Introduction
Inactivating mutations in the BRCA1 (MIM
113705) and BRCA2 (MIM 600185) genes confer a high risk of
developing breast and ovarian cancer (1,2).
Both genes have a contribution of approximately 16% to the risk of
familial breast cancer (3).
Genetic testing for BRCA1 and BRCA2 provides valuable
information for determining the clinical management of patients
with breast/ovarian cancer. However, the data provided are
difficult to interpret due to the identification of many DNA
variants of unknown pathological significance or unclassified
variants (UVs) that hamper genetic counseling in hereditary breast
and ovarian cancer (HBOC) (4).
UVs have the potential to alter protein function by
altering the coding sequence of a transcript, or the level of the
gene transcript by disrupting regulatory regions in promoters,
untranslated regions, exons or introns (5). Such regulatory variants include
those affecting the normal splicing of BRCA1 and
BRCA2, many of which have been shown to be clinically
significant using cDNA studies and multifactorial likelihood
analysis methods that combine bioinformatics, as well as
pathological and clinical information (6,7).
Assessing the impact of UVs on splicing is key to
determining their pathogenicity. The accuracy of pre-mRNA splicing
is determined by the recognition of well known 5′ and 3′ splice
site consensus sequences. However, more discrete elements are also
involved, such as exonic splicing enhancers (ESEs) that enhance
pre-mRNA splicing when present in exons (8). As a result, each UV may potentially
affect normal pre-mRNA splicing and be deleterious through the
disruption of consensus sequences, or through the creation of de
novo sequences or the alteration of splicing regulatory
elements (9). Several
BRCA1 isoforms have been identified in different tissues;
however, their functional significance is not yet fully understood
(10–15).
A recent study on the comprehensive annotation of
BRCA1 splice junctions identified 63 independent alternative
splicing events in RNA samples from healthy control individuals.
Among these, 10 were predominant (Δ1Aq, Δ5, Δ5q, Δ8p, Δ9, Δ9–10,
Δ9-10-11, Δ11q, Δ13p and Δ14p) and represented 5–30% of the
full-length signal; 48 were minor and 5 were non-classifiable
events (16).
Alternative splicing is a highly coordinated
process. Mutations destroying the 5′ and 3′ splice site consensus
sequences may alter the splicing patterns of one or more
transcripts, interrupting the production or function of the encoded
protein (17). A large number of
pathological transcripts of the BRCA1 and BRCA2
genes, deriving from a mutation in the consensus regions, have been
described in the literature (18).
The aim of this study was to identify aberrant
transcript variants resulting from the alternative splicing of
BRCA1 and BRCA2 genes in RNA extracted from blood
lymphocytes from women with a family history of and/or early onset
breast and/or ovarian cancer, in which genomic pathogenic
alterations in BRCA1 and BRCA2 have not been detected
by conventional analysis.
The analysis of all possible transcripts of the
BRCA1/BRCA2 genes may allow us to uncover mRNA splicing
defaults overlooked by conventional protocols and to confirm that
the alternative splicing of the BRCA1 and BRCA2 genes
plays an important role in BRCA1/2-driven tumorigenesis.
This approach has the potential to complete the process of the
characterization of mutations of BRCA1 and BRCA2 in
HBOC.
Materials and methods
Sample acquisition
A total of 13 blood samples were collected from
women with a family history of and/or early-onset breast and/or
ovarian cancer and who tested negative for pathogenic mutations in
the BRCA1 and BRCA2 genes. Sample data details and
characteristics are presented in Table I.
 | Table ISummary of patient data and family
history of cancer. |
Table I
Summary of patient data and family
history of cancer.
| Patient | Personal history
(age at onset, years) | Family history
|
|---|
| No. of breast
cancer cases | No. of ovarian
cancer cases | No. of cases of
other types of cancer |
|---|
| P1 | Br (57) | 4 | – | 1 |
| P2 | Br (34) | 1 | – | 1 |
| P3 | Br (43) | 8 | 1 | 5 |
| P4 | Br (37) | 1 | 1 | 4 |
| P5 | Br (55), Ov
(61) | – | – | 2 |
| P6 | Br (35) | 3 | – | 1 |
| P7 | Br (55) | 3 | 1 | 4 |
| P8 | Br bil (43) | 4 | 1 | 3 |
| P9 | Br (56) | 3 | – | 2 |
| P10 | Br bil (45 and
50) | 4 | – | 9 |
| P11 | Br (25) | – | – | 1 |
| P12 | Br (32) | – | – | 1 |
| P13 | Br bil (38 and
48) | – | – | 6 |
The BRCAPRO (http://www4.utsouthwestern.edu/breast-health/cagene/)
and BOADICEA (https://pluto.srl.cam.ac.uk/cgi-bin/bd2/v2/bd.cgi)
(19,20) programs were used, calculating a
priori mutation carrier risk of >10% for each patient.
Ten blood samples from healthy women, aged 25–45
years, with no family history of breast and/or ovarian cancer, were
used as the controls. All patients and healthy donors were
recruited at the Interdepartmental Centre for Cancer Genetics,
University Hospital of Santa Chiara in Pisa, Italy between 2004 and
2011. Prior to enrollment, informed consent for genetic analysis
was obtained from all patients. All the experiments carried out
complied with the current laws of the country in which they were
performed (Italy).
RNA extraction and cDNA synthesis
All samples were subjected to total RNA extraction
in 2 steps: peripheral blood mononuclear cells (PBMCs) were
isolated from ethylenediaminetetraacetic acid (EDTA)-treated
peripheral blood by standard density gradient centrifugation
(Ficoll 15%; Bio-Rad Medical Diagnostics GmbH, Dreieich, Germany)
and RNA extraction was obtained using a TRI Reagent kit (Molecular
Research Center, Inc., Cincinnati, OH, USA) as recommended by the
manufacturer. Isolated RNA was quantified using a NanoDrop
spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE,
USA) and examined for integrity on a 1.5% agarose/formaldehyde gel
containing 0.5 μg/ml ethidium bromide (RNA Analysis
Notebook; Promega Corp., Madison, WI, USA). First-strand cDNA was
synthesized from at least 1,000 ng of total RNA using an oligo(dT)
primer or random primer and SuperScript III reverse transcriptase
(Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s
instructions.
Reverse transcription-polymerase chain
reaction (RT-PCR)
To perform the amplification of naturally occurring
transcripts of the BRCA1 and BRCA2 genes, we used
multiple combinations of forward and reverse primer pairs to
amplify overlapping regions of the mRNA and to cover the entire
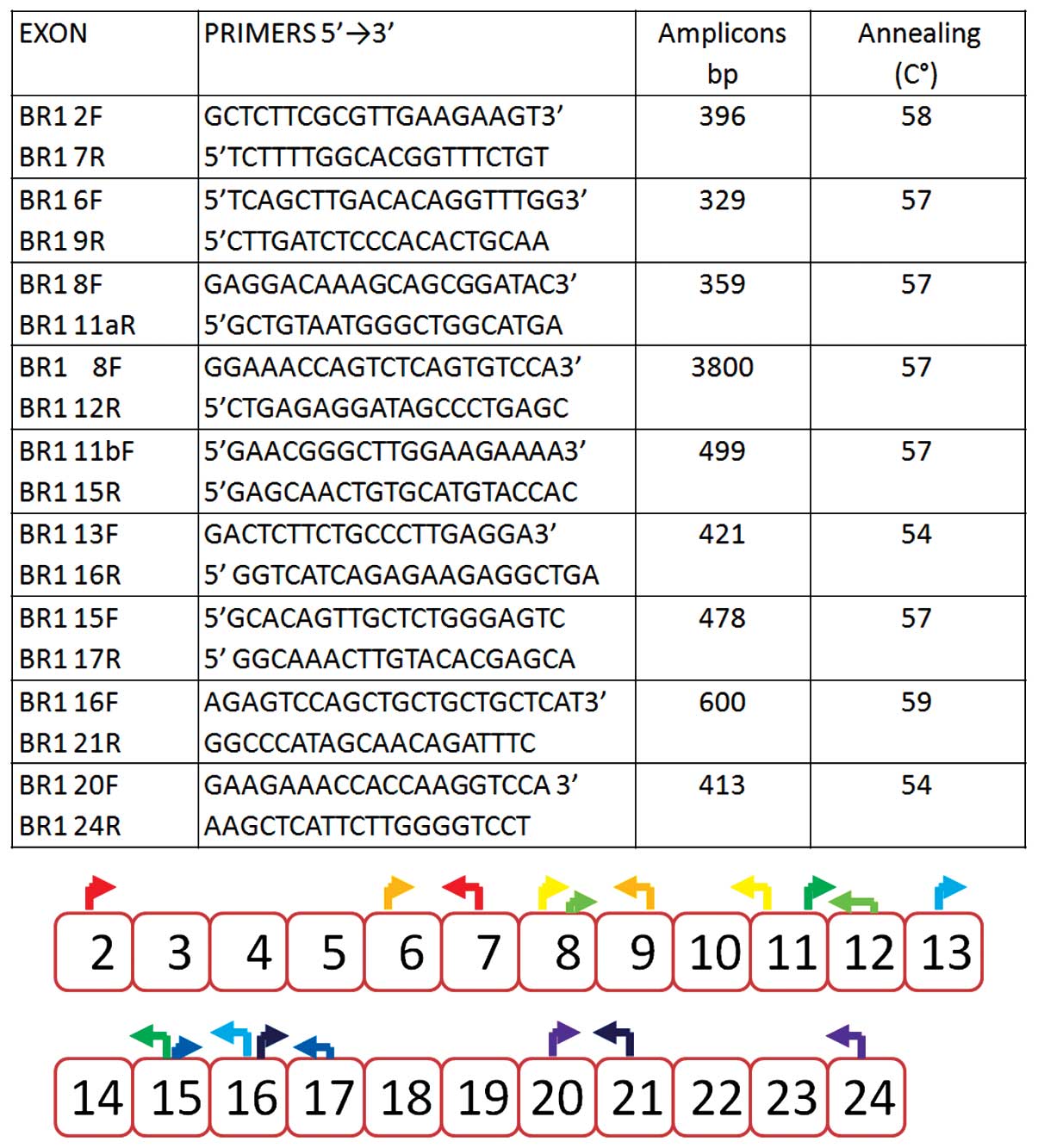
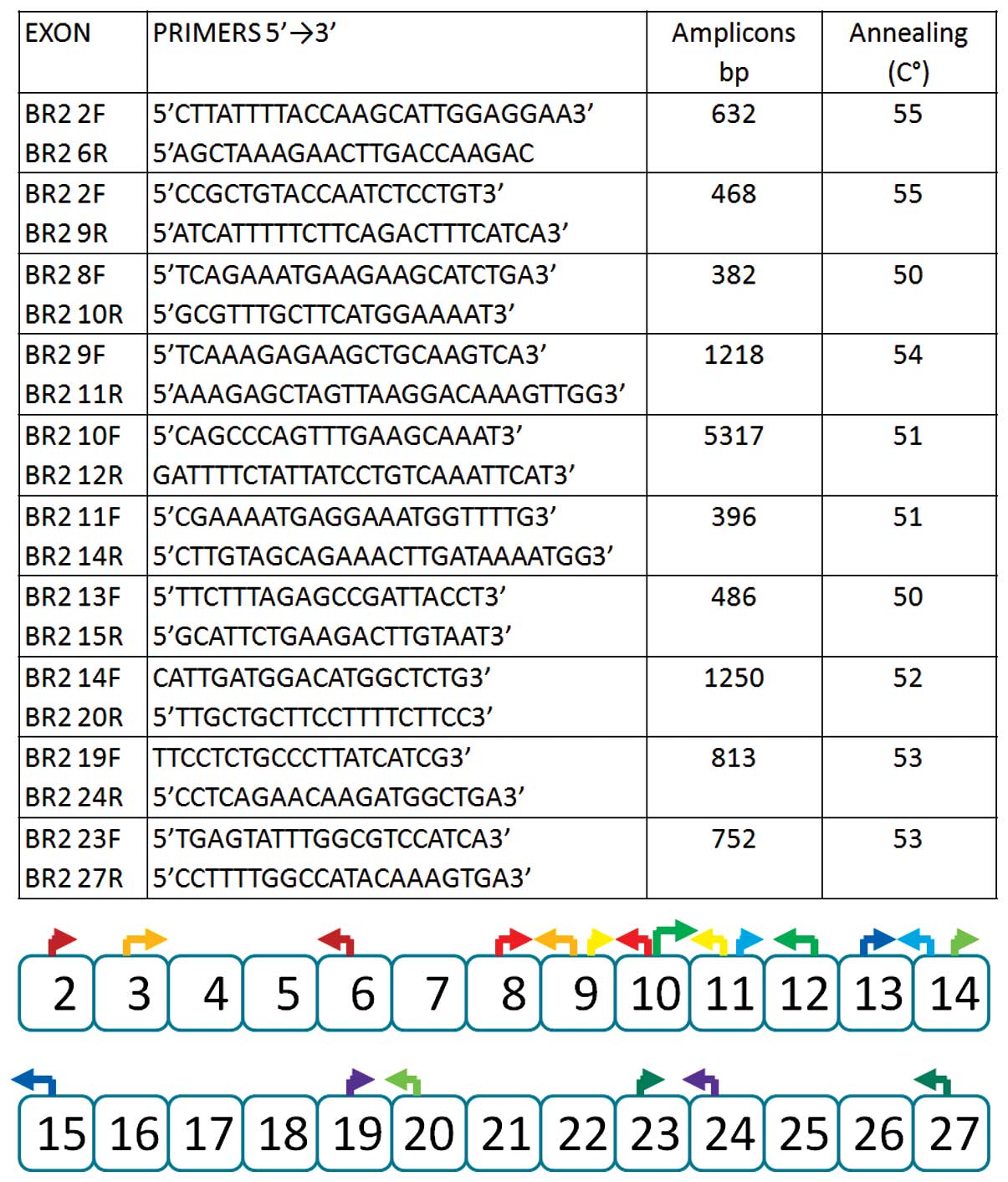
open reading frame (Figs. 1 and
2). For each PCR reaction, 2
μl of cDNA were used. The PCR conditions were as follows: 5
min at 95°C followed by 4–45 cycles at 95°C for 1 min, melting
temperature according to primer pair for 30 sec, 72°C for 1 min
followed by 72°C for 1 min. To encompass multiple exons for large
regions, Long Range PCR (Expand Long Template PCR system; Roche,
Indianapolis, IN, USA) was used. The PCR products, eventually
isolated on agarose gels, were sequenced on both strands using the
BigDye Terminator v3.1 Cycle Sequencing kit and the 3130×l Genetic
Analyzer (both from Life Technologies, Foster City, CA, USA). The
electropherograms were analyzed using SeqScape Software v2.6 (Life
Technologies).
Multiplex ligation-dependent probe
amplification (MLPA)
To exclude large genomic deletions in the
BRCA1 and BRCA2 genes, MLPA (MRC-Holland, Amsterdam,
The Netherlands) was performed as recommended by the manufacturer’s
instructions. The electropherograms were analyzed using GeneScan
(Life Technologies) and Coffalyser software (MRC-Holland).
Results
The aim of this study was to identify alternative
transcripts of the BRCA1 and BRCA2 genes resulting
from aberrant splicing events. The analysis was conducted on 13
total RNA samples extracted from PBMCs from women with a family
history of and/or early-onset breast and/or ovarian cancer, more
specifically, 5 cases of hereditary breast cancer (HBC), 4 cases of
HBOC, 1 case of bilateral carcinoma, 1 case of breast/ovarian
cancer and 2 cases of early-onset breast cancer (Table I.)
All women were affected by breast cancer, 1 women by
breast and ovarian cancer, 3 by bilateral breast cancer and 2 by
early-onset breast cancer. A total of 10 control RNA samples from
healthy women, aged between 25 and 45 years with no family history
of any form of cancer, were also included in this study.
The genomic DNA of each patient was analyzed by
direct sequencing of the entire open reading frame, 5′ and 3′UTRs
and exon/intron junctions of both genes. Approximately 100 bps from
the 5′ and 3′ end of each intron were sequenced. All cases tested
negative for the presence of germline mutations in the BRCA1
and BRCA2 genes. The presence of variants of unknown
pathological significance was also excluded from our analysis. The
presence of large genomic deletions or rearrangements was excluded
by MLPA of DNA extracted from the peripheral blood lymphocytes of
all patients and the controls.
The BRCA1 and BRCA2 mRNA in each
patient was analyzed by dividing the cDNA into 9 amplicons for
BRCA1 and 10 amplicons for BRCA2. The size of the
full-length transcripts of the BRCA1 (5,592 bp) and
BRCA2 (10,987 bp) genes prevents the analysis of the cDNA as
a single amplicon.
Naturally occurring transcripts of
BRCA1
The cDNA of the BRCA1 gene was amplified with
primers localized in exonic sequences so that partially overlapping
amplified products were obtained. The primers were selected in
order to highlight all predominant naturally occurring alternative
splicing isoforms and 28 of the minor transcripts, starting from
exon 2, as previously reported by Colombo et al (16).
Under our experimental conditions, the following
naturally occurring alternative transcripts were observed: the
Δ9–10 transcript was detected in 3 samples, the Δ9 transcript was
detected in 1 patient and Δ14p was detected in all the samples.
Using the long range PCR approach, the following transcripts were
detected: Δ11q (transcript lacking exon 11 except the initial 121
bp) in 10 samples and Δ9-10-11q (transcript lacking exons 9, 10 and
11 except the initial 121 bp) in 2 cases only.
Naturally occurring transcripts of
BRCA2
The cDNA of the BRCA2 gene was amplified with
primers localized in exonic sequences so that partially overlapping
amplified products were obtained. A total of 5 predominant
naturally occurring transcripts have been previously identified:
Δ4, Δ4–7, Δ17–18, Δ18 and Δ20 (7,18,21). Under our experimental conditions,
all 5 alternative transcripts were detected: More specifically, the
Δ4 transcript was detected in 2 patients, the Δ4–7 transcript was
detected in 2 patients, the Δ17–18 transcript was detected in 2
patients and the Δ18 transcript was detected in 7 patients. The Δ20
transcript was also detected in 1 case. All these BRCA1 and BRCA2
naturally occurring transcripts were detected in the controls.
Abberant transcripts
In addition to the above-mentioned predominant
naturally occurring transcripts, we detected 3 aberrant transcripts
in the BRCA1 gene in 2 patients (2 in patient P6 and 1 in
patient P7). No aberrant transcripts were detected in the
BRCA2 gene.
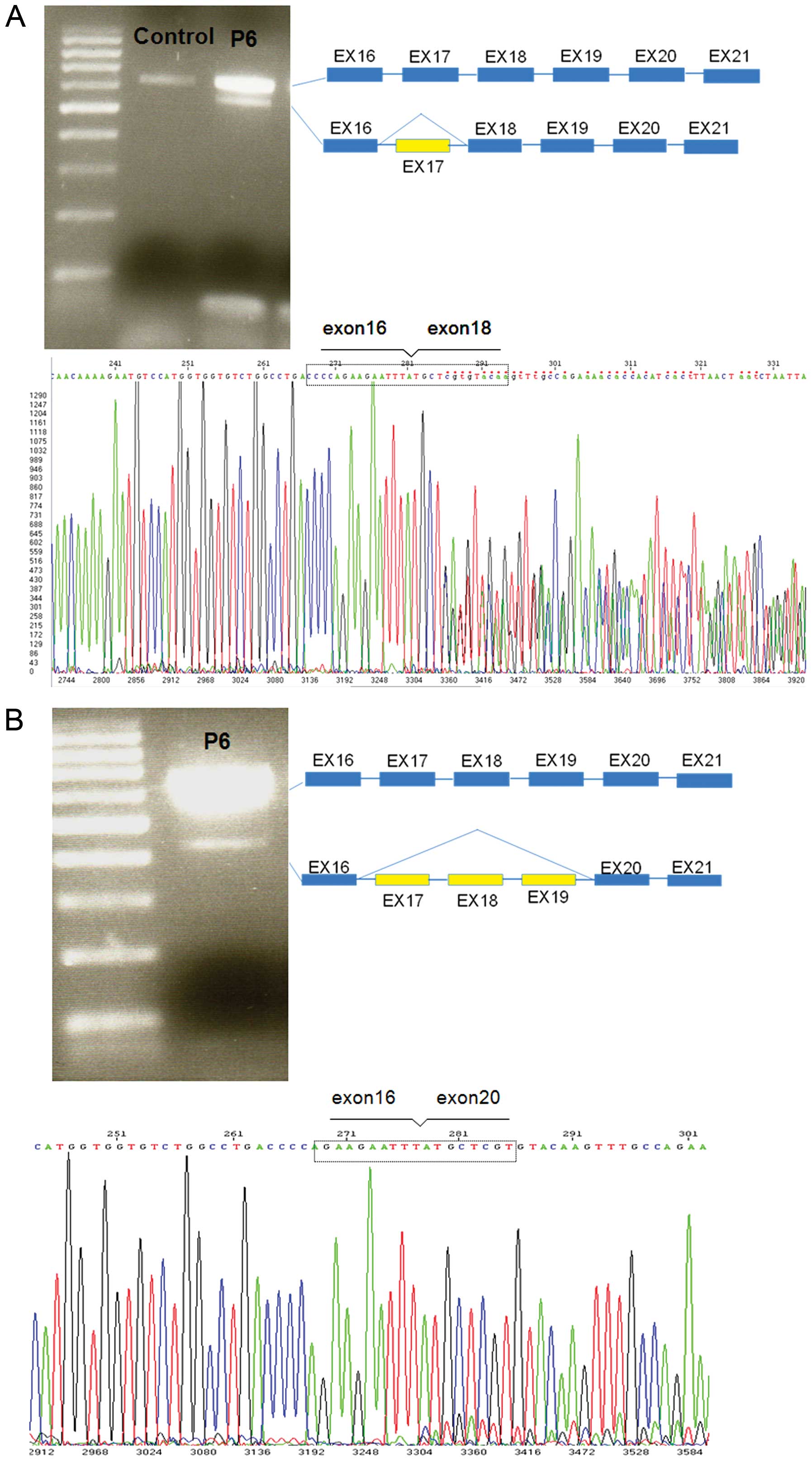
In patient P6, following the amplification of the
cDNA of BRCA1 with primers localized in exons 16 and 21 in
addition to the full-length transcript (600 bp), 2 additional
transcripts of approximately 500 and 400 bp were obtained. These
transcripts were not detected in the 10 healthy control samples.
The aberrant transcripts were gel-purified and then sequenced as
described in the Materials and methods. Sequence analysis allowed
us to detect the presence of a transcript lacking exon 17 and
another transcript lacking exons 17, 18 and 19 (Fig. 3). The transcript containing the
deletion of exon 17 produced an abnormal stop signal at codon 1673
(HGVS codification: p.Val1665Serfs*8) and then a truncated protein
lacking the last 192 amino acids. The transcript containing the
deletion of exons 17, 18 and 19 did not produce an abnormal stop
signal, lost 207 nucleotides and retained the open reading frame,
producing a protein lacking 69 amino acids.
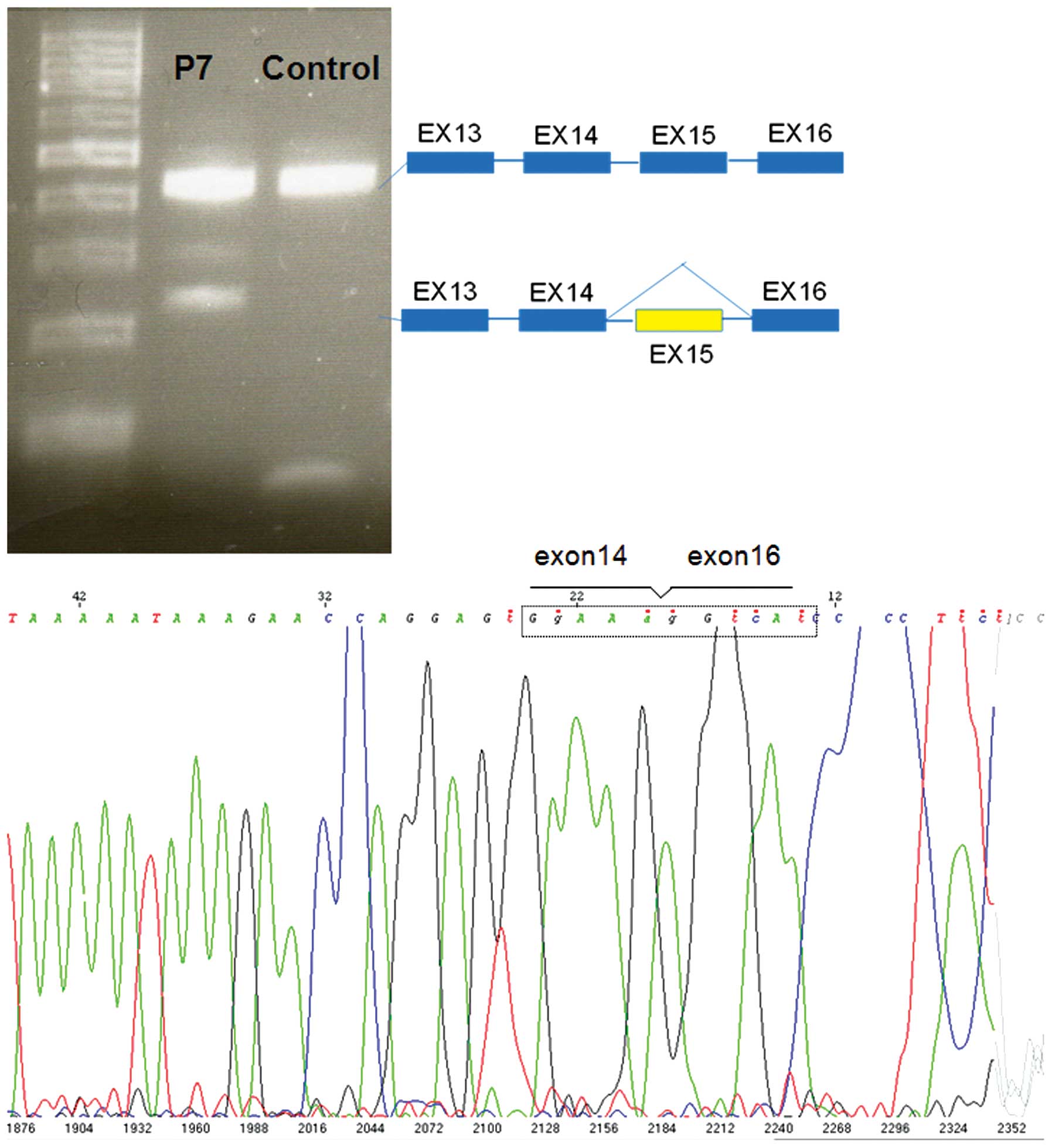
In patient P7, following the amplification of the
cDNA of the BRCA1 gene with primers localized in exon 13 and
16 in addition to the full-length transcript of approximately 420
bp, an additional transcript of approximately 220 bp was obtained
(Fig. 4). Direct sequencing of
this transcript allowed us to detect an exon 15 deletion. The
transcript containing the deletion of exon 15 produced an abnormal
stop signal at codon 1510 (HGVS codification: p.Ser1496Glyfs*14)
and then a truncated protein lackcing the last 405 amino acids.
Phenotype-genotype correlation
Patient P6, at the age of 35 years, developed an
infiltrating ductal carcinoma of the right breast, which was
estrogen receptor-positive, progesterone receptor-negative and
Her2/neu 3+, with loco-regional lymph node metastasis. The
patient’s sister had been diagnosed with breast cancer of a similar
phenotype at the age of 34. These were not the only cases of breast
cancer in the family; the grandmother and maternal aunt (their
mother’s twin sister) had also been afflicted by the disease.
Segregation analysis of the aberrant transcript in this family was
not possible as DNA samples were not available.
Patient P7 was diagnosed with breast cancer at the
age of 55 years. The tumor was an infiltrating breast carcinoma of
the right breast, and was estrogen receptor-positive, progesterone
receptor-negative and Her2/neu-negative, without any lymph node
metastasis. The patient’s mother had been affected by ovarian
cancer at 77 years of age and 3 cases of breast cancer were
reported in the family: 2 sisters of the mother and the mother’s
cousin (36 years of age). On the mother’s side, an uncle had been
affected by malignant melanoma, and an aunt by a brain tumor. These
relatives were not available for co-segregation analysis, but this
aberrant transcript was not detected in the 42-year-old healthy
daughter.
Discussion
This study focused on the possibility that a
proportion of patients with HBOC have mutations in the BRCA1
or BRCA2 genes that affect splicing, but are not detectable
through sequencing of the gene exons or intronic sequences near the
intron-exon boundaries. Several studies have demonstrated that, due
to variations in splice sites, BRCA1/BRCA2 may generate
truncated, non-functional proteins that may be associated with a
predisposition to breast and ovarian cancer (22). However, to the best of our
knowledge, no studies have evaluated the presence of
BRCA1/BRCA2 pathological transcripts in patients without
mutations identified in the canonical splice sites or regulatory
sequences.
Recently, a systematic description of ‘naturally
occurring’ alternative splicing at the BRCA1 locus was conducted by
the Evidence-based Network for the Interpretation of Germline
Mutant Alleles (ENIGMA) consortium (16). This led to the annotation of 63
splicing events, of which 35 were novel findings, even though most
of them are rather minor, and it is likely that some do not qualify
as ‘naturally occurring’ events, suggesting that the
characterization of the full complexity of BRCA1 splicing
requires further investigation (16).
The present study was performed in accordance with a
standard assay design and detection methods formulated by ENIGMA
Consortium members (23) in order
to detect the predominant naturally occurring alternative splicing
isoforms of BRCA1, as reported by Orban et al
(13,14), which were the only data available
at the moment of the study design. There is no standard assay
design for the detection of the predominant naturally occurring
alternative splicing isoforms of BRCA2; thus we referred to
the transcripts described in Ensembl (http://www.ensembl.org/index.html).
Using this strategy, we detected the Δ9, Δ9–10,
Δ11q, Δ9-10-11q and Δ14p BRCA1 isoforms and the Δ4, Δ4–7,
Δ17–18, Δ18 and Δ20 BRCA2 isoforms. In addition these
predominant BRCA1 transcripts, we detected the Δ17 and the
Δ17–19 isoforms in patient P6 and the Δ15 isoform in patient P7. No
minor transcripts were detected for BRCA2.
The Δ17 transcript lost the open reading frame,
leading to the formation of a truncated protein. The Δ17 transcript
was not detected in any of the 10 healthy controls in our study;
however, it has previously been found in 28 out of 103 (27%)
independent control samples or technical replicas in the ENIGMA
Consortium study (16). The Δ17
transcript, due to the genomic deletion of exon 17, has frequently
been detected in Italian families (24) possibly as a result of a
non-homologous rearrangement among ‘Alu repeats’. The Δ17
transcript is not derived from a genomic rearrangement as
demonstrated by MLPA of the patient’s DNA. The Δ17 transcript, even
if not derived from a genomic rearrangement or from aberrant mRNA
maturation due to canonical splice site mutations, it has to be
considered as pathogenetic as it encodes for a truncated
protein.
The Δ17–19 transcript gave rise to a protein with an
in-frame deletion of 69 amino acids. The Δ17–19 isoform was not
detected in either our 10 healthy control samples or the 223
independent control samples and technical replicas in the ENIGMA
Consortium study (16). Although
the Δ17–19 transcript had an in-frame deletion, a recent study
demonstrated that the expression of the human BRCA1
alternative splicing variant, Δ17–19, in the MCF-7 cells resulted
in an impaired assembly of DNA repair complexes and aberrant DNA
damage response (25), thus
contributing to an increased risk of developing cancer in the
carrier subjects.
In patient P7, the BRCA1 alternative
transcript Δ15 was detected. The alternative Δ15 transcript lost
the open reading frame and produced an abnormal stop signal to
position p.Ser1510Glyfs*14, leading to a truncated protein lacking
the last 405 amino acids. To evaluate the presence of Δ15 at the
genomic level, we performed MLPA, which did not reveal any
deletion. This isoform was not detected in our 10 healthy PBMC
control samples or in the 8 PBMC samples analyzed by Colombo et
al (16). They found this
transcript in only 4 of 10 replicas derived from samples of
leukocytes, PHA-stimulated peripheral blood leukocytes and
lymphoblastoid cell lines (they did not specify if these were
independent samples or replicas of the same sample).
In conclusion, the present study on 13 patients with
a family history of breast and/or ovarian cancer detected 3
alternative transcripts, probably pathogenic and not predictable by
genomic screening, in 2 patients. A number of studies have
described these 3 transcripts as consequences of large genomic
rearrangements or mutations in canonical splice sites (24–28). These aberrant alternative
transcripts may undergo nonsense-mediated decay (NMD) as they give
rise to truncated proteins (29);
therefore, the accurate quantification of these alternative
transcripts is crucial to discriminating what is considered
pathological and what is neutral for clinical relevance. The
relative abundance of the minor transcripts detected in this study
may prove to be useful in evaluating their possible role in cancer
predisposition and risk modification.
Acknowledgments
The present study was funded by the Istituto Toscano
Tumori (ITT) grant ‘BRCA1 role in repair of DNA damage: from the
yeast Saccharomyces cerevisiae genetics to human breast
carcinogenesis’ (2009–2012) and the Associazione Italiana per la
Ricerca sul Cancro (AIRC) grant [no. IG2013 N.14477
(2013–2015)].
References
|
1
|
Miki Y, Swensen J, Shattuck-Eidens D,
Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM,
Ding W, et al: A strong candidate for the breast and ovarian cancer
susceptibility gene BRCA1. Science. 266:66–71. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wooster R, Bignell G, Lancaster J, Swift
S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C and Micklem G:
Identification of the breast-cancer susceptibility gene BRCA2.
Nature. 378:789–792. 1995. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stratton MR and Rahman N: The emerging
landscape of breast cancer susceptibility. Nat Genet. 40:17–22.
2008. View Article : Google Scholar
|
|
4
|
Sanz DJ, Acedo A, Infante M, Durán M,
Pérez-Cabornero L, Esteban-Cardeñosa E, Lastra E, Pagani F, Miner C
and Velasco EA: A high proportion of DNA variants of BRCA1 and
BRCA2 is associated with aberrant splicing in breast/ovarian cancer
patients. Clin Cancer Res. 16:1957–1967. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chatterjee S and Pal JK: Role of 5′- and
3′-untranslated regions of mRNAs in human diseases. Biol Cell.
101:251–262. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Farrugia DJ, Agarwal MK, Pankratz VS, et
al: Functional assays for classification of BRCA2 variants of
uncertain significance. Cancer Res. 68:3523–3531. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Walker LC, Whiley PJ, Couch FJ, et al:
Detection of splicing aberrations caused by BRCA1 and BRCA2
sequence variants encoding missense substitutions: implications for
prediction of pathogenicity. Hum Mutat. 31:E1484–E1505. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cartegni L, Chew SL and Krainer AR:
Listening to silence and understanding non-sense: exonic mutations
that affect splicing. Nat Rev Genet. 3:285–298. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Spurdle AB, Couch FJ, Hogervorst FB,
Radice P and Sinilnikova OM: Prediction and assessment of splicing
alterations: implications for clinical testing. Hum Mutat.
29:1304–1313. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lu M, Conzen SD, Cole CN and Arrick BA:
Characterization of functional messenger RNA splice variants of
BRCA1 expressed in nonmalignant and tumor-derived breast cells.
Cancer Res. 56:4578–4581. 1996.PubMed/NCBI
|
|
11
|
Wang H, Shao N, Ding QM, Cui J, Reddy ES
and Rao VN: BRCA1 proteins are transported to the nucleus in the
absence of serum and splice variants BRCA1a, BRCA1b are tyrosine
phosphoproteins that associate with E2F, cyclins and cyclin
dependent kinases. Oncogene. 15:143–157. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wilson CA, Payton MN, Elliott GS, Buaas
FW, Cajulis EE, Grosshans D, Ramos L, Reese DM, Slamon DJ and
Calzone FJ: Differential subcellular localization, expression and
biological toxicity of BRCA1 and the splice variant BRCA1-delta11b.
Oncogene. 14:1–16. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Orban TI and Olah E: Expression profiles
of BRCA1 splice variants in asynchronous and in G1/S synchronized
tumor cell lines. Biochem Biophys Res Commun. 280:32–38. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Orban TI and Olah E: Emerging roles of
BRCA1 alternative splicing. Mol Pathol. 56:191–197. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Maniccia AW, Lewis C, Begum N, et al:
Mitochondrial localization, ELK-1 transcriptional regulation and
growth inhibitory functions of BRCA1, BRCA1a, and BRCA1b proteins.
J Cell Physiol. 219:634–641. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Colombo M, Blok MJ, Whiley P, et al:
Comprehensive annotation of splice junctions supports pervasive
alternative splicing at the BRCA1 locus: a report from the ENIGMA
consortium. Hum Mol Genet. 23:3666–3680. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen M and Manley JL: Mechanisms of
alternative splicing regulation: insights from molecular and
genomics approaches. Nat Rev Mol Cell Biol. 10:741–754.
2009.PubMed/NCBI
|
|
18
|
Tesoriero AA, Wong EM, Jenkins MA, Hopper
JL, Brown MA, Chenevix-Trench G, Spurdle AB and Southey MC;
kConFab: Molecular characterization and cancer risk associated with
BRCA1 and BRCA2 splice site variants identified in multiple-case
breast cancer families. Hum Mutat. 26:4952005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Antoniou AC, Durocher F, Smith P, Simard J
and Easton DF; INHERIT BRCAs program members: BRCA1 and BRCA2
mutation predictions using the BOADICEA and BRCAPRO models and
penetrance estimation in high-risk French-Canadian families. Breast
Cancer Res. 8:R32006. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Antoniou AC, Cunningham AP, Peto J, et al:
The BOADICEA model of genetic susceptibility to breast and ovarian
cancers: updates and extensions. Br J Cancer. 98:1457–1466. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
de Garibay GR, Acedo A, García-Casado Z,
et al: Capillary electrophoresis analysis of conventional splicing
assays: IARC analytical and clinical classification of 31BRCA2
genetic variants. Hum Mutat. 35:53–57. 2014. View Article : Google Scholar
|
|
22
|
Houdayer C, Caux-Moncoutier V, Krieger S,
et al: Guidelines for analysis in molecular diagnosis derived from
a set of 327 combined in silico/in vitro studies on BRCA1 and BRCA2
variants. Hum Mutat. 33:1228–1238. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Whiley PJ, de la Hoya M, Thomassen M, et
al: Comparison of mRNA splicing assay protocols across multiple
laboratories: recommendations for best practicein standardized
clinical testing. Clin Chem. 60:341–352. 2014. View Article : Google Scholar
|
|
24
|
Montagna M, Santacatterina M, Torri A,
Menin C, Zullato D, Chieco-Bianchi L and D’Andrea E: Identification
of a 3 kb Alu-mediated BRCA1 gene rearrangement in two
breast/ovarian cancer families. Oncogene. 18:4160–4165. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sevcik J, Falk M, Macurek L, et al:
Expression of human BRCA1Delta17-19 alternative splicing variant
with a truncated BRCT domain in MCF-7 cells results in impaired
assembly of DNA repair complexes and aberrant DNA damage response.
Cell Signal. 25:1186–1193. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mazoyer S: Genomic rearrangements in the
BRCA1 and BRCA2 genes. Hum Mutat. 25:415–422. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bonnet C, Krieger S, Vezain M, et al:
Screening BRCA1 and BRCA2 unclassified variants for splicing
mutations using reverse transcription PCR on patient RNA and an ex
vivo assay based on a splicing reporter minigene. J Med Genet.
45:438–446. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gutiérrez-Enríquez S, Coderch V, Masas M,
Balmaña J and Diez O: The variants BRCA1 IVS6-1G>A and BRCA2
IVS15+1G>A lead to aberrant splicing of the transcripts. Breast
Cancer Res Treat. 117:461–465. 2009. View Article : Google Scholar
|
|
29
|
Perrin-Vidoz L, Sinilnikova OM,
Stoppa-Lyonnet D, Lenoir GM and Mazoyer S: The nonsense-mediated
mRNA decay pathway triggers degradation of most BRCA1 mRNAs bearing
premature termination codons. Hum Mol Genet. 11:2805–2814. 2002.
View Article : Google Scholar : PubMed/NCBI
|