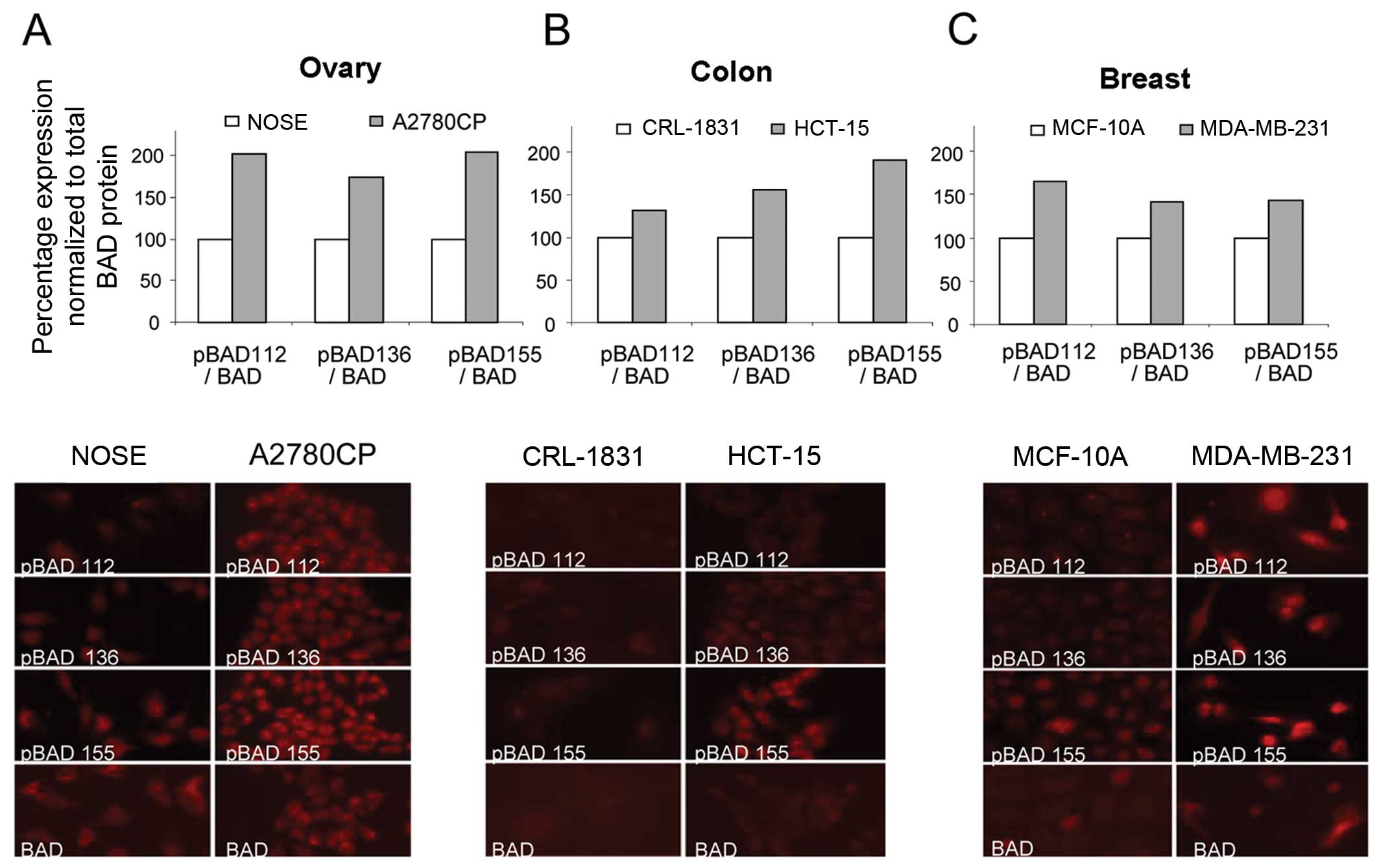
cancer cells. We analyzed the expression levels of pBAD (Ser-112,
-136 and -155), as well as total BAD, by immunofluorescence,
comparing a normal (immortalized) cell line with a cancer cell line
from several tissue types, including ovarian (NOSE vs. A2780CP)
(Fig. 2A), colon (CRL-1831 vs.
HCT-15) (Fig. 2B) and breast
(MCF-10A vs. MBA-231) (Fig. 2C).
Compared to the immortalized normal cells, the cancer cell lines
showed an increase in the percentage of pBAD relative to the total
BAD protein levels.
PP2C levels are associated with
cancer
PP2C is a key phosphatase that influences BAD
protein phosphorylation status (13). We previously demonstrated that
decreased PP2C levels were associated with increased
chemoresistance in ovarian cancer and endometrial cancer cells, as
well as primary ovarian cancer samples (3,4,14).
In this study, to determine whether PP2C levels may play a role in
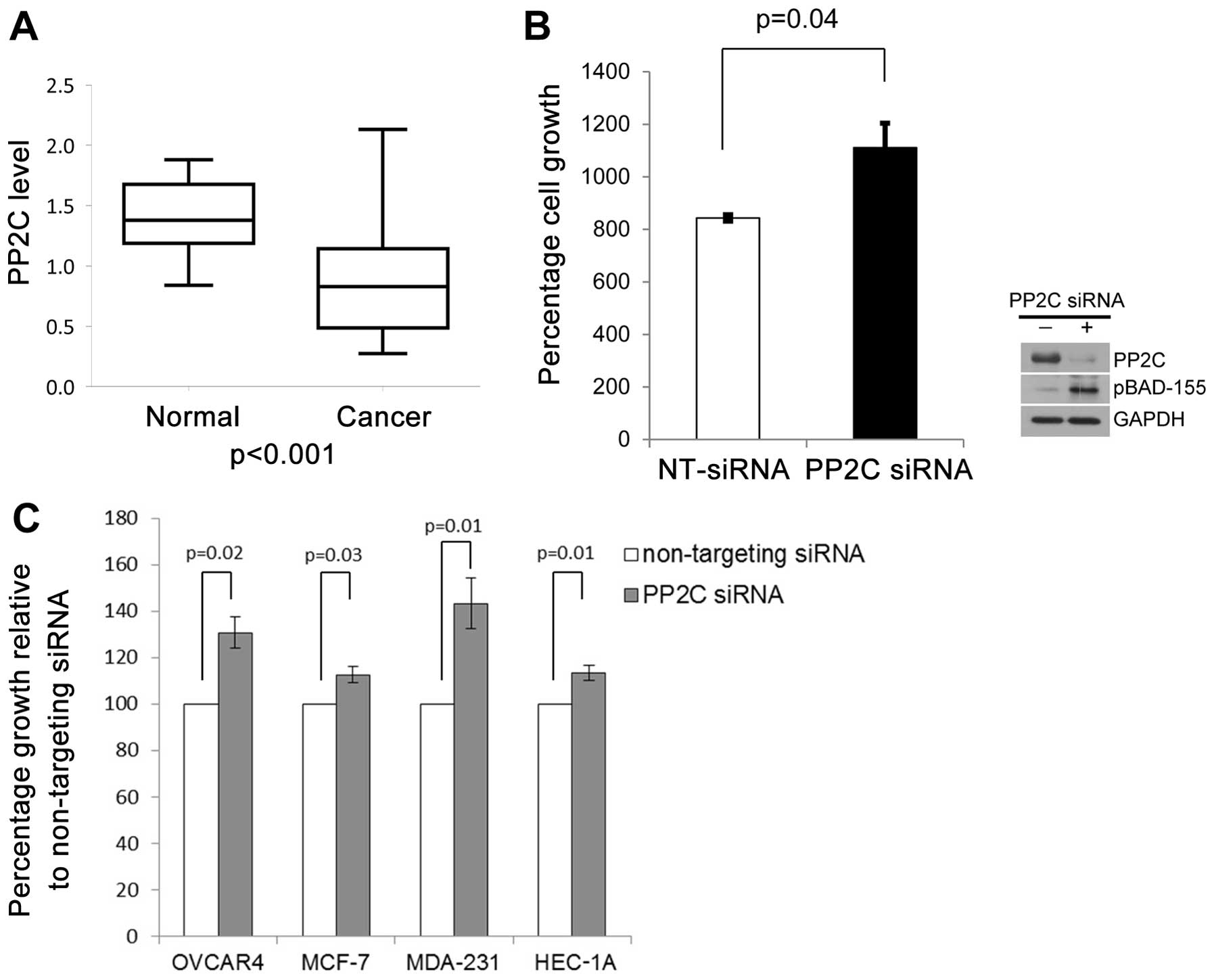
ovarian carcinogenesis, we evaluated the PP2C mRNA levels by
RT-qPCR in a dataset of 9 normal ovarian surface epithelial samples
and 67 primary ovarian cancer samples. We found that the mean
relative expression of PP2C was 0.864 in the ovarian cancer patient
samples (95% CI, 0.763–0.965, n=67) and 1.403 (95% CI, 1.188–1.618,
n=9) in the normal ovarian epithelial samples (P<0.001)
(Fig. 3A). No statistically
significant differences were observed between PP2C expression and
clinical variables, including response to therapy (complete vs.
incomplete response), disease-free survival (long vs. short) and
overall survival (Table II).
Patients with complete response showed a higher level of PP2C
expression than those with incomplete response, and patients who
had undergone optimal debulking showed a higher level of PP2C than
patients who had suboptimal debulking. However, neither comparison
reached statistical significance (Table II).
 | Table IIPP2C relative expression in normal
ovary and ovarian cancer samples. |
Table II
PP2C relative expression in normal
ovary and ovarian cancer samples.
| Group | Relative mean PP2C
expression | 95% CI | P-value |
|---|
| Normal ovary | 1.403 | 1.188–1.618 | <0.001 |
| All ovarian
cancers | 0.864 | 0.763–0.965 | |
| CR | 0.867 | 0.726–1.008 | 0.413 |
| IR | 0.765 | 0.569–0.961 | |
| Optimally
debulked | 0.875 | 0.732–0.963 | 0.34 |
| Sub-optimally
debulked | 0.771 | 0.604–0.938 | |
| Short DFS | 0.893 | 0.724–1.063 | 0.51 |
| Long DFS | 0.819 | 0.68–0.959 | |
| Short overall
survival | 0.81 | 0.676–0.944 | 0.36 |
| Long overall
survival | 0.876 | 0.7–1.052 | |
To further explore a role for PP2C in human cancer
development, we evaluated whether decreased PP2C levels may provide
a growth advantage to normal ovary epithelial cells. We evaluated
the cell growth rates following the depletion of PP2C by siRNA in
the immortalized normal ovarian surface epithelial cells, IVAN.
Depletion of PP2C by siRNA in the IVAN cells resulted in increased
cell growth rates for up to 5 days when compared to the cells
transfected with non-targeting negative control siRNA (Fig. 3B). Increased cell growth rates
were accompanied by an upregulation of pBAD, Ser-155 levels
(Fig. 3B). To determine whether
PP2C levels may influence the progression of cancer, we evaluated
the effects of the depletion of PP2C on the growth rates of i)
ovarian cancer cells (OVCAR4), ii) breast cancer cells (MCF-7 and
MDA-MB-231), and iii) endometrial cancer cells (HEC-1A). Similar to
the IVAN cells, the depletion of PP2C by siRNA provided a growth
advantage to all cancer cell lines examined (Fig. 3C). As shown in Fig. 3C, a significant increase in cell
growth at 72 h after the depletion of PP2C was observed in the
cancer cell lines, OVCAR4 (P=0.02), MCF-7 (P=0.03), MDA-MB-231
(P=0.01) and HEC-1A (P=0.01).
Discussion
The evasion of apoptotic signaling is a hallmark of
cancer cells (18). Since Bcl-2
family proteins are critical determinants of cellular apoptosis and
survival, we evaluated the role of the BAD-mediated apoptotic
pathway as a determinant of cancer development and progression. We
previously developed a BAD-mediated apoptotic pathway gene
expression signature using PCA that summarized the overall
expression of the BAD pathway and found that the expression of the
BAD pathway was associated with the development of ovarian cancer
chemoresistance (4). In this
study, we evaluated whether the BAD-mediated apoptotic pathway also
influences carcinogenesis. Using PCA modeling, we evaluated the
associations between BAD-mediated apoptotic pathway expression and
carcinogenesis using a series of clinico-genomic datasets
comprising normal and cancer tissues, including cancers of the
breast, colon and endometrium. We revealed differences in
BAD-mediated apoptotic pathway expression between the normal tissue
and cancer samples. Moreover, we observed a correlation between the
BAD-mediated apoptotic pathway expression score and the transition
from normal tissue to pre-cancer/pre-invasive cancer, to invasive
cancer, suggesting that the BAD-mediated apoptotic pathway
influences the development and progression of several solid tumor
types.
Bcl-2 family proteins determine cell survival by
both differential expression and post-translational modifications.
The pro-apoptotic activity of BAD is inhibited by phosphorylation
at the Ser-112, -136 and -155 sites. The post-translational
modification of BAD represents a key control step between cell
survival and apoptosis. Thus, the phosphorylation of these serine
residues is required to prevent BAD-induced apoptosis (5,19).
We found the percentage of pBAD to be higher in cancer cells of the
ovary, breast and colon than corresponding immortalized normal cell
lines. Furthermore, we demonstrated that PP2C, a BAD phosphatase at
Ser-155, is expressed at a higher level in normal ovaries than in
ovarian cancers. The phosphorylation of BAD at Ser-155 is known to
contribute to cancer cell survival in vitro (20). Our present findings demonstrate
that the depletion of PP2C, resulting in increased levels of pBAD
at Ser-155, confers a significant growth advantage to both
immortalized normal and cancer cell lines. We also observed a trend
toward higher in vivo levels of PP2C in chemosensitive
cancers vs. chemoresistant ovarian cancers, although this did not
reach statistical significance. The range of PP2C expression was
wider in the ovarian cancer samples than in the normal ovary
samples, thus suggesting that additional factors may influence the
phosphorylation status of BAD and thus the potential for
oncogenesis.
In conclusion, our results suggest that the
subversion of BAD-mediated apoptosis may be an important step in
human cancer development and progression. It may also be an
important mechanism through which cancer cells acquire increased
growth potential. Further elucidation of the interactions between
various members of the BAD-mediated apoptotic pathway may lead to
the identification of novel targets for molecular therapy and
biomarker development.
Acknowledgments
Opinions, interpretations, conclusions and
recommendations are those of the authors and are not necessarily
endorsed by the funding agencies. We would like to thank Rasa
Hamilton (Moffitt Cancer Center) for providing editorial
assistance. This study was supported in part, by Moffitt’s Total
Cancer Care® Protocol (TCC). TCC is enabled, in part, by
the generous support of the DeBartolo Family, and we thank the many
patients who so graciously provided data and tissue to the TCC
Consortium. This study also received valuable assistance from the
Cancer Informatics Core at Moffitt Cancer Center, a National Cancer
Institute-designated Comprehensive Cancer Center, supported under
NIH grant P30 CA-76292. This study was also supported in part by
National Cancer Institute Grant R21 CA-110499-01A2, the Ocala Royal
Dames for Cancer Research Inc., the Phi Beta Psi Sorority, the
Hearing the Ovarian Cancer Whisper, Jacquie Liggett Foundation, the
Ovarian Cancer Research Fund and the US Army Medical Research and
Materiel Command under Award no. DAMD17-02-2-0051. J.M. Lancaster
has an advisory relationship and honoraria with Amgen, as well as
research funding from Vermillion.
References
|
1
|
Marone M, Scambia G, Mozzetti S, et al:
bcl-2, bax, bcl-XL, and bcl-XS expression in normal and neoplastic
ovarian tissues. Clin Cancer Res. 4:517–524. 1998.PubMed/NCBI
|
|
2
|
Llambi F and Green DR: Apoptosis and
oncogenesis: give and take in the BCL-2 family. Curr Opin Genet
Dev. 21:12–20. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chon HS, Marchion DC, Xiong Y, et al: The
BCL2 antagonist of cell death pathway influences endometrial cancer
cell sensitivity to cisplatin. Gynecol Oncol. 124:119–124. 2012.
View Article : Google Scholar
|
|
4
|
Marchion DC, Cottrill HM, Xiong Y, et al:
BAD phosphorylation determines ovarian cancer chemosensitivity and
patient survival. Clin Cancer Res. 17:6356–6366. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tan Y, Demeter MR, Ruan H and Comb MJ: BAD
Ser-155 phosphorylation regulates BAD/Bcl-XL interaction and cell
survival. J Biol Chem. 275:25865–25869. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hayakawa J, Ohmichi M, Kurachi H, et al:
Inhibition of BAD phosphorylation either at serine 112 via
extracellular signal-regulated protein kinase cascade or at serine
136 via Akt cascade sensitizes human ovarian cancer cells to
cisplatin. Cancer Res. 60:5988–5994. 2000.PubMed/NCBI
|
|
7
|
Zha J, Harada H, Yang E, Jockel J and
Korsmeyer SJ: Serine phosphorylation of death agonist BAD in
response to survival factor results in binding to 14-3-3 not
BCL-X(L). Cell. 87:619–628. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang E, Zha J, Jockel J, Boise LH,
Thompson CB and Korsmeyer SJ: Bad, a heterodimeric partner for
Bcl-XL and Bcl-2, displaces Bax and promotes cell death. Cell.
80:285–291. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hirai I and Wang HG:
Survival-factor-induced phosphorylation of Bad results in its
dissociation from Bcl-x(L) but not Bcl-2. Biochem J. 359:345–352.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
del Peso L, González-García M, Page C,
Herrera R and Nunez G: Interleukin-3-induced phosphorylation of BAD
through the protein kinase Akt. Science. 278:687–689. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lizcano JM, Morrice N and Cohen P:
Regulation of BAD by cAMP-dependent protein kinase is mediated via
phosphorylation of a novel site, Ser155. Biochem J. 349:547–557.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou XM, Liu Y, Payne G, Lutz RJ and
Chittenden T: Growth factors inactivate the cell death promoter BAD
by phosphorylation of its BH3 domain on Ser155. J Biol Chem.
275:25046–25051. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Klumpp S, Selke D and Krieglstein J:
Protein phosphatase type 2C dephosphorylates BAD. Neurochem Int.
42:555–560. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bansal N, Marchion DC, Bicaku E, et al:
BCL2 antagonist of cell death kinases, phosphatases, and ovarian
cancer sensitivity to cisplatin. J Gynecol Oncol. 23:35–42. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Youle RJ and Strasser A: The BCL-2 protein
family: opposing activities that mediate cell death. Nat Rev Mol
Cell Biol. 9:47–59. 2008. View
Article : Google Scholar
|
|
16
|
Ma XJ, Salunga R, Tuggle JT, et al: Gene
expression profiles of human breast cancer progression. Proc Natl
Acad Sci USA. 100:5974–5979. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jolliffe IT: Principal Component Analysis.
2nd edition. Springer; New York: pp. p4882002
|
|
18
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: the next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Datta SR, Katsov A, Hu L, et al: 14-3-3
proteins and survival kinases cooperate to inactivate BAD by BH3
domain phosphory-lation. Mol Cell. 6:41–51. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Virdee K, Parone PA and Tolkovsky AM:
Phosphorylation of the pro-apoptotic protein BAD on serine 155, a
novel site, contributes to cell survival. Curr Biol. 10:R8832000.
View Article : Google Scholar : PubMed/NCBI
|