Introduction
Bronchopulmonary dysplasia (BPD) is a serious
respiratory complication, affecting premature infants. In recent
years, the administration of antenatal steroids and postnatal
pulmonary surfactants, coupled with the advancements in rescue
technologies for very low birth weight and extremely low birth
weight infants, has meant that the pathological characteristics of
BPD have changed so now the disease is characterized as an
alveolarization disorder with microvascular dysplasia (1). The reported incidence of BPD is 42%
for premature infants with a birth weight between 501–750 g and 25%
for those with a birth weight between 751–1,000 g (2). Patients with BPD often have an
increased risk of hospital readmission as a result of developing
respiratory disorders after discharge and a greater probability of
requiring drug intervention (3),
and they are also more susceptible to nervous system developmental
disorders involving recognition, speech, motion and behavioural
problems (4).
The lungs arise from the anterior foregut endoderm,
and normal human lung development is commonly divided into five
stages: the embryonic stage (4–7 weeks post-conception),
pseudoglandular stage (5–17 weeks post-conception), canalicular
stage (16–24 weeks post-conception), saccular stage (24–35 weeks
post-conception) and alveolar stage (36 weeks post-conception to 2
years postnatal). During the final stage, the secondary septa
divide the saccular units, increasing the alveolar number and
reducing the alveolar size. The majority of the gas exchange
surface is formed during this stage. The alveolar stage is a
critical stage in the development of BPD (5,6).
It has been demonstrated that the disruption of the
formation of secondary septa and arrested alveolarization in BPD
may be caused by the abnormal accumulation of extracellular matrix
components (including elastins) and angiodysplasia (7,8).
These studies have only partially elucidated the mechanisms
responsible for alveolarization arrest in BPD and thus, further
investigations are required to fully elucidate the mechanisms
responsible for the development of BPD in order to establish
effective treatment methods.
MicroRNAs (miRNAs or miRs) are a group of endogenous
non-coding RNAs that are 22–25 nucleotides (nt) in length, and
their 5′-seed sequence specifically binds to the 3′-UTR base
sequence of target mRNAs to cause the degradation or translational
suppression of target mRNAs, and thus they play a role in
regulating gene expression at the post-transcriptional level
(9). Studies have demonstrated
that miRNAs participate widely in various life processes, such as
cell differentiation, proliferation and apoptosis, as well as organ
development, tumorigenesis and immune response, and are highly
conserved among species (10,11). It has been confirmed that miRNA
expression during lung development changes dynamically, indicating
that miRNAs participate in the regulation of lung development
(12). miRNAs have also been
found to play an important role in the development of lung diseases
such as asthma, idiopathic pulmonary fibrosis and lung cancer
(13–15).
Studies have revealed the functions of miRNAs in
embryonic lung development. miR-17 and its homologous genes miR-20a
and miR-106b were found to have high levels of expression during
the pseudoglandular stage of lung development (16). In mice with an overexpression of
the miR-17–92 cluster, the proliferation of pulmonary epithelial
progenitors takes place, but cell differentiation is suppressed,
demonstrating an abnormal lethal phenotype (17). The miR-17 family alters E-cadherin
expression and distribution, as well as β-catenin activity by
regulating STAT3 and MAPK14, which eventually increases the number
and area of terminal branches (18). Navarro et al (19) found that let-7 expression was
downregulated in the pseudoglandular stage of lung development.
Let-7 suppresses the Ras-Raf-MAP kinase pathway and thus, causes a
decrease in cell viability and the proliferation rate. Bhaskaran
et al (20) found the
highest expression level of miR-127 during the late stage of rat
embryonic lung development (E21, saccular stage) and that miR-127
expression shifted from mesenchymal cells to epithelial cells
during embryonic lung development. In lung tissue cultures, the
overexpression of miR-127 resulted in a defect in pulmonary branch
formation. Zhang et al (21) observed that A549 cell
proliferation was significantly suppressed following transfection
with miR-127. Yet, little is known about the role of miRNAs in the
regulation of postnatal lung development and in the development of
BPD.
Thus, in the present study, we investigated the
mechanisms through which miRNAs regulate lung development after
birth, as well as the role of miRNAs in the development of
bronchopulmonary dysplasia (BPD) using a rat model of BPD induced
by hyperoxia.
Materials and methods
Animal models
All animal procedures were approved by the Committee
on the Ethics of Animal Experiments of China Medical University,
Shenyang, China. All surgeries were performed under chloral hydrate
anesthesia, and all efforts were made to minimize animal
suffering.
Full-term newborn Wistar rats (purchased from the
Experimental Animal Center of Shengjing Hospital of China Medical
University, Shenyang, China) were randomly assigned to either the
model group (n=45) or the control group (n=45) within 12 h after
birth. The neonatal rats in the model group were kept in a glass
chamber with an oxygen concentration between 60–85%, monitored
continuously by an oxygen analyzer. The newborn rats in the control
group inhaled fresh air (21% oxygen). All other conditions and
control factors were the same as those of the model group. The
nursing rats were exchanged between the 2 groups every 24 h to
avoid oxygen toxicity.
Lung tissue preparation
On postnatal days 3, 7 and 14, 15 pups from both
groups were anesthetized by intraperitoneal injection of 10%
chloral hydrate. The chest of the rats was then opened, and whole
lungs were collected. The inferior lobe of the right lung was fixed
in 4% paraformaldehyde for hematoxylin and eosin (H&E)
staining. The remaining lung tissue was preserved in liquid
nitrogen: the left lungs were used for mRNA detection and
microarray analysis, while the right lungs were used for western
blot analysis. From each group and at each time point, 10 inferior
lobes from the right lungs (one from each litter) were randomly
selected for morphological evaluation. Five left lungs were
randomly selected for the measurement of mRNA expression, while 3
left lungs were selected for microarray analysis and 5 right lungs
were selected for the measurement of protein expression.
Lung histology
The inferior lobes from the right lungs were fixed
in 4% paraformaldehyde for 24 h, and then dehydrated in gradient
ethanol, vitrified in xylene and embedded in paraffin. The
paraffin-embedded sections (4-µm-thick) were stained with
H&E. Morphological changes were evaluated using an optical
microscope (×40 magnification; AX70+U-PHOTO; Olympus, Tokyo,
Japan). Ten fields were randomly selected for analysis from each
section. The radial alveolar count (RAC) was counted with a method
developed by Emery and Mithal, as described in our previous study
(22), to assess the level of
alveolarization. The counting was carried out by 2 independent
pathologists who were blinded to the experimental design.
Western blot analysis
The protein expression levels were determined using
rabbit polyclonal anti-proliferating cell nuclear antigen (PCNA)
antibody (ab2426, 1:200 dilution) and anti-platelet endothelial
cell adhesion molecule-1 (PECAM-1, also known CD31) antibody
(ab32457, 1:1,000 dilution) (both from Abcam, Cambridge, UK).
Briefly, total protein extracted from lung tissue was quantified
using a BCA protein assay kit, 20 µl of protein was then
loaded onto a 12% SDS-PAGE gel and transferred onto polyvinylidene
difluoride membranes (Millipore, Billerica, MA, USA). The membranes
were blocked with 5% skimmed milk for 1 h, then incubated with
anti-PCNA or anti-CD31 antibody diluted in PBS overnight at 4°C.
After being washed in Tris-buffered saline + 1% Tween-20 (TBST),
the membranes were incubated for 2 h with horseradish
peroxidase-conjugated secondary antibody and then imaged using
enhanced chemiluminescence reagents. The density of the protein
bands was analyzed using ImageJ software. GAPDH was used as an
internal control.
RNA extraction
Total RNA was isolated following the manufacturer's
instructions using TRIzol reagent (Invitrogen, Carlsbad, CA, USA)
and the miRNeasy mini kit (Qiagen, Hilden, Germany). It was then
qualified and quantified using a NanoDrop spectrophotometer
(ND-1000; NanoDrop Technologies, Wilmington, DE, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
A total of 1 µg of RNA was
reverse-transcribed into cDNA using the PrimeScript™ RT reagent kit
(Takara Biotechnology, Dalian, China), according to the
manufacturer's instructions. qPCR was performed on a LightCycler
(Applied Biosystems 7500 Fast Real-Time PCR System; Applied
Biosystems, Carlsbad, CA, USA). Appropriate primers were designed
using Primer Premier 5.0 software (Premier Biosoft International,
Palo Alto, CA, USA) and the primer sequences are listed in Table I. The reaction was carried out at
95°C for 30 sec, 60°C for 34 sec, and 72°C for 1 min for 40 cycles.
The relative transcript levels were normalized to GAPDH and
evaluated using the ΔΔCT method.
 | Table ISequences of primers used for
RT-qPCR. |
Table I
Sequences of primers used for
RT-qPCR.
| Gene name | Primer
sequences |
|---|
| CD31 | F:
CTGGGAGGTATCGAATGGGC |
| R:
CCCGAGACTGAGGAATGACG |
| PCNA | F:
TAAGGGCTGAAGATAATGCTGAT |
| R:
CCTGTTCTGGGATTCCAAGTT |
| GAPDH | F:
AGACAGCCGCATCTTCTTGT |
| R:
CTTGCCGTGGGTAGAGTCAT |
miRNA microarray
We used the seventh generation of the miRCURY™ LNA
array (v.18.0) (Exiqon, Vedbaek, Denmark), which covered all rat
miRNAs in the miRBase 18.0, to detect the expression of miRNAs in
rat lungs.
RNA labeling
Total RNA was labeled with the miRCURY™ Hy3™/Hy5™
Power labeling kit (Exiqon) according to the manufacturer's
instructions. Briefly, 1 µg of RNA was added to the mixture
of 1.0 µl of CIP buffer and 1.0 µl of CIP (Exiqon),
incubated for 30 min at 37°C and terminated by incubation for 5 min
at 95°C. Subsequently, 3.0 µl of labeling buffer, 1.5
µl of fluorescent label (Hy3™), 2.0 µl of DMSO and
2.0 µl of T4 RNA ligase were added to the mixture. The
labeling reaction was incubated for 1 h at 16°C and terminated by
incubation for 15 min at 65°C.
Array hybridization
The Hy3™-labeled samples were hybridized with the
miRCURY™ LNA array (v.18.0), according to the manufacturer's
instructions. A total of 25 µl of samples, mixed with the
equivalent amount of hybridization buffer, was first denatured for
2 min at 95°C, incubated on ice for 2 min, and then hybridized to
the microarray for 16–20 h at 56°C. The 12-Bay Hybridization system
(Nimblegen Systems, Inc., Madison, WI, USA) controls active mixing
action and temperature, enabling uniform hybridization and an
enhanced signal. Subsequently, the slides were washed several times
with the wash buffer kit (Exiqon), dried by centrifugation at 400
rpm for 5 min, and then scanned using an Axon GenePix 4000B
microarray scanner (Axon Instruments, Foster City, CA, USA).
Data analysis
Grid alignment and data extraction were carried out
using GenePix Pro 6.0 software (Axon Instruments). Replicated
miRNAs were averaged, and miRNAs with intensities of ≥30 were
selected from all the samples for calculation of the normalization
factor. Data were normalized with a median normalization.
Significantly differentially expressed miRNAs between the 2 groups
were then confirmed with volcano plot filtering. Hierarchical
clustering was accomplished with MEV software (v4.6, TIGR).
miRNA target prediction
The significantly differentially expressed miRNAs
were ran through 3 online databases, miRBase (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/),
miRanda (http://www.microrna.org/microrna/home.do) and miRDB
(http://mirdb.org/miRDB/) for target prediction.
The overlapping section was obtained from these databases.
Statistical analysis
We used SPSS17.0 software for statistical analysis.
Values are presented as the means ± standard deviation (SD);
statistical significance was determined using the Student's t-test.
A P-value of <0.05 was considered to indicate a statistically
significant difference.
Results
Histology
On postnatal day 3, the alveolar wall in the model
group was slightly thicker than that of the control group (Fig. 1A and B). By day 7, the number of
alveoli had significantly decreased, and the formation of the
secondary septa was reduced; the alveolar size was also slightly
increased in the model group (Fig. 1C
and D). On day 14, the following changes between the 2 groups
became more apparent: thickening of the alveolar wall, a decrease
in the number of alveoli, reduced numbers of secondary septa, and
alveolar structural abnormalities (Fig. 1E and F). Compared with the control
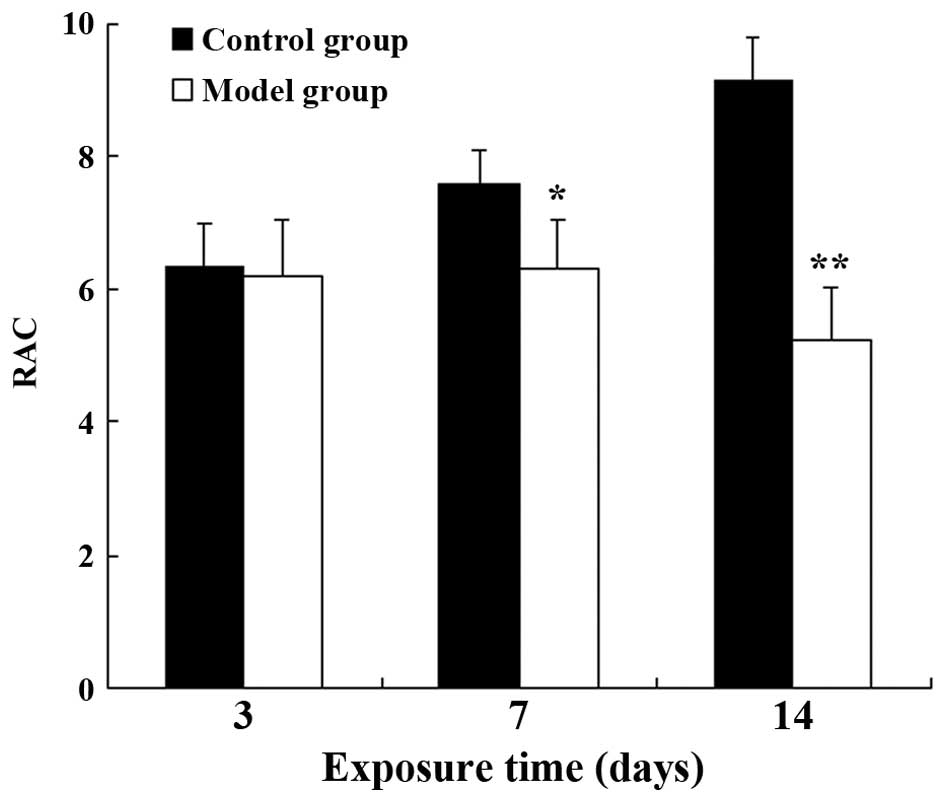
group, the RAC value in the BPD model group had markedly decreased
by postnatal day 7 (P<0.05), and the difference between the 2
groups was even greater by day 14 (P<0.01) (Fig. 2). These results indicated that the
formation of secondary septa was impaired and that alveolarization
was suppressed in the rats in the model group, indicative of
BPD.
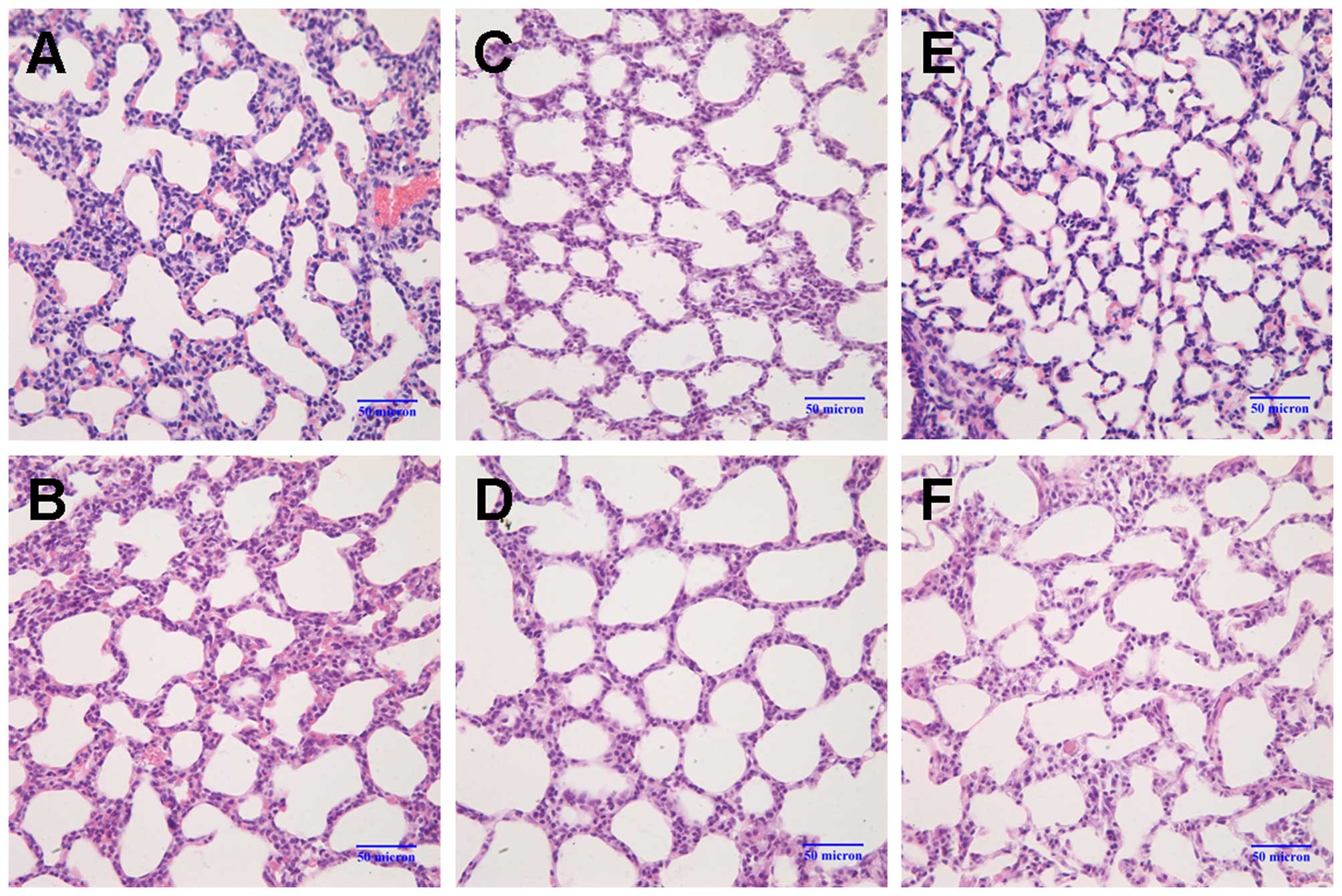 | Figure 1Changes in lung morphology. Lung
sections were stained with H&E and images were captured under a
light microscope (magnification, ×40). (A andB) On postnatal day 3,
the alveolar walls were slightly thicker in the model group (M3d)
compared to the control group (C3d). (C and D) On postnatal day 7,
the number of alveoli had markedly decreased, secondary septa
formation was reduced, and the alveolar size was slightly increased
in the model group (M7d) compared to the control group (C7d). (E
and F) On postnatal day 14, the following changes between the 2
groups became more apparent: thickening of the alveolar wall,
decrease in the number of alveoli, reduced numbers of secondary
septa, and alveolar structural abnormalities (control group, C14d;
model group, M14d). Model group refers to the bronchopulmonary
dysplasia (BPD) group. |
Expression of PCNA and CD31 in rat lung
tissue
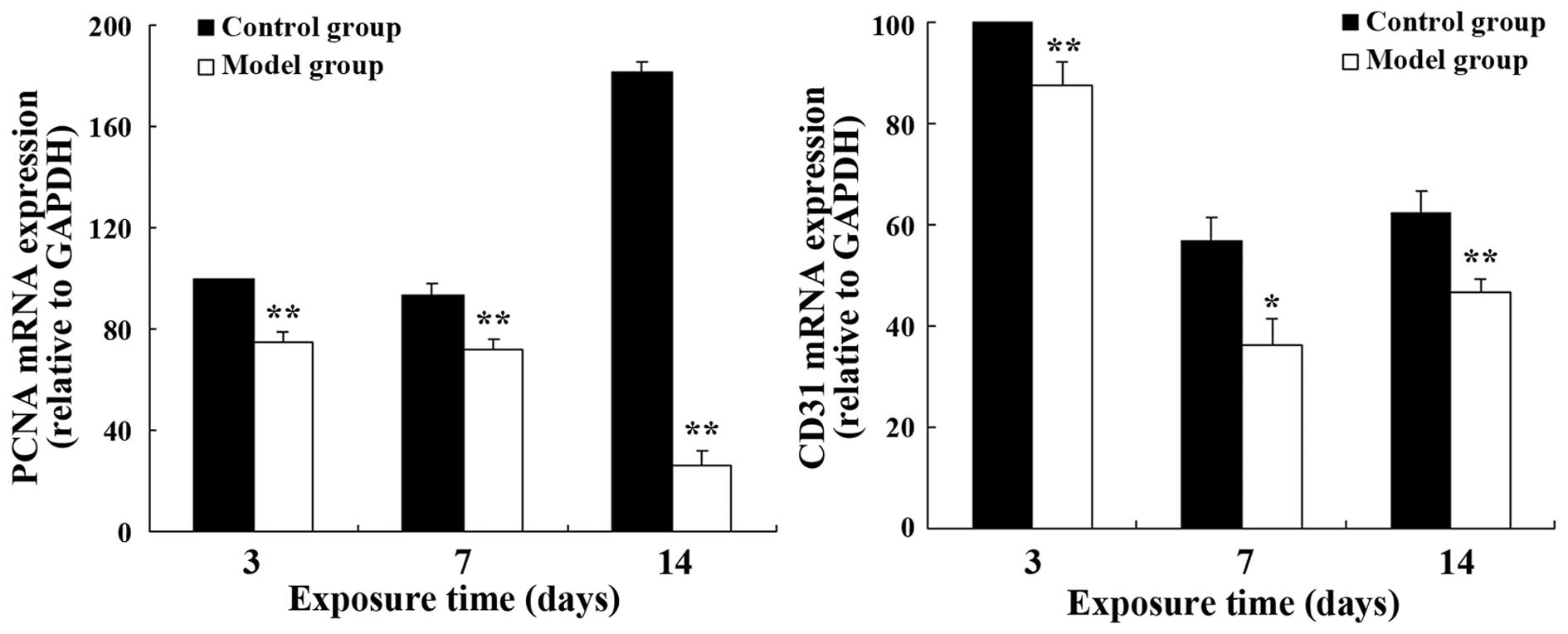
The mRNA and protein expression levels of PCNA and
CD31 in the model group were significantly lower than those in the
control group on postnatal days 3, 7 and 14 (CD31: P<0.05 on day
7; P<0.01 at the other time points; PCNA: P<0.05 at all time
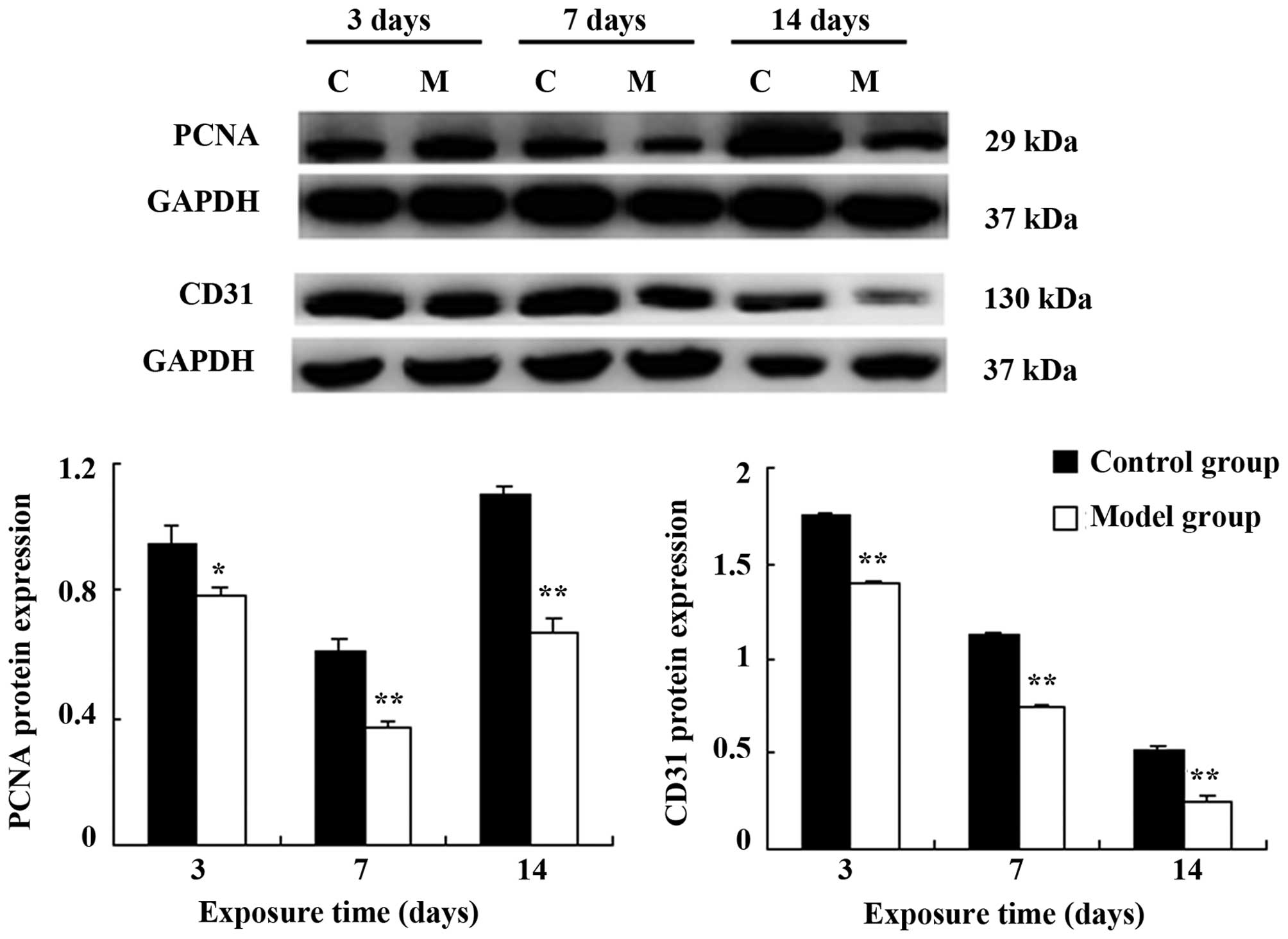
points) (Fig. 3). A similar
effect was observed in the protein expression levels (PCNA:
P<0.05 on day 3; P<0.01 at the other time points; CD31:
P<0.05 at all time points) (Fig.
4).
miRNA expression profiles in rat lung
tissue
A total of 50 differentially expressed miRNAs passed
the fold change and t-test filter between the 2 groups (fold change
>2.0; P<0.05), and the upregulated and downregulated miRNAs
are listed in Tables II and
III, respectively.
 | Table IIDifferentially expressed and
upregulated miRNAs at 3 time points between the 2 groups. |
Table II
Differentially expressed and
upregulated miRNAs at 3 time points between the 2 groups.
M3d vs. C3d
| M7d vs. C7d
| M14d vs. C14d
|
|---|
| Name | FC | Name | FC | Name | FC |
|---|
| rno-miR-490-3p | 2.05 |
rno-miR-3584-3p | 6.38 | rno-miR-34c-3p | 2.25 |
|
rno-miR-1193-3p | 5.06 | | | rno-let-7b-5p | 2.25 |
| | | |
rno-miR-3068-5p | 7.76 |
| | | | rno-miR-872-5p | 6.49 |
| | | | rno-miR-183-5p | 2.05 |
| | | | rno-miR-33-5p | 3.80 |
| | | | rno-miR-182 | 2.70 |
| | | | rno-miR-322-3p | 5.31 |
| | | | rno-miR-340-3p | 3.05 |
| | | | rno-miR-142-3p | 2.42 |
| | | | rno-miR-141-5p | 5.63 |
| | | | rno-miR-96-5p | 5.12 |
| | | | rno-let-7f-5p | 5.09 |
| | | | rno-miR-15b-5p | 7.81 |
| | | |
rno-miR-449a-5p | 4.84 |
| | | | rno-miR-22-5p | 6.62 |
| | | | rno-miR-362-3p | 49.02 |
| | | |
rno-miR-301a-3p | 4.08 |
| | | | rno-miR-365-3p | 3.03 |
 | Table IIIDifferentially expressed and
downregulated miRNAs at 3 time points between the 2 groups. |
Table III
Differentially expressed and
downregulated miRNAs at 3 time points between the 2 groups.
M3d vs. C3d
| M7d vs. C7d
| M14d vs. C14d
|
|---|
| Name | FC | Name | Name | FC | Name |
|---|
| rno-miR-377-3p | 0.14 | rno-miR-542-5p | 0.43 |
rno-miR-181c-3p | 0.30 |
| | rno-miR-99a-3p | 0.35 | rno-miR-465-5p | 0.37 |
| | rno-miR-139-5p | 0.26 | rno-miR-382-5p | 0.24 |
| |
rno-miR-208a-3p | 0.32 |
rno-miR-208a-3p | 0.15 |
| | rno-miR-33-5p | 0.43 | rno-miR-351-5p | 0.25 |
| |
rno-miR-190a-5p | 0.40 | rno-miR-503-3p | 0.25 |
| | rno-miR-335 | 0.39 | rno-miR-127-3p | 0.18 |
| | rno-miR-708-3p | 0.35 |
rno-miR-664-2-5p | 0.30 |
| | rno-miR-15b-5p | 0.45 | rno-miR-298-5p | 0.17 |
| | rno-miR-674-3p | 0.45 |
rno-miR-376a-3p | 0.49 |
| | rno-miR-188-5p | 0.18 | rno-miR-186-5p | 0.35 |
| | | | rno-miR-134-3p | 0.35 |
| | | | rno-miR-92a-3p | 0.48 |
| | | |
rno-miR-378a-3p | 0.40 |
| | | | rno-miR-541-5p | 0.19 |
| | | | rno-miR-154-5p | 0.10 |
We identified 1 downregulated miRNA (miR-377-3p) and
2 upregulated miRNAs (miR-490-3p and miR-1193-3p) on postnatal day
3 in the model group compared to the control group (M3d vs. C3d);
11 downregulated miRNAs (miR-542-5p, miR-99a-3p, miR-139-5p,
miR-208a-3p, miR-33-5p, miR-190a-5p, miR-335, miR-708-3p,
miR-15b-5p, miR-674-3p and miR-188-5p) and 1 upregulated miRNA
(miR-3584-3p) on day 7 in the model group compared to the control
group (M7d vs. C7d); and 16 downregulated miRNAs (miR-181c-3p,
miR-465-5p, miR-382-5p, miR-208a-3p, miR-351-5p, miR-503-3p,
miR-127-3p, miR-664-2-5p, miR-298-5p, miR-376a-3p, miR-186-5p,
miR-134-3p, miR-92a-3p, miR-378a-3p, miR-541-5p and miR-154-5p) and
19 upregulated miRNAs (miR-34c-3p, let-7b-5p, miR-3068-5p,
miR-872-5p, miR-183-5p, miR-33-5p, miR-182, miR-322-3p, miR-340-3p,
miR-142-3p, miR-141-5p, miR-96-5p, let-7f-5p, miR-15b-5p,
miR-449a-5p, miR-22-5p, miR-362-3p, miR-301a-3p and miR-365-3p) on
day 14 in the model group compared to the control group (M14d vs.
C14d).
We also identified miRNAs with a change in
expression during septation in both groups by making comparisons of
the 2 groups (model and control group) between days 14 and 3 (FC≥2,
P<0.05). We found that compared with the control group on day 3
(C3d), 27 miRNAs were upregulated, and 20 were downregulated in the
control group on day 14 (C14d); compared with the model group on
day 3 (M3d), 19 miRNAs were upregulated, whereas 26 were
downregulated in the model group on day 14 (M14d). The upregulated
and downregulated miRNAs in the 2 groups on day 14 compared to day
3 are listed in Table IV.
 | Table IVDifferentially expressed miRNAs of
the 2 groups during septation. |
Table IV
Differentially expressed miRNAs of
the 2 groups during septation.
C14d vs. C3d
| M14d vs. M3d
|
|---|
Up
| Down
| Up
| Down
|
|---|
| Name | FC | Name | FC | Name | FC | Name | FC |
|---|
| rno-miR-425-5p | 2.85 | rno-miR-542-5p | 0.34 | rno-miR-31a-5p | 19.28 | rno-miR-665 | 0.47 |
| rno-miR-21-3p | 6.39 |
rno-miR-218a-5p | 0.39 | rno-let-7b-5p | 2.38 | rno-miR-542-5p | 0.39 |
|
rno-miR-181a-5p | 4.52 | rno-miR-3571 | 0.22 | rno-miR-10a-5p | 2.31 | rno-miR-329-3p | 0.19 |
|
rno-miR-181c-3p | 2.71 | rno-miR-139-5p | 0.11 |
rno-miR-146b-5p | 2.44 | rno-miR-668 | 0.31 |
| rno-miR-465-5p | 6.44 | rno-miR-33-5p | 0.28 | rno-miR-128-3p | 7.30 |
rno-miR-466c-3p | 0.21 |
| rno-miR-23a-3p | 2.92 |
rno-miR-24-1-5p | 0.34 | rno-miR-194-5p | 2.86 | rno-miR-3571 | 0.25 |
|
rno-miR-208a-3p | 3.03 | rno-miR-342-3p | 0.31 | rno-miR-221-3p | 6.03 | rno-miR-434-3p | 0.39 |
| rno-miR-351-5p | 2.74 | rno-miR-503-5p | 0.28 | rno-miR-224-5p | 5.53 | rno-miR-136-5p | 0.44 |
| rno-miR-674-5p | 2.63 | rno-miR-142-3p | 0.50 | rno-miR-193-3p | 2.17 | rno-miR-431 | 0.13 |
| rno-miR-503-3p | 3.73 | rno-let-7e-5p | 0.29 | rno-miR-30d-3p | 4.32 |
rno-miR-344b-2-3p | 0.38 |
|
rno-miR-664-2-5p | 8.32 | rno-miR-448-5p | 0.28 | rno-miR-34b-5p | 2.37 | rno-miR-322-3p | 0.38 |
| rno-miR-298-5p | 3.45 | rno-miR-96-5p | 0.26 |
rno-miR-24-2-5p | 2.55 |
rno-miR-466b-2-3p | 0.27 |
| rno-miR-186-5p | 4.30 | rno-miR-7a-5p | 0.36 | rno-miR-222-3p | 3.78 | rno-miR-335 | 0.40 |
| rno-miR-25-3p | 3.07 | rno-miR-15b-5p | 0.12 | rno-miR-324-3p | 3.64 | rno-miR-127-3p | 0.27 |
|
rno-miR-106b-3p | 2.24 |
rno-miR-449a-5p | 0.20 | rno-miR-34a-5p | 2.69 | rno-miR-3572 | 0.40 |
| rno-miR-30d-3p | 3.72 | rno-miR-22-5p | 0.18 | rno-miR-100-5p | 2.26 |
rno-miR-376a-3p | 0.16 |
| rno-miR-222-3p | 4.03 | rno-miR-455-5p | 0.18 |
rno-let-7a-1-3p/rno-let-7c-2-3p | 3.45 | rno-miR-410-3p | 0.06 |
| rno-let-7e-3p | 3.39 | rno-miR-362-3p | 0.02 | rno-miR-143-5p | 2.21 | rno-miR-503-5p | 0.38 |
| rno-miR-30a-3p | 3.34 |
rno-miR-301a-3p | 0.24 |
rno-miR-146a-5p | 2.47 | rno-miR-667-3p | 0.31 |
|
rno-miR-125a-3p | 4.42 | rno-miR-365-3p | 0.48 | | | rno-miR-185-3p | 0.24 |
| rno-miR-22-3p | 2.26 | | | | | rno-miR-134-3p | 0.35 |
|
rno-miR-30c-1-3p | 8.68 | | | | |
rno-miR-466b-1-3p | 0.25 |
|
rno-miR-181b-5p | 4.92 | | | | | rno-miR-466d | 0.39 |
|
rno-miR-664-1-5p | 2.79 | | | | | rno-miR-541-5p | 0.15 |
| rno-miR-339-5p | 3.88 | | | | | rno-miR-742-3p | 0.23 |
| rno-miR-143-5p | 3.91 | | | | | rno-miR-3596c | 0.34 |
|
rno-miR-378a-3p | 2.09 | | | | | | |
Hierarchical clustering was carried out to
illustrate the distinguishable miRNA expression profiles between
the 2 groups at different time points. The heatmap is shown in
Fig. 5.
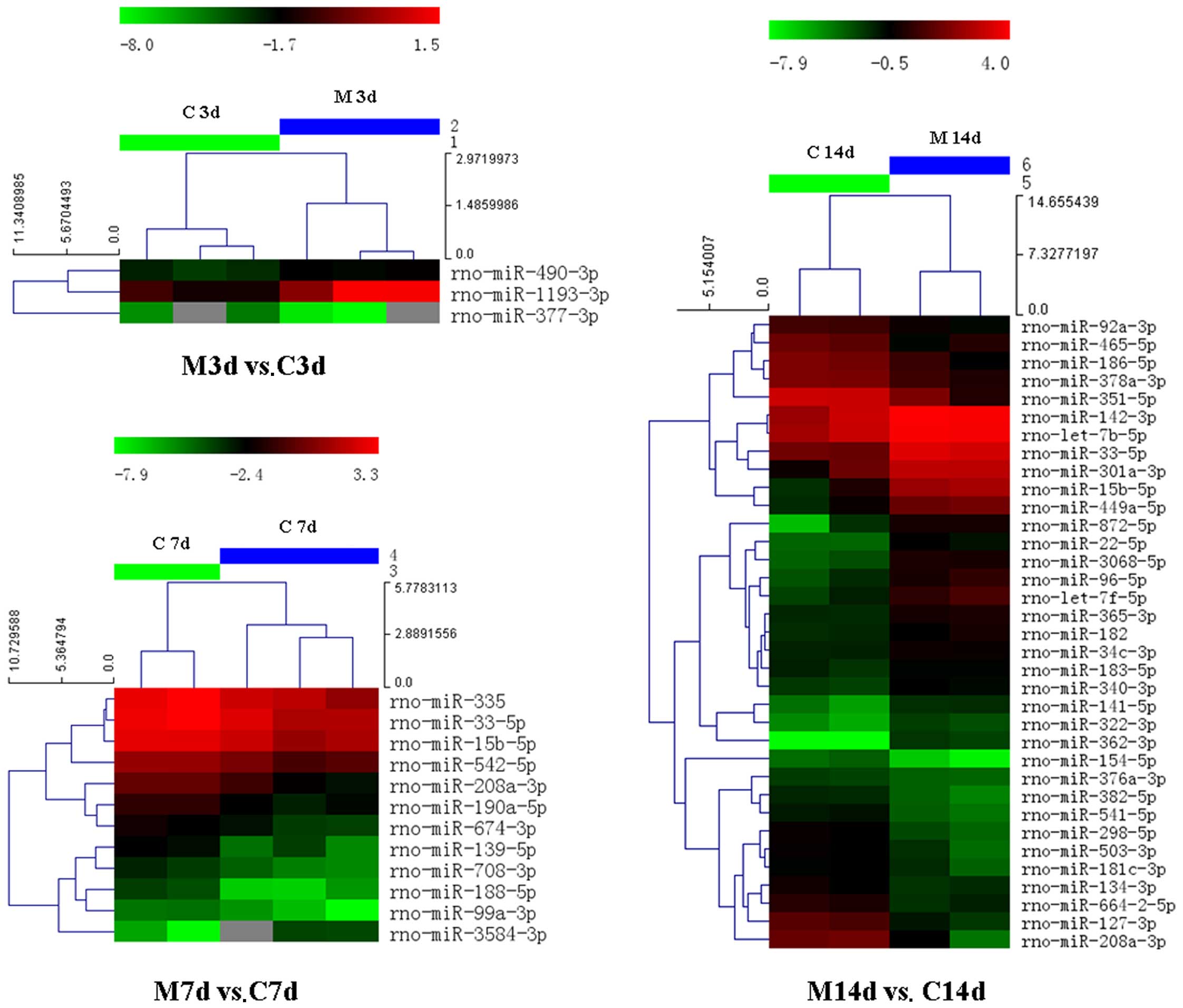 | Figure 5Hierarchical clustering of the 2
groups (M, model group; C, control group) on days 3, 7 and 14. The
heatmap shows the result of the two-way hierarchical clustering of
miRNAs and samples. Each row represents a miRNA and each column
represents a sample. The miRNA clustering tree is shown on the
left, and the sample clustering tree appears at the top. The color
scale shown at the top illustrates the relative expression level of
an miRNA in the certain slide: red color denotes a high relative
expression level; green color denotes a low relative expression
level. M3d, model group on day 3; C3d, control group on day 3; M7d,
model group on day 7; C7d, control group on day 7; M14d, model
group on day 14; C14d, control group on day 14. |
Target prediction
The target genes for 36, 38 and 46 miRNAs were
successfully predicted using the miRBase, miRanda and miRDB
databases, respectively. Each miRNA had one or more predicted
target genes. The overlapping section of the 3 databases contained
target genes for 32 miRNAs (Table
V).
 | Table VTarget prediction: the overlapping of
miRBase, miRanda and miRDB. |
Table V
Target prediction: the overlapping of
miRBase, miRanda and miRDB.
| miRNA_name | Gene symbol |
|---|
| rno-let-7b-5p | Adrb3, Apbb3,
Ikbkap |
| rno-let-7f-5p | Adrb2, Adrb3,
Apbb3, Ptafr |
| rno-miR-127-3p | Odf4, Sept7 |
| rno-miR-139-5p | Btg3, Fbxo9,
Galnt3, Gclc, Gdf10, Gpr56, Nxph1, Sell, Serpini1, Slc25a3, Syn2,
Tspan3, Ube2f |
| rno-miR-142-3p | Cask, Serinc1,
Stx12, Tsen34 |
| rno-miR-154-5p | Aldh1a2, Cdca4,
Chrm2, Cops2, Tmem35 |
| rno-miR-15b-5p | B4galt7, Btrc,
Ccdc19, Cdc25a, Cops2, Glud1, Inhbc, Itpr1, Kcnc2, MGC114483, Mkks,
Mlycd, Ppp1r11, Pth, RGD1308059, RGD1311739, Rnf10, Srpr, Usp14,
Wbp11, Wee1, Wipi2 |
| rno-miR-182 | Adcy6, Anxa11,
Arf4, Ccr5, Cd38, Cttn, Dazap2, Dnajb9, Gyg1, Il2rg, LOC500148,
Lphn2, Pc, RGD1310861, Rnf44, Rtn4, Stk19, Tmem50b, Txnl1 |
| rno-miR-183-5p | Ap3m1, Btg1,
Katna1, Ppp2cb, Ppp2r1a, Rcn2, Snx1, Spry2, Tmpo, Zfp451 |
| rno-miR-186-5p | Aldh6a1, Clcn3,
Drg1, Dusp13, Eif4b, Fbxo30, Hhex, Irgm, LOC257650, Paqr3 Prkaa2,
Psph, Sema4a, Serinc3, Sycp1, Tmco1, Ttc12, Xrcc4 |
| rno-miR-188-5p | Dcps, Pde4a,
Psmc3ip, Ptprr, RGD1359108, Tmem30a, Tomm70a, Ube2i |
|
rno-miR-190a-5p | Ambn, Ctbs,
Epb4.1l5, Neurod1, Nlgn1, Stard3nl |
|
rno-miR-208a-3p | Slc39a3, Srr |
| rno-miR-22-5p | Arhgef2, Ndrg2,
Rdh11, Rgs1, Vrk1 |
| rno-miR-298-5p | Arhgdia, Arl3,
Atp6v0e1, Btrc, Cd14, Eral1, Mustn1, Vdr |
|
rno-miR-301a-3p | Arfip1, Cbfb,
Clcn4-2, Dynll2, Enpp5, Esm1, Gadd45a, Il6st, Irf1, Laptm4a, Manba,
Ndel1, Pparg, Rab30, RGD621098, Slfn8, Snx7, St18, St8sia3, Tdrd7,
Wee1 |
| rno-miR-322-3p | Arhgap17, Cox8a,
Dcun1d3, Dspp, Galnt11, Gpm6b, Lcp2, Ly49i6, Mycn, Ndufs1,
Nit2 |
| rno-miR-33-5p | Cdc42bpa, Crot,
Gria3, Hadhb, Hsd17b12, Kcns3, Lap3, Lum, Mdm4, Nr4a2, Slc39a8,
St18, Tph2 |
| rno-miR-335 | Apeh, C4bpa, Chfr,
Cln8, Cyp2u1, Il7, Klhl25, LOC499331, Ndfip1, Neu2, Nnt, Ikbkap,
Rab11b |
| rno-miR-340-3p | Sept9 |
| rno-miR-34c-3p | Kif22, Lias,
Sfrp4 |
| rno-miR-351-5p | Ahrr, Fcgr3a,
Galnt14, Itga7, Prim2, RGD1359378, Rhot2, Slc35a4, Taz |
| rno-miR-365-3p | Adm, Arrb2, Crbn,
Eltd1, Ing3, Pebp1, RGD1305235, Ublcp1 |
|
rno-miR-378a-3p | Camlg, Kcna4,
Sbds |
| rno-miR-382-5p | Dld, Eef1a1, Pkia,
RGD1311863, Slc6a8, Smpx, Uqcrc2 |
|
rno-miR-449a-5p | Capn8, Eno3,
Fbxo30, Gmfb, Ldha, Mycn, Rai14, Stag3, Tmem109, Tmem22 |
| rno-miR-541-5p | Ly49i6, Rnd3 |
| rno-miR-674-3p | Emb, LOC500118,
Lrrc4, Plcl1, Plp2, Slc12a2 |
| rno-miR-708-3p | Ctgf, Fmr1, Nedd9,
Slfn8, Vim |
| rno-miR-872-5p | Azin1, Cntf,
Sqstm1 |
| rno-miR-92a-3p | Adcy3, Adm, Ccnh,
Cd69, Col1a2, Dkk3, Dnajc4, Fmr1, Gata2, Gria3, Ibsp, Klf4, Nelf,
Pigv, Ptpro, RGD1308059, Ugp2, Wrnip1 |
| rno-miR-96-5p | Btg4, Klhl7,
MGC105560, Morf4l2, Ppp3r1, RGD1310794, ST7 |
Discussion
BPD is a common respiratory disease affecting
premature infants. In recent years, due to the increasing rescue
success rates for very low birth weight and extremely low birth
weight infants, as well as the wide application of pulmonary
surfactants, extremely premature infants with a gestational age
lower than 28 weeks have become the major group experiencing BPD.
These infants were born towards the end of the canalicular stage or
at the saccular stage of lung development; therefore, the entire
alveolarization and microvascularization process occurred
postnatally. This is different from the 'old BPD', as suggested by
Northway et al (23),
which mainly manifested with 'acute lung injury, pneumonedema,
extensive airway epithelial metaplasia, pulmonary fibrosis, and
airway and vascular smooth muscle hyperplasia'. The 'new BPD', in
these extremely premature infants is characterized by 'alveolar
hypoplasia and impaired pulmonary vascular development' (1).
In the present study, we found that the RAC value in
the BPD model group was markedly decreased by postnatal day 7
(P<0.05) as compared with the control group, and that the
difference between the 2 groups was even greater by day 14
(P<0.01). This indicated that the formation of secondary septa
was impaired and that alveolarization was suppressed in the model
of BPD. The results from western blot analysis and RT-qPCR for PCNA
expression revealed that the expression levels in the model group
were lower than those in the control group by postnatal day 3, and
this difference persisted and reached a peak by day 14. This
suggests that cell proliferation may be suppressed in BPD.
Similarly, CD31 expression was lower in the model group than in the
control group at all time points, indicating that vascular
development may be suppressed in the model group.
The role of miRNAs in BPD has been previously
investigated using microarray assays (21,24,25), and the results were partially
consistent with ours. For example, Bhaskaran et al (24) and Zhang et al (21) found that the expression level of
miR-335 was downregulated, and Zhang et al also observed the
upregulation of miR-449a and let-7f expression. There are few
studies available which have investigated the association between
miRNAs and BPD. Bhaskaran et al (24) confirmed that one of the miR-150
targets was glycoprotein nonmetastatic melanoma protein b (GPNMB),
which may participate in the regulation of angiogenesis. Narasaraju
et al (26) further
confirmed the regulatory role of miR-150 in angiogenesis in BPD.
Dong et al (25) found
that miR-29 expression was upregulated in BPD and suggested that
miR-29 may suppress lung development by inhibiting Ntrk2. However,
we failed to detect these 2 miRNAs, possibly due to the different
animal species and modeling method used. Most of the other miRNAs
that had expression changes have not been previously reported in
studies of BPD to the best of our knowledge.
In rats, alveolarization occurs mainly during
postnatal days 5–14. By postnatal day 3, alveolarization has not
yet begun, so we observed only a few differentially expressed
miRNAs between the 2 groups. To date, downregulated miR-377-3p and
upregulated miR-1193-3p have not been reported, to the best of our
knowledge. Gu et al found that the overexpression of
miR-490-3p in the A549 cell line had an effect on the targeted
downregulation of CCND1 expression. As CCND1 plays an important
role in the G1/S transition, the cell cycle was arrested in the G1
phase, and thus, cell proliferation was suppressed (27).
In our study, alveolarization reached a peak 7 days
after birth when the difference in miRNA expression between the 2
groups was significant. The upregulation of miR-3584-3p has not yet
been reported to the best of our knowledge. The expression of the
following miRNAs was downregulated: miR-542-5p, miR-99a-3p,
miR-139-5p, miR-208a-3p, miR-33-5p, miR-190a-5p, miR-335,
miR-708-3p, miR-15b-5p, miR-674-3p and miR-188-5p. Bray et
al (28) found that
miR-542-5p had a tumor inhibitory effect on neuroblastoma. In
colorectal cancer, esophageal cancer and breast cancer, the
mechanisms of action of miR-139-5p as a tumor inhibitory factor
have also been reported (29–32). Wang et al (33) observed the downregulated
expression of miR-335 in NSCLC tissues and an increased apoptotic
rate in the miR-335-transfected cell line. In the same study,
miR-355 was shown to suppress the invasive ability of tumor cells
through the targeted regulation of Bcl-w and SP1. Other
downregulated miRNAs have not yet been reported to the best of our
knowledge.
By postnatal day 14, alveolarization was almost
complete, and the manifestations of BPD were also observed in the
lung tissues of the model group. In addition, differences in the
miRNA expression levels between the 2 groups were increasingly
apparent. Now, it is widely recognized that the pulmonary
microvascularization stage begins on postnatal day 14, during which
the double layers of the microvascular network in the normal
secondary septa will fuse into one layer. Therefore, differentially
expressed miRNAs on day 14 may also be associated with the failure
of microvascularization caused by the absence of secondary septa in
the BPD model group. Jiang et al found that downregulated
miR-127-3p inhibited the proliferation of tumor cells in malignant
glioma by suppressing SKI and activating the TGF-β signaling
pathway, and promoted metastasis through the targeted inhibition of
SEPT7 (34,35). Zha et al found that
miR-134-3p inhibited tumor metastasis in hepatocellular carcinoma
through the targeted regulation of ITGB1 (36). The decreased expression of
miR-378a-3p in colorectal cancer has been shown to increase the
phosphorylated ERK1/2 protein level (37), and the downregulated expression of
miR-378a-3p is also present in rhabdomyosarcoma (38). Both these studies suggest that
miR-378a-3p suppresses tumor growth by inhibiting the expression of
the target gene, IGF1R. Of the upregulated miRNAs, miR-34c-3p
caused the arrest of glioma cells in the S phase, reduced the
number of cells in the G0/G1 phase, and induced cell apoptosis
(39). miR-34c-3p caused SiHa
cells to arrest in the S phase and to undergo apoptosis, and
inhibited metastasis and invasion in cervical cancer (40). Similarly, let-7b-5p inhibited the
proliferation of multiple myeloma cells through the targeted
regulation of IGF1R and promoted cell cycle arrest and apoptosis
(41). Zhu et al (42) found that hsa-miR-182 was highly
expressed in pulmonary adenocarcinoma, which led to the inhibition
of cell proliferation through the downregulation of RASA1. Stenvold
et al (43) found that the
expression of miR-182 was well correlated with FGF2, HIF2α and
MMP-7 in NSCLC, indicating its possible role in angiogenesis.
Patients with squamous cell carcinoma (SCC) and stage II patients,
expressing high levels of miR-182 had a better prognosis. Yang
et al (44) observed the
role of miR-182 in the targeted regulation of FOXO3 and found that
in early-stage lung cancer, Sp1 stimulated miR-182 expression and
led to the downregulated expression of FOXO3, thus allowing tumor
growth. In advanced lung cancer, Sp1 and miR-182 were both
downregulated, while FOXO3 expression was upregulated, leading to
lung cancer metastasis. Wang et al (45) found that PDCD4 was also one of the
target genes for miR-182. miR-182 was upregulated in the lung
cancer cell lines, A549 and SPC-A-1, which resulted in a decrease
in PDCD4 expression, thus weakening the inhibitory effect of PDCD4
on tumor growth. Zhang et al (46) observed that miR-182 inhibited
tumor cell proliferation and promoted tumor cell apoptosis through
the targeted regulation of the cortactin (CTTN) gene. In the
miR-182-transfected A549 cell line, most cells were arrested in
G0/G1 phase, and the percentages of cells in the S and G2/M phase
were greatly decreased. Sun et al (47) found that RGS17 was a target gene
of miR-182 and that the overexpression of transfected miR-182
inhibited RGS17 mRNA transcription and thus, suppressed tumor cell
proliferation, leading to the recurrence of cell adherence. Lei
et al (48) observed that
the high expression of TGF-β1 in NSCLC upregulated miR-142-3p
expression, and the latter targeted TGF-βR1, thus inhibiting its
expression, weakening SMAD3 phosphorylation, and promoting tumor
growth. Carraro et al (49) found that miR-142-3p kept the
balance between the proliferation and differentiation of
mesenchymal cells during lung development. miR-142 participated in
the regulation of the Wnt-CTNNB1 pathway (Apc binds to CTNNB1 to
induce its ubiquitination and degradation) by binding to Apc mRNA,
and its downregulation promoted the differentiation of
parabronchial smooth muscle ancestral cells. In colorectal cancer
cells, miR-96-5p decreased cyclin D1 expression but increasesd
p27-CDKN1A expression through the targeted regulation of KRAS, thus
slowing cell growth (50).
miR-96-5p regulated GPC1 to inhibit the proliferation of pancreatic
cancer cells (51). Other miRNAs
have not yet been reported to the best of our knowledge.
During septation, compared with the control group on
day 3 (C3d), 27 miRNAs were upregulated, and 20 were downregulated
in the control group on day 14 (C14d); compared with the model
group on day 3 (M3d), 19 miRNAs were upregulated, while 26 were
downregulated in the model group on day14 (M14d) (Table IV). Several miRNAs were shared by
the model and control groups, including highly upregulated
miR-143-5p and miR-222-3p and markedly downregulated miR-542-5p and
miR-503-5p. These data demonstrated similarities between septation
in the 2 groups during, suggesting that these miRNAs play an
important role during septation, despite the cause of the injury
related to hyperoxia.
Three online databases were used to predict the
target genes of 32 differentially expressed miRNAs, according to
the base sequence complementation principle, and some miRNAs even
helped to predict several target genes (Table V). However, the prediction results
were not able to indicate the actual presence of these targeted
regulations in vivo. Regarding the regulation of miRNA in
other diseases, the target genes of some miRNAs have been confirmed
experimentally, for example, Sept7 is the target gene of miR-127-3p
(40), while CTTN is the target
gene of rno-miR-182 (51). Some
target genes that were confirmed in previous studies were not found
in our results, possibly because we only selected the intersection
set of results from three databases, so some target molecules may
have been missed. In the future, we may construct luciferase
reporter vectors for the miRNA of interest to further validate the
targeted relationships between miRNA and the predicted genes.
In conclusion, the majority of the miRNAs we found
still play an unknown role in the development of BPD, and some of
them have not been previously reported to the best of our
knowledge. Currently available data also revealed that the
regulation of miRNA is complex regarding life processes. Further
study is necessary to understand the effects of miRNA in the
development of BPD.
In this study, we screened possible miRNAs that
participated in the development of BPD during the alveolar
septation phase using microarray assays and identified the
important role of miRNAs in the development of normal lungs and
BPD. In the future, we aim to further investigate the functions of
these miRNAs, in order to broaden our understanding of the
pathogenesis of BPD.
Acknowledgments
This study was supported by grant from the Natural
Science Foundation of China (no. 81471489).
References
|
1
|
Hilgendorff A, Reiss I, Ehrhardt H,
Eickelberg O and Alvira CM: Chronic lung disease in the preterm
infant. Lessons learned from animal models. Am J Respir Cell Mol
Biol. 50:233–245. 2014.
|
|
2
|
Bhandari A and Bhandari V: Pitfalls,
problems, and progress in bronchopulmonary dysplasia. Pediatrics.
123:1562–1573. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bhandari A and McGrath-Morrow S: Long-term
pulmonary outcomes of patients with bronchopulmonary dysplasia.
Semin Perinatol. 37:132–137. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Anderson PJ and Doyle LW:
Neurodevelopmental outcome of bronchopulmonary dysplasia. Semin
Perinatol. 30:227–232. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Herriges M and Morrisey EE: Lung
development: orchestrating the generation and regeneration of a
complex organ. Development. 141:502–513. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Warburton D, El-Hashash A, Carraro G,
Tiozzo C, Sala F, Rogers O, De Langhe S, Kemp PJ, Riccardi D,
Torday J, et al: Lung organogenesis. Curr Top Dev Biol. 90:73–158.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hadchouel A, Franco-Montoya ML and
Delacourt C: Altered lung development in bronchopulmonary
dysplasia. Birth Defects Res A Clin Mol Teratol. 100:158–167. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Madurga A, Mizíková I, Ruiz-Camp J and
Morty RE: Recent advances in late lung development and the
pathogenesis of bronchopulmonary dysplasia. Am J Physiol Lung Cell
Mol Physiol. 305:L893–L905. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Du T and Zamore PD: microPrimer: the
biogenesis and function of microRNA. Development. 132:4645–4652.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dong J, Jiang G, Asmann YW, Tomaszek S,
Jen J, Kislinger T and Wigle DA: MicroRNA networks in mouse lung
organogenesis. PLoS One. 5:e108542010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tay HL, Plank M, Collison A, Mattes J,
Kumar RK and Foster PS: MicroRNA: potential biomarkers and
therapeutic targets for allergic asthma? Ann Med. 46:633–639. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lino Cardenas CL, Kaminski N and Kass DJ:
Micromanaging microRNAs: using murine models to study microRNAs in
lung fibrosis. Drug Discov Today Dis Models. 10:e145–e151. 2013.
View Article : Google Scholar
|
|
15
|
Joshi P, Middleton J, Jeon YJ and Garofalo
M: MicroRNAs in lung cancer. World J Methodol. 4:59–72. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lu Y, Thomson JM, Wong HY, Hammond SM and
Hogan BL: Transgenic overexpression of the microRNA miR-17-92
cluster promotes proliferation and inhibits differentiation of lung
epithelial progenitor cells. Dev Biol. 310:442–453. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ventura A, Young AG, Winslow MM, Lintault
L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone
JR, et al: Targeted deletion reveals essential and overlapping
functions of the miR-17 through 92 family of miRNA clusters. Cell.
132:875–886. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Carraro G, El-Hashash A, Guidolin D,
Tiozzo C, Turcatel G, Young BM, De Langhe SP, Bellusci S, Shi W,
Parnigotto PP and Warburton D: miR-17 family of microRNAs controls
FGF10-mediated embryonic lung epithelial branching morphogenesis
through MAPK14 and STAT3 regulation of E-Cadherin distribution. Dev
Biol. 333:238–250. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Navarro A, Marrades RM, Viñolas N, Quera
A, Agustí C, Huerta A, Ramirez J, Torres A and Monzo M: MicroRNAs
expressed during lung cancer development are expressed in human
pseudoglandular lung embryogenesis. Oncology. 76:162–169. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bhaskaran M, Wang Y, Zhang H, Weng T,
Baviskar P, Guo Y, Gou D and Liu L: MicroRNA-127 modulates fetal
lung development. Physiol Genomics. 37:268–278. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang X, Peng W, Zhang S, Wang C, He X,
Zhang Z, Zhu L, Wang Y and Feng Z: MicroRNA expression profile in
hyperoxia-exposed newborn mice during the development of
bronchopulmonary dysplasia. Respir Care. 56:1009–1015. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang H, Fu J, Xue X, Yao L, Qiao L, Hou A,
Jin L and Xing Y: Epithelial-mesenchymal transitions in
bronchopulmonary dysplasia of newborn rats. Pediatr Pulmonol.
49:1112–1123. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Northway WH Jr, Rosan RC and Porter DY:
Pulmonary disease following respirator therapy of hyaline-membrane
disease. Bronchopulmonary dysplasia. N Engl J Med. 276:357–368.
1967. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bhaskaran M, Xi D, Wang Y, Huang C,
Narasaraju T, Shu W, Zhao C, Xiao X, More S, Breshears M and Liu L:
Identification of microRNAs changed in the neonatal lungs in
response to hyperoxia exposure. Physiol Genomics. 44:970–980. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dong J, Carey WA, Abel S, Collura C, Jiang
G, Tomaszek S, Sutor S, Roden AC, Asmann YW, Prakash YS and Wigle
DA: MicroRNA-mRNA interactions in a murine model of
hyperoxia-induced bronchopulmonary dysplasia. BMC Genomics.
13:2042012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Narasaraju T, Shukla D, More S, Huang C,
Zhang L, Xiao X and Liu L: Role of microRNA-150 and glycoprotein
nonmetastatic melanoma protein B in angiogenesis during
hyperoxia-induced neonatal lung injury. Am J Respir Cell Mol Biol.
52:253–261. 2015. View Article : Google Scholar
|
|
27
|
Gu H, Yang T, Fu S, Chen X, Guo L and Ni
Y: MicroRNA-490-3p inhibits proliferation of A549 lung cancer cells
by targeting CCND1. Biochem Biophys Res Commun. 444:104–108. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bray I, Tivnan A, Bryan K, Foley NH,
Watters KM, Tracey L, Davidoff AM and Stallings RL: MicroRNA-542-5p
as a novel tumor suppressor in neuroblastoma. Cancer Lett.
303:56–64. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Song M, Yin Y, Zhang J, Zhang B, Bian Z,
Quan C, Zhou L, Hu Y, Wang Q, Ni S, et al: MiR-139-5p inhibits
migration and invasion of colorectal cancer by downregulating AMFR
and NOTCH1. Protein Cell. 5:851–861. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shen K, Mao R, Ma L, Li Y, Qiu Y, Cui D,
Le V, Yin P, Ni L and Liu J: Post-transcriptional regulation of the
tumor suppressor miR-139-5p and a network of miR-139-5p-mediated
mRNA interactions in colorectal cancer. FEBS J. 281:3609–3624.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu R, Yang M, Meng Y, Liao J, Sheng J, Pu
Y, Yin L and Kim SJ: Tumor-suppressive function of miR-139-5p in
esophageal squamous cell carcinoma. PLoS One. 8:e770682013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Krishnan K, Steptoe AL, Martin HC,
Pattabiraman DR, Nones K, Waddell N, Mariasegaram M, Simpson PT,
Lakhani SR, Vlassov A, et al: miR-139-5p is a regulator of
metastatic pathways in breast cancer. RNA. 19:1767–1780. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang H, Li M, Zhang R, Wang Y, Zang W, Ma
Y, Zhao G and Zhang G: Effect of miR-335 upregulation on the
apoptosis and invasion of lung cancer cell A549 and H1299. Tumour
Biol. 34:3101–3109. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jiang H, Hua D, Zhang J, Lan Q, Huang Q,
Yoon JG, Han X, Li L, Foltz G, Zheng S and Lin B: MicroRNA-127-3p
promotes glioblastoma cell migration and invasion by targeting the
tumor-suppressor gene SEPT7. Oncol Rep. 31:2261–2269.
2014.PubMed/NCBI
|
|
35
|
Jiang H, Jin C, Liu J, Hua D, Zhou F, Lou
X, Zhao N, Lan Q, Huang Q, Yoon JG, et al: Next generation
sequencing analysis of miRNAs: MiR-127-3p inhibits glioblastoma
proliferation and activates TGF-beta signaling by targeting SKI.
OMICS. 18:196–206. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zha R, Guo W, Zhang Z, Qiu Z, Wang Q, Ding
J, Huang S, Chen T, Gu J, Yao M and He X: Genome-wide screening
identified that miR-134 acts as a metastasis suppressor by
targeting integrin β1 in hepatocellular carcinoma. PLoS One.
9:e876652014. View Article : Google Scholar
|
|
37
|
Li H, Dai S, Zhen T, Shi H, Zhang F, Yang
Y, Kang L, Liang Y and Han A: Clinical and biological significance
of miR-378a-3p and miR-378a-5p in colorectal cancer. Eur J Cancer.
50:1207–1221. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Megiorni F, Cialfi S, McDowell HP, Felsani
A, Camero S, Guffanti A, Pizer B, Clerico A, De Grazia A, Pizzuti
A, et al: Deep Sequencing the microRNA profile in rhabdomyosarcoma
reveals downregulation of miR-378 family members. BMC Cancer.
14:8802014. View Article : Google Scholar
|
|
39
|
Wu Z, Wu Y, Tian Y, Sun X, Liu J, Ren H,
Liang C, Song L, Hu H, Wang L and Jiao B: Differential effects of
miR-34c-3p and miR-34c-5p on the proliferation, apoptosis and
invasion of glioma cells. Oncol Lett. 6:1447–1452. 2013.PubMed/NCBI
|
|
40
|
López JA and Alvarez-Salas LM:
Differential effects of miR-34c-3p and miR-34c-5p on SiHa cells
proliferation apoptosis, migration and invasion. Biochem Biophys
Res Commun. 409:513–519. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xu H, Liu C, Zhang Y, Guo X, Liu Z, Luo Z,
Chang Y, Liu S, Sun Z and Wang X: Let-7b-5p regulates proliferation
and apoptosis in multiple myeloma by targeting IGF1R. Acta Biochim
Biophys Sin (Shanghai). 46:965–972. 2014. View Article : Google Scholar
|
|
42
|
Zhu YJ, Xu B and Xia W: Hsa-miR-182
downregulates RASA1 and suppresses lung squamous cell carcinoma
cell proliferation. Clin Lab. 60:155–159. 2014.PubMed/NCBI
|
|
43
|
Stenvold H, Donnem T, Andersen S, Al-Saad
S, Busund LT and Bremnes RM: Stage and tissue-specific prognostic
impact of miR-182 in NSCLC. BMC Cancer. 14:1382014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yang WB, Chen PH, Hsu T, Fu TF, Su WC,
Liaw H, Chang WC and Hung JJ: Sp1-mediated microRNA-182 expression
regulates lung cancer progression. Oncotarget. 5:740–753. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang M1, Wang Y, Zang W, Wang H, Chu H, Li
P, Li M, Zhang G and Zhao G: Downregulation of microRNA-182
inhibits cell growth and invasion by targeting programmed cell
death 4 in human lung adenocarcinoma cells. Tumour Biol. 35:39–46.
2014. View Article : Google Scholar
|
|
46
|
Zhang L, Liu T, Huang Y and Liu J:
microRNA-182 inhibits the proliferation and invasion of human lung
adenocarcinoma cells through its effect on human cortical
actin-associated protein. Int J Mol Med. 28:381–388.
2011.PubMed/NCBI
|
|
47
|
Sun Y, Fang R, Li C, Li L, Li F, Ye X and
Chen H: Hsa-miR-182 suppresses lung tumorigenesis through
downregulation of RGS17 expression in vitro. Biochem Biophys Res
Commun. 396:501–507. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lei Z, Xu G, Wang L, Yang H, Liu X, Zhao J
and Zhang HT: MiR-142-3p represses TGF-beta-induced growth
inhibition through repression of TGFbetaR1 in non-small cell lung
cancer. FASEB J. 28:2696–2704. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Carraro G, Shrestha A, Rostkovius J,
Contreras A, Chao CM, El Agha E, Mackenzie B, Dilai S, Guidolin D,
Taketo MM, et al: miR-142-3p balances proliferation and
differentiation of mesenchymal cells during lung development.
Development. 141:1272–1281. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ress AL, Stiegelbauer V, Winter E,
Schwarzenbacher D, Kiesslich T, Lax S, Jahn S, Deutsch A,
Bauernhofer T, Ling H, et al: MiR-96-5p influences cellular growth
and is associated with poor survival in colorectal cancer patients.
Mol Carcinog. Sep 25–2014.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Li C, Du X, Tai S, Zhong X, Wang Z, Hu Z,
Zhang L, Kang P, Ji D, Jiang X, et al: GPC1 regulated by miR-96-5p,
rather than miR-182-5p, in inhibition of pancreatic carcinoma cell
proliferation. Int J Mol Sci. 15:6314–6327. 2014. View Article : Google Scholar : PubMed/NCBI
|