Introduction
Osteoarthritis (OA) is a musculoskeletal disorder
(1) accounting for 3% of total
number of years of living with a disability, particularly in the
elderly (2,3). Knee OA is the most prevalent type of
OA (4). It is characterized by
the progressive degeneration and structural disorder of the
articular cartilage, resulting in loss of joint space, accompanied
by marginal and central new bone formation (5).
Over the past 30 years, a number of cytokines have
been identified to be involved in OA, particularly interleukin
(IL)-1 and IL-6 (6). Oncostatin M
(OSM) is a glycoprotein which belongs to the IL-6 family of
cytokines. It was firstly purified and biochemically characterized
for its anti-proliferative effects on the A375 human melanoma cell
line (7). An increasing amount of
evidence has indicated that OSM participates in a variety of
biological activities, such as differentiation, inflammation,
development and the enhancement of metastatic capacity (8–11).
Osteoblasts and stromal cells isolated from the
subchondral femoral heads of patients with OA express OSM (12). Mice transfected with the OSM gene
have been shown to develop prominent characteristics of arthritis,
such as joint inflammation, bone cell apoptosis, chondrophyte
formation and the depletion of articular cartilage proteoglycans
(13). However, the exact role of
OSM in the development of OA and the underlying mechanism are not
yet fully understood. To the best of our knowledge, this study is
the first to detect the expression of OSM in the synovial tissue of
patients with knee OA. Furthermore, a new signaling pathway was
found to participate in the effects of OSM on MC3T3-E1 cell
proliferation and differentiation. This study may broaden our
understanding of the mechanisms behind the role of OSM in the
development of knee OA.
Materials and methods
Sample collection
The present study was approved by the Ethics
Committee of Beijing Shijitan Hospital, Capital Medical University
and all the patients provided informed written consent prior to
participation in the study. The OA synovial tissue of the knee
joint was obtained from 32 patients with OA who underwent total
knee replacement and arthroscopy. Normal control synovial tissue of
the knee joint was obtained from 12 patients with a discoid
meniscus. The tissues were immediately stored at −80°C until use in
western blot analysis and reverse transcription-quantitative
polymerase chain reaction (RT-qPCR).
Cell culture and transfection
The mouse osteoblast cell line, MC3T3-E1, was
purchased from the American Type Culture Collection (Manassas, VA,
USA). The cells were maintained in Dulbecco’s modified Eagle’s
medium (DMEM) supplemented with 10% heat-inactivated fetal bovine
serum (FBS) (both from Gibco-BRL Life Technologies, Grand Island,
NY, USA) at 37°C in a humidified atmosphere with 5% CO2.
OSM was purchased from R&D Systems (Minneapolis, MN, USA) and
was diluted with phosphate-buffered saline (PBS) into the indicated
concentrations. The full-length cDNA fragment of the mouse Dll1
gene was amplified and cloned into the pEGFP-C1 vector (Clontech
Laboratories, Inc., Mountain View, CA, USA) at the BglII and
BamHI sites to generate the Dll1 overexpression vector. The
Dll1 overexpression vector was transfected into the MC3T3-E1 cells
using Lipofectamine 2000 (Invitrogen Life Technologies, Grand
Island, NY, USA) following the manufacturer’s instructions.
Measurement of alkaline phosphatase (ALP)
activity
ALP activity was determined in the MC3T3-E1 cells in
U/ml sample using an Alkaline Phosphatase Activity Colorimetric
Assay kit (BioVision Inc., Mountain View Milpitas, CA, USA)
according to the manufacturer’s instructions. Briefly, the cells
were homogenized in the assay buffer and centrifuged at 13,000 x g
for 3 min. Different volume of samples were added into a 96-well
plate and assay buffer was used to increase the total volume to 80
μl. A total of 50 μl of the 5 mM pNPP solution was
added to each well followed by incubation for 60 min at room
temperature. A standard curve was created by the addition of 1 mM
pNPP 0, 4, 8, 12, 16 and 20 μl into a 96-well plate in
duplicate to generate 0, 4, 8, 12, 16 and 20 nmol/well pNPP
standard followed by the addition of 10 μl of ALP enzyme
solution to each well containing the pNPP standard. Subsequenlty,
the reactions were terminated by the addition of 20 μl Stop
Solution and the optical density (OD) was measured at 405 nm using
a microplate reader (Ascent 354; Thermo Labsystems, Waltham, MA,
USA).
MTT assay
The MC3T3-E1 cells were seeded into a 96-well plate
and allowed to grow for 24 h. Following treatment with various
concentrations of OSM (5–100 ng/ml) for the appropriate periods of
time, 50 μl MTT solution (Beyotime, Shanghai, China) were
added to the well followed by incubation at 37°C for 4 h. Formazin
granules were dissolved with 150 μl DMSO (Sigma, St. Louis,
MO, USA) and the OD at 570 nm was measured using a microplate
reader (Ascent 354; Thermo Labsystems).
RT-qPCR
Total RNA was isolated using TRIzol reagent
(Invitrogen Life Technologies, Carlsbad, CA, USA). A total of 2
μg of RNA was reverse transcribed using a cDNA synthesis kit
(RevertAid™ First Strand cDNA Synthesis kit; Fermentas, Vilnius,
Lithuania). Quantitative (real-time) PCR was performed using the
SYBR PCR Master Mix (Applied Biosystems, Foster City, CA, USA) on
the Applied Biosystems 7900HT Fast Real-Time PCR system. The
relative expression level was calculated using the comparative Ct
method.
Western blot analysis
Total protein was separated using ice-cold lysis
buffer [1 mM EDTA, 20 mM Tris-HCl (pH 7.5), 10 mg/ml soybean
trypsin inhibitor, 15 mM CHAPS, 0.05% Tween-20 and 10 mM PMSF] and
subjected to 10% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis. The separated proteins were transferred onto a
PVDF membrane (EMD Millipore Corp., Billerica, MA, USA). The
membrane was then blocked with 5% BSA at 4°C overnight and
incubated with antibodies against OSM (Cat. no. sc-50296), Notch
homolog 1 (Notch1; Cat. no. sc-9170), Hes family bHLH transcription
factor 1 (Hes1; Cat. no. sc-25392) (Santa Cruz Biotechnology, Inc.,
Santa Cruz, CA, USA), osteocalcin (OCN; Cat. no. ab93876),
Delta-like 1 (Dll1; Cat. no. ab10554) (Abcam, Cambridge, MA, USA)
and glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Cat. no.
10494-1-AP) (Proteintech Group Inc., Chicago, IL, USA) at 37°C for
2 h. After washing with PBS, the membrane was incubated with
horseradish peroxidase (HRP)-labeled antibody (Cat. no. sc-2004) at
37°C for 1 h. Immunoreactive bands were visualized using a
chemiluminescence-based detection system (ECL Western Blotting
Detection kit; Pierce Biotechnology, Inc., USA).
Statistical analysis
Data were shown as the means ± SD of at least 3
independent studies. The Student’s t-test was used to assess
differences between 2 groups. P-values <0.05 were considered to
indicate statistically significant differences.
Results
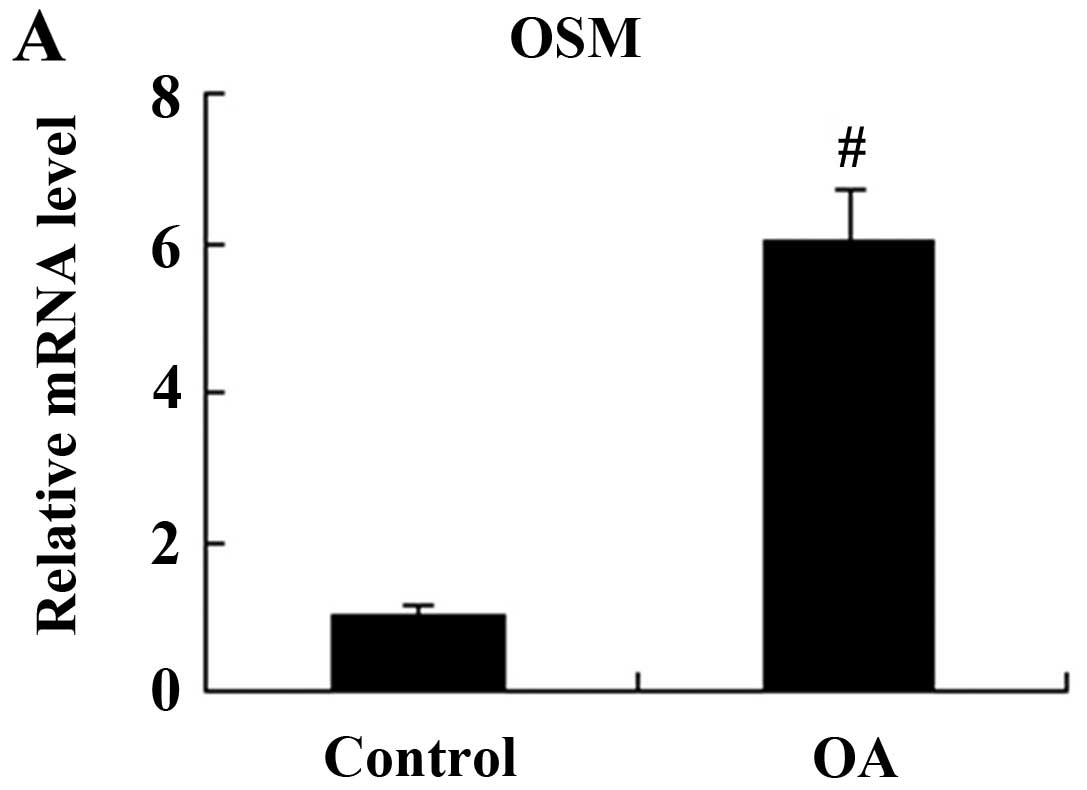
Expression of OSM in the synovial tissue
of the knee joint
OSM mRNA and protein expression in the synovial
tissue of the knee joint was detected by RTqPCR and western blot
analysis. The synovial tissue of the knee joint obtained from
patients with discoid menisci was used as a control (Fig. 1). The relative mRNA expression
level of OSM was significantly increased in the patients with OA
compared with the controls (P<0.01; Fig. 1A). The results from western blot
analysis revealed that the OSM protein expression level was also
higher in the synovial tissue of patients with knee OA compared to
that of the controls (P<0.01; Fig.
1B).
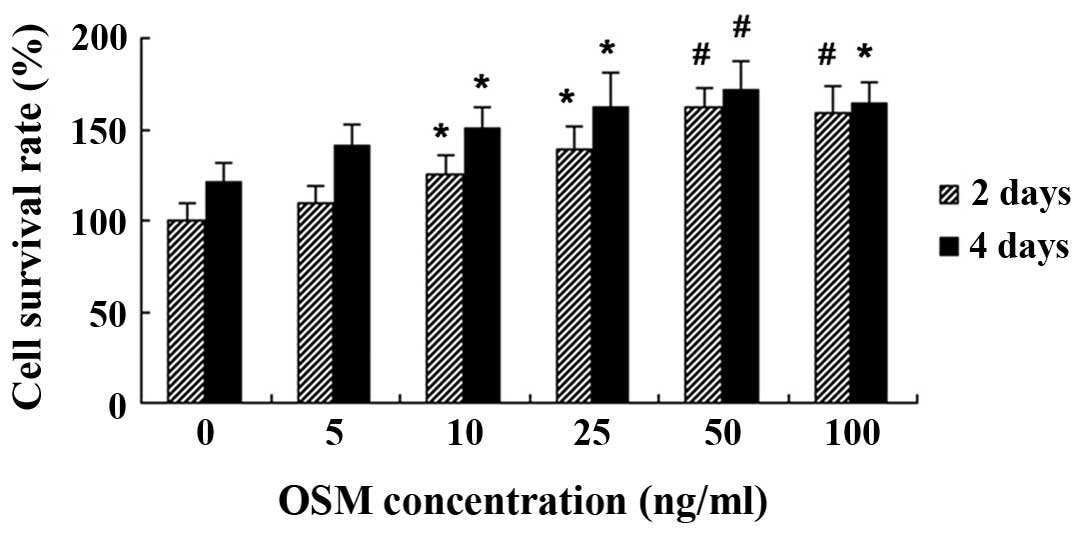
Effect of OSM on MC3T3-E1 cell
proliferation
Following 2 and 4 days of treatment with various
concentrations of OSM, MC3T3-E1 cell proliferation was measured by
MTT assay. We found that treatment with 5 ng/ml OSM did not affect
cell viability following 2 and 4 days of treatment (P>0.05).
However, treatment with OSM at 10–100 ng/ml significantly increased
MC3T3-E1 cell viability following 2 and 4 days of incubation
(Fig. 2).
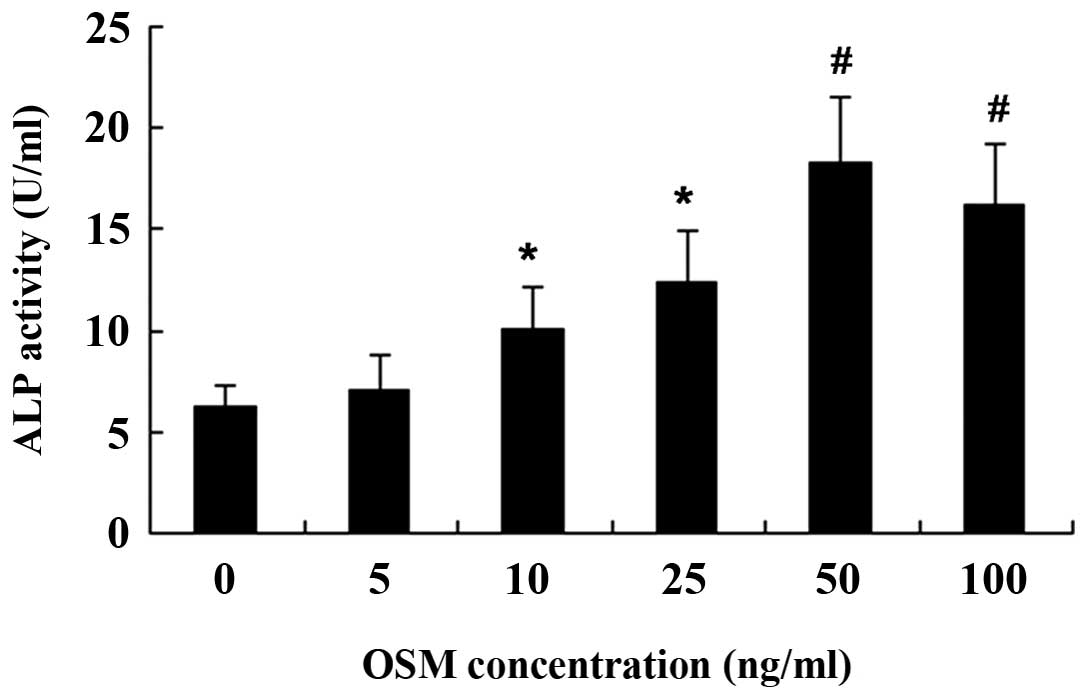
Effect of OSM on MC3T3-E1 cell
differentiation
The MC3T3-E1 cells were incubated for 3 days with
various concentrations of OSM, and the early- and middle-stage
osteogenic markers, ALP activity and OCN expression, respectively,
were then examined. Treatment with OSM did not induce ALP activity
and OCN at the concentration of 5 ng/ml. Treatment with OSM at
10–100 ng/ml induced ALP activity with the maximum effect (peak
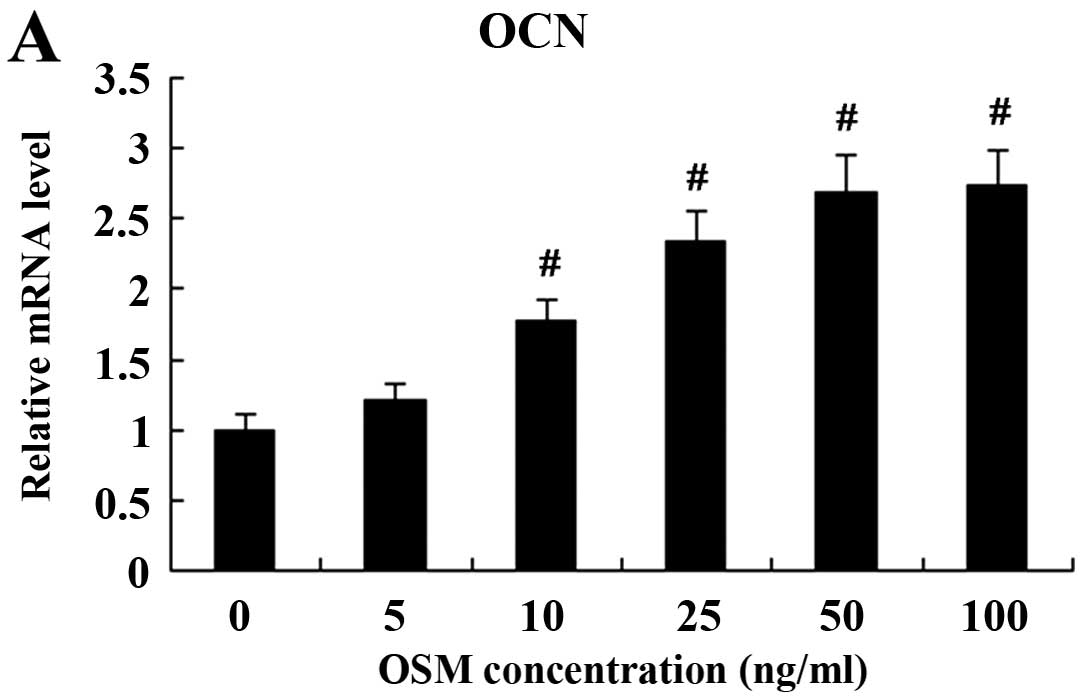
value) being observed at the dose of 50 ng/ml (Fig. 3). OCN mRNA and protein expressin
were also induced following treatment with increasing
concentrations of OSM. The increase in its expression was observed
with OSM at the dose of 10–100 ng/ml (Fig. 4).
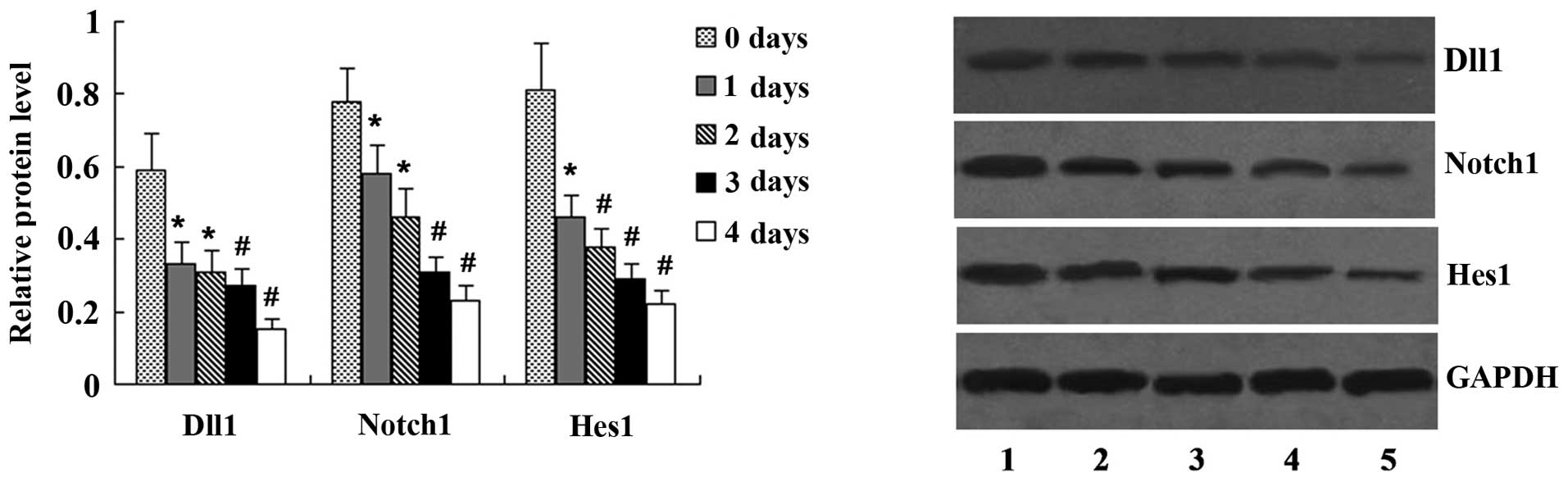
Effect of OSM on the expression of Notch
signaling molecules
The MC3T3-E1 cells were incubated with 50 ng/ml OSM
for 1–4 days, and the expression of Dll1, Notch1 and Hes1 was
determined by western blot analysis. OSM at the concentration of 50
ng/ml inhibited the expression of Dll1, Notch1 and Hes1 following
treatment for 1–4 days compared with the untreated cells (Fig. 5).
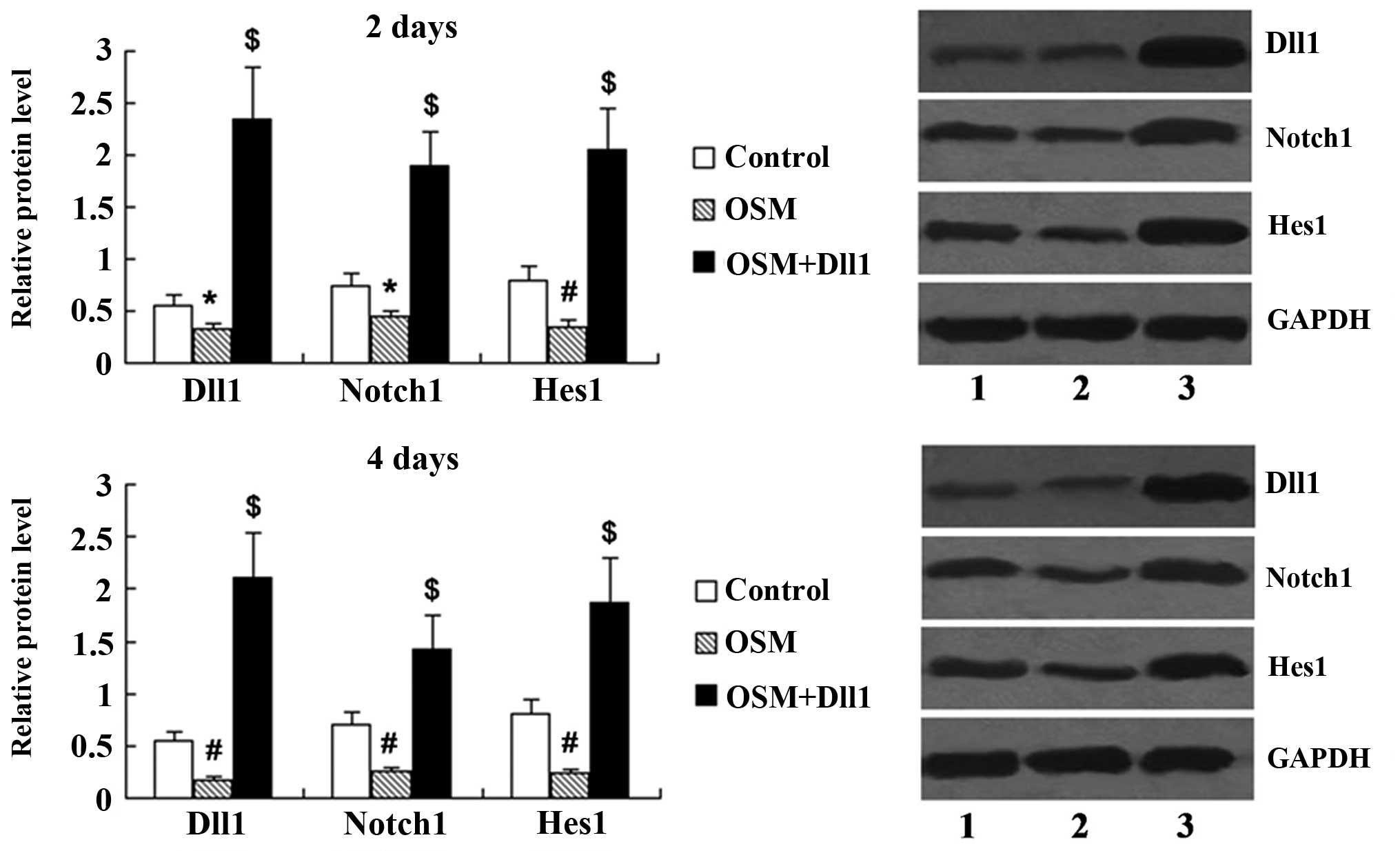
Activation of Notch signaling attenuates
the effects of OSM on MC3T3-E1 cell proliferation
Following transfection with the Dll1 overexpression
vector for 48 h, the expression levels of Dll1, Notch1 and Hes1 in
the MC3T3-E1 cells treated with OSM for 2 and 4 days were
determined by western blot analysis. The protein expression of Dll1
was significantly upregulated in the OSM + Dll1 group compared with
the OSM group (P<0.01; Fig.
6). Notch1 and Hes1 expression was also significantly increased
in the MC3T3-E1 cells transfected with the Dll1 overexpression
vector (P<0.01).
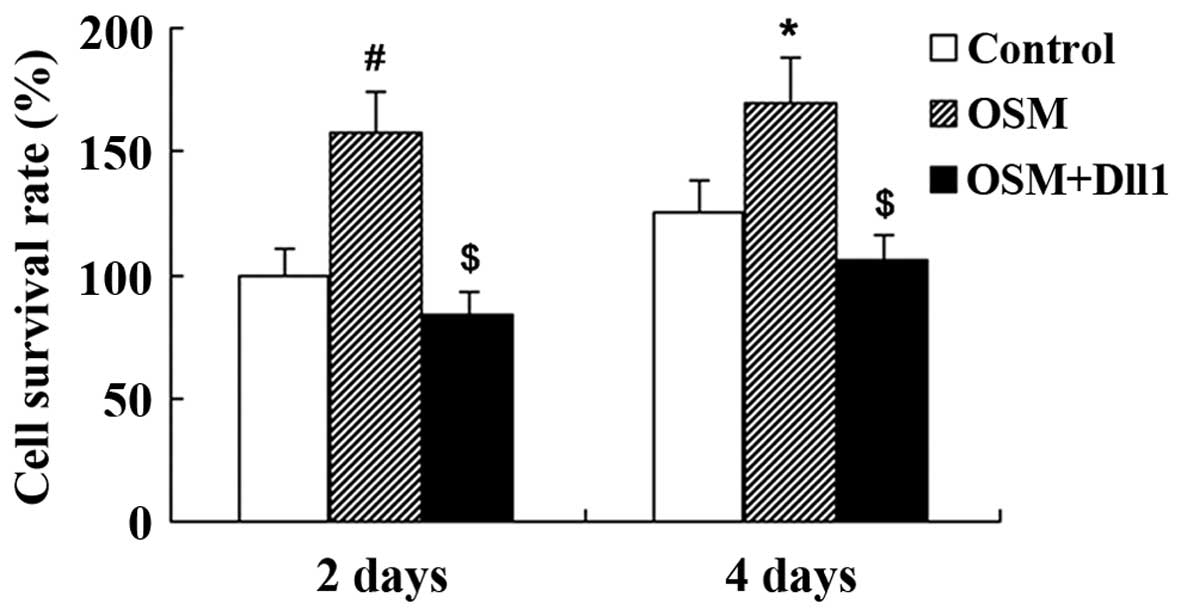
We then investigated whether the activation of Notch
signaling affects the OSM-induced MC3T3-E1 cell proliferation. Cell
viability was assessed by MTT assay following treatment with 50
ng/ml OSM. We found that cell viability was significantly increased
in the OSM-treated group compared with the control group
(P<0.01). However, the activation of Notch signaling led to a
decrease in OSM-induced cell proliferation; cell viability was
significantly decreased in the OSM + Dll1 group compared with the
OSM group (P<0.01; Fig.
7).
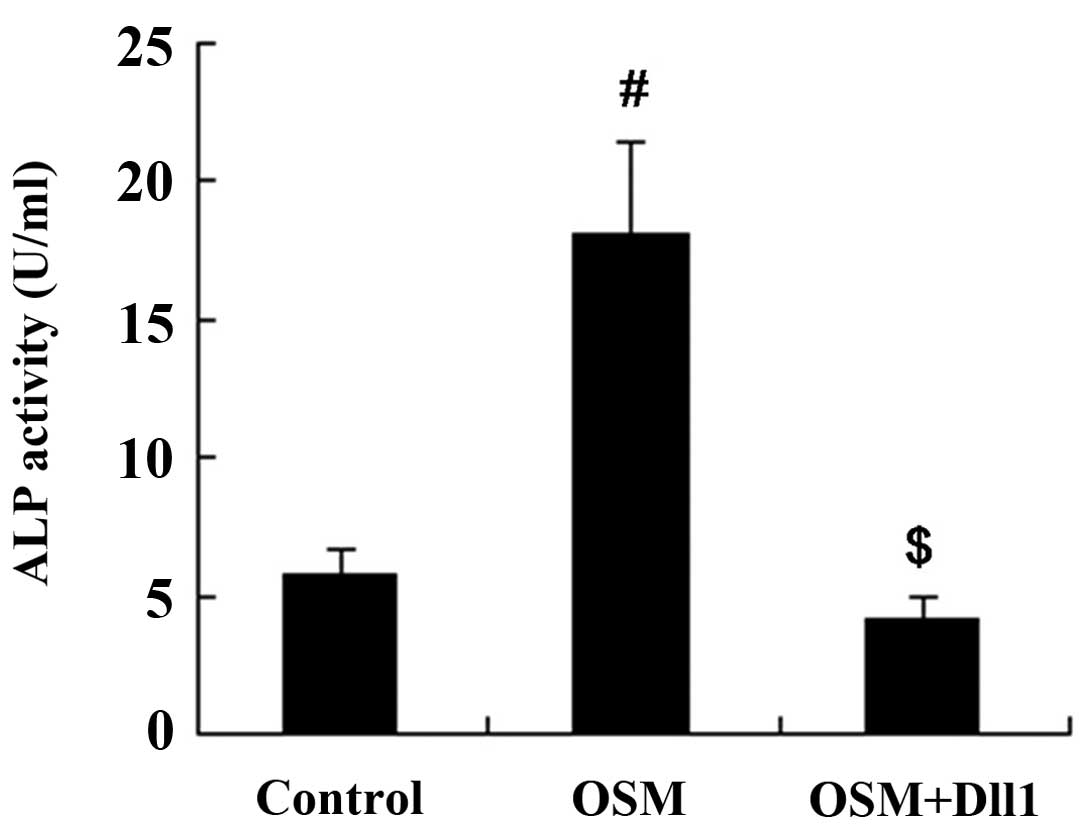
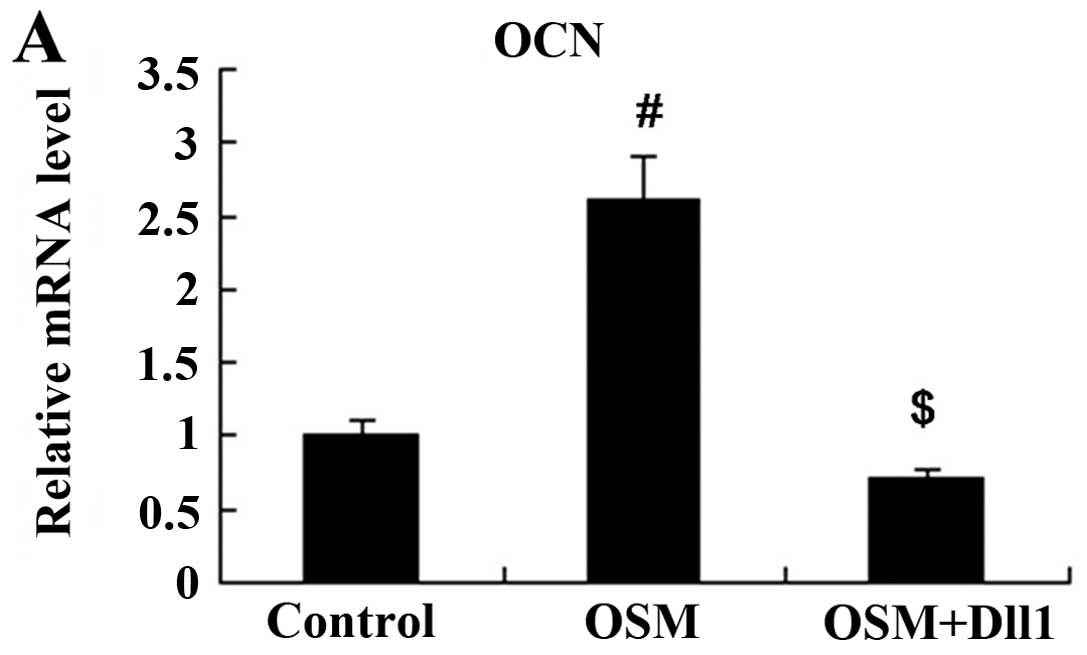
Activation of Notch signaling attenuates
the effects of OSM on MC3T3-E1 cell differentiation
The MC3T3-E1 cells were treated with 50 ng/ml OSM
for 3 days following transfection with the Dll1 overexpression
vector. Subsequently, the cells were harvested for the measurement
of ALP activity and OCN expression. The results revealed that both
ALP activity and OCN expression in the MC3T3-E1 cells were
increased following treatment with OSM. However, the activation of
Notch signaling attenuated the effects of OSM on MC3T3-E1 cell
differentiation; compared with the OSM group, both ALP activity and
OCN expression were decreased in the OSM + Dll1 group (P<0.01;
Figs. 8 and 9).
Discussion
Previous studies have suggested that OSM contributes
to the pathogenesis of OA. It has been demonstrated that OSM
over-expression in the mouse knee joint induces changes to the
joint that resemble OA, including cartilage destruction and
periosteal bone formation similar to osteophytes (14,15). In the present study, we found that
OSM was detected in the synovial tissue of the knee joint, and the
expression level of OSM was higher in patients with knee OA
compared with the controls. This suggests that OSM contributes to
the development of knee OA.
OSM has been shown to stimulate osteoblast
differentiation by stromal cells and to reduce the ability of
stromal cells to differentiate into adipocytes (16,17). It also drives the formation of
osteoclasts, particularly under pathological conditions. It has
been documented that osteoblasts play essential roles in bone
remodeling in arthritis (18). In
the present study, we performed in vitro experiments using
MC3T3-E1 cells (mouse osteoblasts) to investigate the role of OSM
on MC3T3-E1 cell proliferation and differentiation. It was
demonstrated that OSM affected MC3T3-E1 cell proliferation in a
concentration-dependent manner. The results of specific ALP
activity and OCN, as indicators of osteogenic differentiation
revealed that OSM induced MC3T3-E1 differentiation. These findings
demonstrate that OSM induces bone formation by increasing
osteoblast cell proliferation and differentiation.
Notch signaling is a highly conserved pathway
(19) regulated by interactions
between neighboring cells. It is crucial for the regulation of a
number of cellular processes, including proliferation,
differentiation, apoptosis and cell death during embryogenesis, as
well as the development and renewal of adult tissues (20,21). Notch signaling has also been
implicated in regulating articular cartilage homeostasis during
adult life (22,23). It has been demonstrated that
several Notch signaling molecules are abundantly expressed in OA
(23), and Notch signaling is
activated in OA cartilage (24).
Our findings suggest a connection between OSM and Notch signaling.
We found that the expression of Notch ligand, receptor and target
gene, including Dll1, Notch1 and Hes1 was decreased following
treatment with OSM in a time-dependent manner in the MC3T3-E1
cells. We then investigated the hypothesis that Notch signaling
plays a role in the effects of OSM on MC3T3-E1 cell proliferation
and differentiation.
Dll1 is able to activate the Notch1 receptor,
leading to the activation of endogenous Hes1 genes (25) and several studies have
demonstrated the inhibitory effects of Notch1 on osteoblastic cell
differentiation (26–28). In the present study, the Dll1
overexpression vector was used to activate Notch signaling and the
results revealed that the activation of Notch signaling attenuated
the effects of OSM on MC3T3-E1 cell proliferation and
differentiation.
In conclusion, elevated levels of OSM in the
synovial tissue may induce bone formation by increasing osteoblast
cell proliferation and differentiation. OSM exerts an inhibitory
effect to Notch signaling, and the OSM-induced MC3T3-E1
proliferation and differentiation may be reversed by the activation
of Notch signaling. It may prove useful if future directions in the
research and treatment of OA focus on the upstream and downstream
molecules of Notch signaling that modulate the initiation and
progression of OA.
References
|
1
|
Quilty B, Tucker M, Campbell R and Dieppe
P: Physiotherapy, including quadriceps exercises and patellar
taping, for knee osteoarthritis with predominant patello-femoral
joint involvement: Randomized controlled trial. J Rheumatol.
30:1311–1317. 2003.PubMed/NCBI
|
|
2
|
Guccione AA, Felson DT, Anderson JJ,
Anthony JM, Zhang Y, Wilson PW, Kelly-Hayes M, Wolf PA, Kreger BE
and Kannel WB: The effects of specific medical conditions on the
functional limitations of elders in the Framingham Study. Am J
Public Health. 84:351–358. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Woolf AD and Pfleger B: Burden of major
musculoskeletal conditions. Bull World Health Organ. 81:646–656.
2003.
|
|
4
|
Parmet S, Lynm C and Glass RM: JAMA
patient page. Osteoarthritis of the knee. JAMA. 289:10682003.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lin CW, Taylor D, Bierma-Zeinstra SM and
Maher CG: Exercise for osteoarthritis of the knee. Phys Ther.
90:839–842. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fernandes JC, Martel-Pelletier J and
Pelletier JP: The role of cytokines in osteoarthritis
pathophysiology. Biorheology. 39:237–246. 2002.PubMed/NCBI
|
|
7
|
Zarling JM, Shoyab M, Marquardt H, Hanson
MB, Lioubin MN and Todaro GJ: Oncostatin M: A growth regulator
produced by differentiated histiocytic lymphoma cells. Proc Natl
Acad Sci USA. 83:9739–9743. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tanaka M and Miyajima A: Oncostatin M, a
multifunctional cytokine. Rev Physiol Biochem Pharmacol. 149:39–52.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Brounais B, David E, Chipoy C, et al: Long
term oncostatin M treatment induces an osteocyte-like
differentiation on osteosarcoma and calvaria cells. Bone.
44:830–839. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jorcyk CL, Holzer RG and Ryan RE:
Oncostatin M induces cell detachment and enhances the metastatic
capacity of T-47D human breast carcinoma cells. Cytokine.
33:323–336. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Brounais B, Chipoy C, Mori K, Charrier C,
Battaglia S, Pilet P, Richards CD, Heymann D, Rédini F and
Blanchard F: Oncostatin M induces bone loss and sensitizes rat
osteosarcoma to the antitumor effect of Midostaurin in vivo. Clin
Cancer Res. 14:5400–5409. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lisignoli G, Piacentini A, Toneguzzi S,
Grassi F, Cocchini B, Ferruzzi A, Gualtieri G and Facchini A:
Osteoblasts and stromal cells isolated from femora in rheumatoid
arthritis (RA) and osteoarthritis (OA) patients express IL-11,
leukaemia inhibitory factor and oncostatin M. Clin Exp Immunol.
119:346–353. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
de Hooge AS, van de Loo FA, Bennink MB,
Arntz OJ, Fiselier TJ, Franssen MJ, Joosten LA, Van Lent PL,
Richards CD and van den Berg WB: Growth plate damage, a feature of
juvenile idiopathic arthritis, can be induced by adenoviral gene
transfer of oncostatin M: A comparative study in gene-deficient
mice. Arthritis Rheum. 48:1750–1761. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Langdon C, Kerr C, Hassen M, Hara T,
Arsenault AL and Richards CD: Murine oncostatin M stimulates mouse
synovial fibroblasts in vitro and induces inflammation and
destruction in mouse joints in vivo. Am J Pathol. 157:1187–1196.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
de Hooge AS, van de Loo FA, Bennink MB, de
Jong DS, Arntz OJ, Lubberts E, Richards CD and vandDen Berg WB:
Adenoviral transfer of murine oncostatin M elicits periosteal bone
apposition in knee joints of mice, despite synovial inflammation
and up-regulated expression of interleukin-6 and receptor activator
of nuclear factor-kappa B ligand. Am J Pathol. 160:1733–1743. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Walker EC, McGregor NE, Poulton IJ,
Pompolo S, Allan EH, Quinn JM, Gillespie MT, Martin TJ and Sims NA:
Cardiotrophin-1 is an osteoclast-derived stimulus of bone formation
required for normal bone remodeling. J Bone Miner Res.
23:2025–2032. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Walker EC, McGregor NE, Poulton IJ, et al:
Oncostatin M promotes bone formation independently of resorption
when signaling through leukemia inhibitory factor receptor in mice.
J Clin Invest. 120:582–592. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lisignoli G, Toneguzzi S, Pozzi C,
Piacentini A, Riccio M, Ferruzzi A, Gualtieri G and Facchini A:
Proinflammatory cytokines and chemokine production and expression
by human osteoblasts isolated from patients with rheumatoid
arthritis and osteoarthritis. J Rheumatol. 26:791–799.
1999.PubMed/NCBI
|
|
19
|
Richards GS and Degnan BM: The dawn of
developmental signaling in the metazoa. Cold Spring Harb Symp Quant
Biol. 74:81–90. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Calvi LM, Adams GB, Weibrecht KW, et al:
Osteoblastic cells regulate the haematopoietic stem cell niche.
Nature. 425:841–846. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yoon K and Gaiano N: Notch signaling in
the mammalian central nervous system: Insights from mouse mutants.
Nat Neurosci. 8:709–715. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sassi N, Laadhar L, Driss M,
Kallel-Sellami M, Sellami S and Makni S: The role of the Notch
pathway in healthy and osteoarthritic articular cartilage: From
experimental models to ex vivo studies. Arthritis Res Ther.
13:2082011. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Karlsson C, Brantsing C, Egell S and
Lindahl A: Notch1, Jagged1, and HES5 are abundantly expressed in
osteoarthritis. Cells Tissues Organs. 188:287–298. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sassi N, Laadhar L, Mahjoub M, Driss M,
Zitouni M, Benromdhane K, Makni S and Sellami S: Expression of
Notch family members in cultured murine articular chondrocytes.
Biotech Histochem. 84:313–320. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jarriault S, Le Bail O, Hirsinger E,
Pourquié O, Logeat F, Strong CF, Brou C, Seidah NG and Israël A:
Delta-1 activation of notch-1 signaling results in HES-1
transactivation. Mol Cell Biol. 18:7423–7431. 1998.PubMed/NCBI
|
|
26
|
Sciaudone M, Gazzerro E, Priest L, Delany
AM and Canalis E: Notch 1 impairs osteoblastic cell
differentiation. Endocrinology. 144:5631–5639. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zamurovic N, Cappellen D, Rohner D and
Susa M: Coordinated activation of notch, Wnt, and transforming
growth factor-beta signaling pathways in bone morphogenic protein
2-induced osteogenesis. Notch target gene Hey1 inhibits
mineralization and Runx2 transcriptional activity. J Biol Chem.
279:37704–37715. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bai S, Kopan R, Zou W, Hilton MJ, Ong CT,
Long F, Ross FP and Teitelbaum SL: NOTCH1 regulates
osteoclastogenesis directly in osteoclast precursors and indirectly
via osteoblast lineage cells. J Biol Chem. 283:6509–6518. 2008.
View Article : Google Scholar
|