Introduction
Osteoporosis (OP), a major health concern of
post-menopausal women and the elderly, is a common skeletal
disorder characterized by low bone mass and microarchitectural
deterioration of bone tissue with increased susceptibility to
fracture (1,2). Advanced age, gender and
immobilization are major risk factors for developing OP besides a
series of other contributors, such as diminished gender steroid
production in elderly individuals and following menopause (3,4).
Emerging evidence has indicated that OP is
associated with stem cell defects (5,6),
including osteoblast progenitors [mesenchymal stem cells (MSCs)]
residing in the bone marrow (BM) (7). Bone marrow-derived mesenchymal stem
cells (BMSCs), a class of multipotent and self-renewing cells that
give rise to differentiated progeny when implanted into appropriate
tissues, have emerged as a particularly appealing option over the
last decade. Pittenger et al found that BMSCs, the precursor
cells of adipocytes and osteoblasts (8), play an important role in bone
physiology and partly participate in the pathophysiology of OP.
Furthermore, as previoulsy demonstrated the bone mass and strength
of osteoporotic rats and mice can be restored through direct BMSC
injection by local or systemic transplantation (9–11).
BMSCs are more sensitive and more effective towards
osteogenic differentiation than adipose tissue-derived stem cells
(ADSCs) (12), another emerging
type of stem cells for treating OP, which is relatively abundant
and can be easily harvested through simple methods, such as
lipoaspiration or surgical resection (13).
A previous study demonstrated that the osteogenic
differentiation ability of BMSCs is lower in post-menopausal women
suffering from OP compared to pre-menopausal women (14). Furthermore, in another previous
study, an augmented volume of adipose tissue was found in the BM of
post-menopausal women, indicating that the ability of BMSCs to
differentiate into adipocytes was enhanced (7). Theoretically, there is an inverse
association between osteogenic and adipogenic lineage commitment
and differentiation; that is to say, differentiation towards an
adipocytic phenotype occurs at the cost of an osteoblast
phenotype.
In the present study, we examined the initiation and
adipogenic differentiation of cultured BMSCs at the cellular level.
We hypothesized that ectopic viral integration site-1 (Evi1), a key
regulator in the process of the adipogenic differentiation of
3T3-L1 preadipocytes (15), would
lead to the adipogenic differentiation of BMSCs in vitro. To
the best of our knowledge, the expression of Evi1 in BMSCs has not
been reported to date. We hypothesized that the suppression of Evi1
by RNA interference (RNAi) would mediate the expression of specific
osteogenic and adipogenic genes and become a drug target for the
treatment of OP.
Materials and methods
Isolation of BMSCs and cell culture
BMSCs were isolated and cultured in vitro and
passaged using a cell adherent culture method from the BM of
Sprague-Dawley (SD) rats, as previously described (16). Briefly, rat BM was obtained
according to the research guidelines and under the approval of
Shanxi Medical University (Shanxi, China). Healthy specific
pathogen-free (SPF) grade 4-week-old SD rats (weighing 100–120 g)
were sacrificed by an intraperitoneal injection of pentobarbital
sodium. The bilateral femurs and tibias were harvested under
aseptic conditions and all soft tissues were removed. Metaphyses
from both ends were resected and BM cells were collected by
flushing the BM cavity in a sterile petri dish with Dulbecco's
modified Eagle's medium (DMEM; Gibco, Shanghai, China) containing
10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA) and
antibiotics (100 U/ml penicillin and 100 µg/ml
streptomycin). Cells isolated from each rat were inoculated in two
25 cm2 flasks. The medium was first changed 24 h after
inoculation to remove the unattached cells, and then twice each
week until confluence was achieved. The BMSCs were cultured at 37°C
in a 5% CO2-supplemented incubator and were capable of
differentiating into adipogenic and osteogenic cells under specific
induction, as previously described (17).
The differentiation of cultured BMSCs was monitored
with Oil Red O staining (on day 16) for terminal adipocyte
differentiation and with alizarin red staining (on day 16) for
terminal osteoblast differentiation.
Cells from the 3rd passage were used for the
experiments. BMSC adipogenesis was induced at confluence with the
use of an induction medium of 10% FBS in DMEM supplemented with
penicillin/streptomycin (P/S), 200 µM indomethacin, 1
µM dexamethasone, 0.5 mM isobutylmethylxanthine and 0.5
µg/ml insulin (all from Sigma, St. Louis, MO, USA) for 21
days, and the medium was changed twice each week. RNA and protein
were isolated within the first 10 days. Transient transfections
were performed with the use of adenovirus.
Vector construction and transfection of
the cells
Short hairpin RNA (shRNA) encoding DNA sequences
were synthesized by Invitrogen Life Technologies (Carlsbad, CA,
USA) and constructed into adenoviral plasmids, and adenoviruses
were prepared according to previously described procedures
(18). The sequence of shRNA
against luciferase was 5′-CTTACGCTGAGTACTTCGA-3′; shRNA against
Evi1 was (shEvi1-1) 5′-GGAAGCAACATGGAAACAA-3′ and (shEvi1-2)
5′-GCAGTGAGGTCTGCCATAA-3′. We created three adenoviral vectors,
termed Ad-shEvi1-1 (KD1), Ad-shEvi1-2 (KD2) and AD-shSc,
respectively. KD1 and KD2 encoding Evi1 shRNA were used in the
experimental groups. AD-shScr expressing scrambled shRNA was used
in the mock group and the control group was untransfected
cells.
The cells were inoculated into 60-mm dishes in 10%
FBS-DMEM and cultured until nearly 90% confluence and were then
incubated in complete serum-free medium for 12 h. Subsequently, the
cells were treated with shEvi1 or shScr (scrambled) for 6–8 h and
the medium was then changed to adipogenic induction medium, which
was changed every 3 days. Untransfected cells were used as
controls. RNA and protein were isolated on days 1, 3, 5 and 7. The
transfection efficiency was analyzed by quantifying GFP expression
using an inverted fluorescence microscope (LX70; Olympus, Tokyo,
Japan) at 24 h after transduction. The interference efficiency was
analyzed using the comparative 2−ΔΔCT method with
β-actin as an endogenous control on day 3 in each group. Detailed
information on the shRNA sequences is provided in Table I.
 | Table INucleotide sequences of the shRNAs for
RNA interference. |
Table I
Nucleotide sequences of the shRNAs for
RNA interference.
| Name | shRNA primer sequence
(5′→3′) | Size (bp) |
|---|
| shEvi1-2 | F:
GATCCGCAGTGAGGTCTGCCATAATTCAAGAGATTATGGCAGACCTCACTGCTTTTTTG | 59 |
| R:
AATTCAAAAAAGCAGTGAGGTCTGCCATAATCTCTTGAATTATGGCAGACCTCACTGCG | 59 |
| shEvi1-1 | F:
GATCCGGAAGCAACATGGAAACAATTCAAGAGATTGTTTCCATGTTGCTTCCTTTTTTG | 59 |
| R:
AATTCAAAAAAGGAAGCAACATGGAAACAATCTCTTGAATTGTTTCCATGTTGCTTCCG | 59 |
Analysis of cultured cells by flow
cytometry
An analysis of cell surface molecules was performed
on passage 3 cultures of rat BMSCs by flow cytometry with the
following procedure: the medium was removed from the flasks and the
cell layers were then washed twice with PBS and detached with 0.05%
trypsin-EDTA (HyClone, Logan, UT, USA) for 2 min at room
temperature. The BMSCs were collected by centrifugation (179 × g,
at room temperature) and washed in flow cytometry buffer (BD
Biosciences, Franklin Lakes, NJ, USA) consisting of 2% bovine serum
albumin and 0.1% sodium azide in PBS. Subsequently, the cells were
incubated with fluorescein-5-isothiocyanate (FITC)-conjugated
monoclonal antibodies against CD44 (553133), CD45 (561867) and CD90
(561969), and phycoerythrin (PE)-conjugated antibody against CD34
(550619) (all from BD Biosciences). Following incubation with the
antibodies for 30 min at 4°C, the cells were washed with flow
cytometry buffer. The washed cells were pelleted and resuspended in
flow cytometry buffer containing 1% paraformaldehyde for 20 min.
Non-specific fluorescence was determined using equal aliquots of
the cell preparation that were incubated with anti-mouse monoclonal
antibodies. Data were acquired and analyzed on a FACSCalibur flow
cytometer with CellQuest software (BD Biosciences).
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the cells using TRIzol
(Takara Bio, Inc., Tokyo, Japan) according to the manufacturer's
instructions. RNA concentrations were assessed with the optical
density at 260 nm (OD260) using a kinetic biospectrometer
(Eppendorf AG, Hamburg, Germany). Reverse transcription reactions
were completed using 300–500 ng of total RNA as input for the High
Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster
City, CA, USA). cDNA was generated from the total RNA using
PrimeScript RT Master Mix (5X) (Takara Bio, Inc.) according to the
manufacturer's instructions. To quantify mRNA expression,
SYBR® Premix Ex Taq (2X) (Takara Bio, Inc.) was used.
Briefly, each of the resulting eight reverse transcription pools
containing cDNA template was diluted, mixed with Taq and loaded
into each of the 8 fill ports on the TaqMan array. The card was
centrifuged once for 1 min at 331 × g to distribute samples to the
multiple wells on the array, then sealed to prevent well-to-well
contamination. Finally, the cards were processed and analyzed in
the CFX96™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA,
USA) with an initial denaturation step at 95°C for 30 sec followed
by 40 cycles of denaturation for 5 sec at 95°C, annealing and
extension for 30 sec at 60°C. We compared the relative mRNA levels
by the comparative 2−ΔΔCT method with β-actin as an
endogenous control for RT-qPCR in each group on days 1, 3, 5 and 7.
Detailed information on PCR, including the primer sequences, is
provided in Table II.
 | Table IINucleotide sequences of the primers
used for RT-qPCR. |
Table II
Nucleotide sequences of the primers
used for RT-qPCR.
| Gene | Primer sequence
(5′→3′) | Melting
temperature | Product size
(bp) |
|---|
| β-actin | F:
GTGGAGTGCCCAAGCACCA | 52 | 213 |
| R:
CTCTAATGTCACGACGATTTC | 52 | |
| Evi1 | F:
AGGAGAAGAACCCTGTGGCTA | 57.8 | 280 |
| R:
GAGTCCTAAAAGCGCTGTCC | 57.45 | |
| OCN | F:
AATAGACTCCGGCGCTACCT | 57.45 | 229 |
| R:
AGCTGTGCCGTCCATACTTT | 55.4 | |
| OPN | F:
AGCCATGAGTCAAGTCAGCT | 55.85 | 260 |
| R:
ACTCGCCTGACTGTCGATAG | 57.8 | |
| BSP | F:
GCACGGTTGAGTATGGGGAA | 57.45 | 260 |
| R:
TGCACCTTCCTGAGTTGAGC | 57.45 | |
| LPL | F:
AGCTGACCAGTTATGGCACC | 57.8 | 238 |
| R:
ATCCTGACCCTCGTAGCCTT | 57.45 | |
| PPARγ2 | F:
GCTGCAGCGCTAAATTCATCT | 58.66 | 195 |
| R:
GGGAGTGGTCATCCATCACAG | 59.76 | |
Protein isolation and western blot
analysis
The cells were plated into 60-mm dishes in 10%
FBS-DMEM and cultured until nearly 90% confluence. Subsequently,
they were washed twice with ice-cold PBS and then harvested with
cell lysis buffer containing Protease Inhibitor Cocktail
(Complete™; Roche Diagnostics GmbH, Mannheim, Germany) and 1 mM
phenylmethylsulfonyl fluoride (PMSF). Upon centrifugation at 13,000
x g for 5 min at 4°C, the supernatants were collected and the total
protein concentrations were determined using the BCA Protein Assay
kit (Beyotime, Shanghai, China) following the manufacturer's
instructions. SDS-PAGE (10%) gels were prepared and 5–10 mg/lane of
cellular proteins were loaded. The resolved proteins were
transferred onto a polyvinylidene difluoride (PVDF) membrane and
incubated (1:200 dilution) with primary antibodies [bone
sialoprotein (BSP; SC-292394), osteocalcin (OCN; SC-30044) and
lipoprotein lipase (LPL; SC-32885): rabbit polyclonal antibody;
osteopontin (OPN; SC-21742) and peroxisome proliferator-activated
receptor γ2 (PPARγ2; SC-7273): mouse monoclonal antibody; Santa
Cruz Biotechnology, Inc., Santa Cruz, CA, USA] and incubated
(1:1,000 dilution) with β-actin (BM0627; mouse monoclonal antibody;
Wuhan Boster Biological Technology, Ltd., Hubei, China) following
the manufacturer's instructions. Following incubation (1:5,000
dilution) with HRP-conjugated secondary antibodies (goat
anti-rabbit or goat anti-mouse IgG; Wuhan Boster Biological
Technology, Ltd.), the blotting bands were visualized using the ECL
Chemiluminescence kit (Thermo Fisher Scientific, Inc., Waltham, MA,
USA) and quantified using the ChemiDoc™ XRS+ Imaging System
(Bio-Rad).
Statistical analysis
Statistical analysis was performed using SPSS 13.0
software. Significant differences between the study groups were
determined using statistical methods [e.g., one-way analysis of
variance (ANOVA) or the Student's t-test where appropriate]. A
P-value <0.05 was considered to indicate a statistically
significant difference.
Results
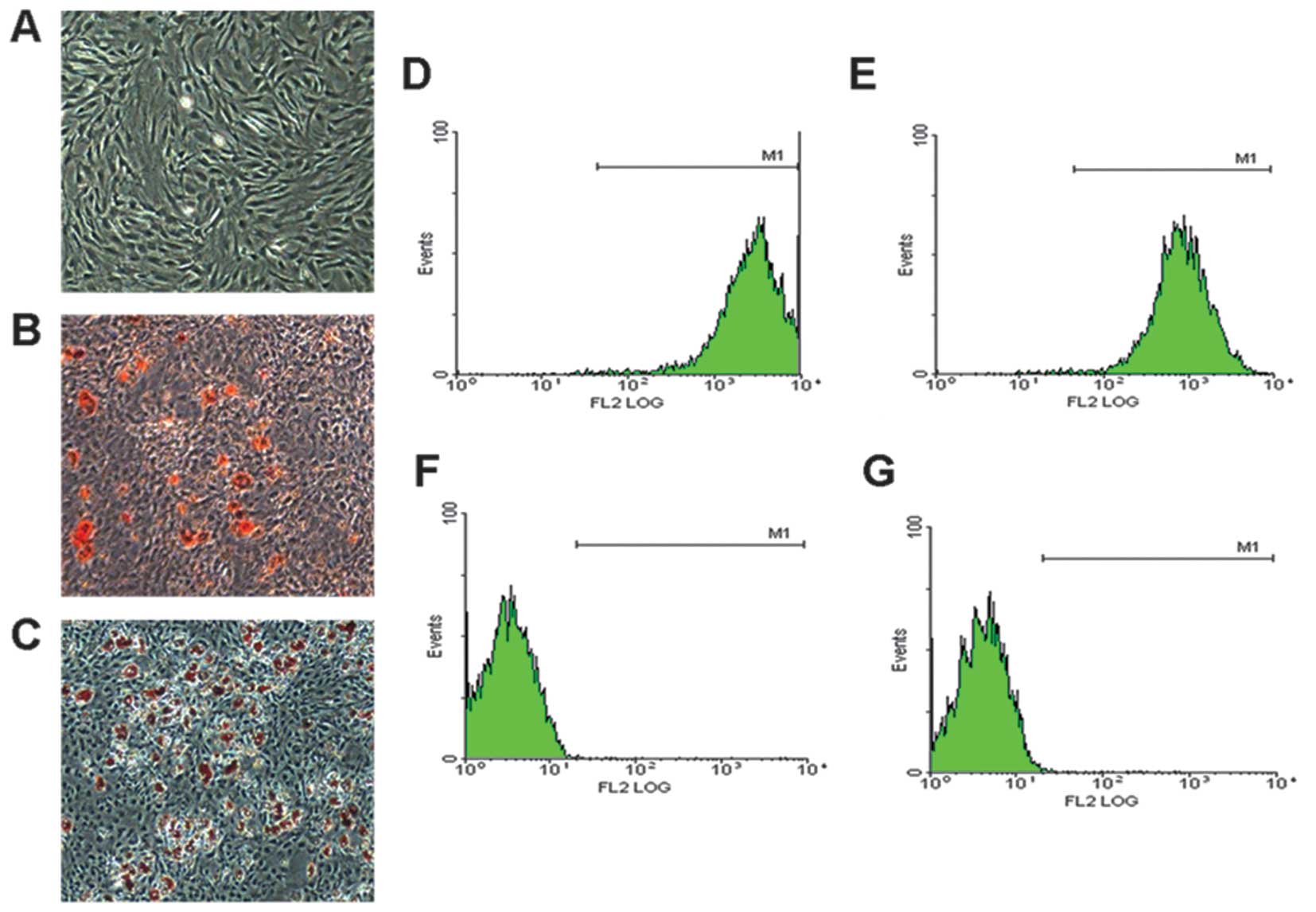
BMSC morphology and characteristics
The BMSCs were obtained from the BM of 4-week-old SD
rats maintained a fibroblast-like morphology under monolayer
culture conditions (Fig. 1A), and
were capable of differentiating into adipocytes (Fig. 1B) and osteogenic cells (Fig. 1C) under specific induction. Flow
cytometric analysis revealed that the BMSC populations were
positive for the MSC markers, CD90 and CD44 (Fig. 1D and E), but were negative for the
hematopoietic markers, CD34 and CD45 (Fig. 1F and G). These results
demonstrated that we obtained the correct rat BMSCs.
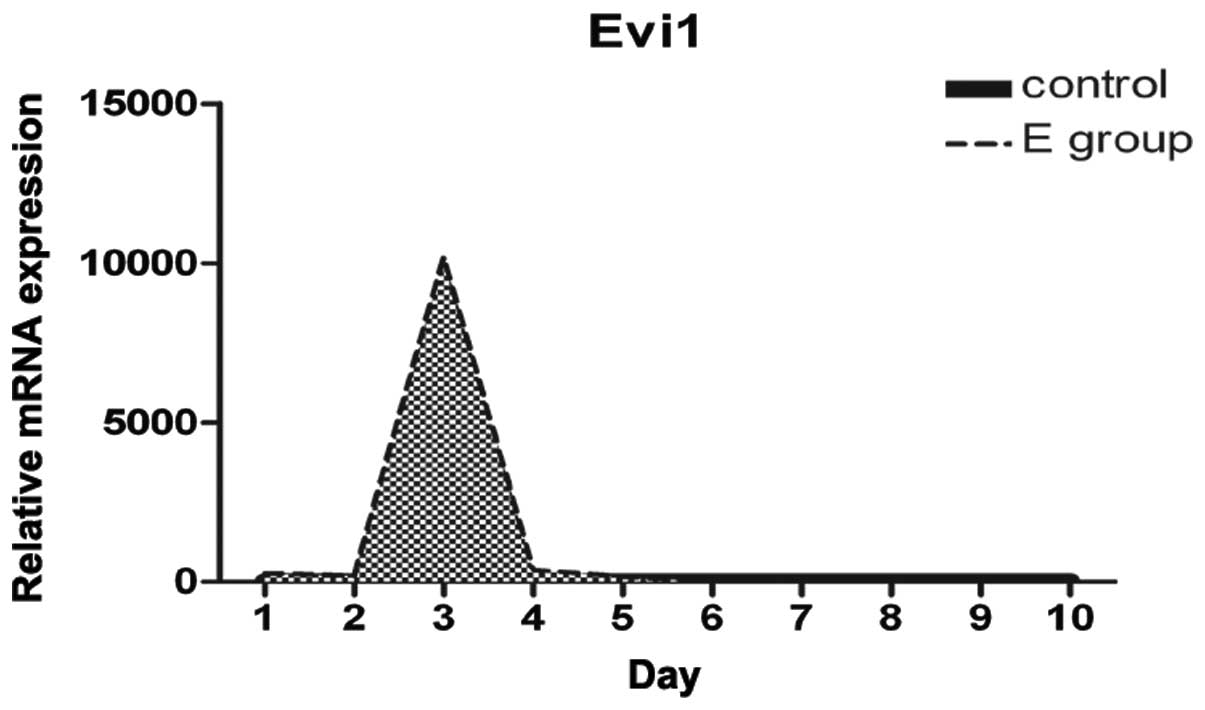
Increased Evi1 expression at the onset of
BMSC adipogenesis
We examined the differences and changes occurring in
gene expression profiles between rat BMSCs during the process of
adipogenesis and normal BMSCs over the first 10 days. Evi1
expression was dynamically regulated during adipocyte
differentiation. Evi1 mRNA levels were low in the normal BMSCs
(control), with a peak in expression being observed at confluence
and early after hormone induction, and with moderate expression
levels during the later stages of differentiation (Fig. 3). Evi1 expression was
significantly higher in the rat BMSCs undergoing adipogenic
differentiation than in the normal BMSCs. This finding suggests
that the increased Evi1 expression in differentiating BMSCs during
the early stages plays a critical role in the adipogenic
process.
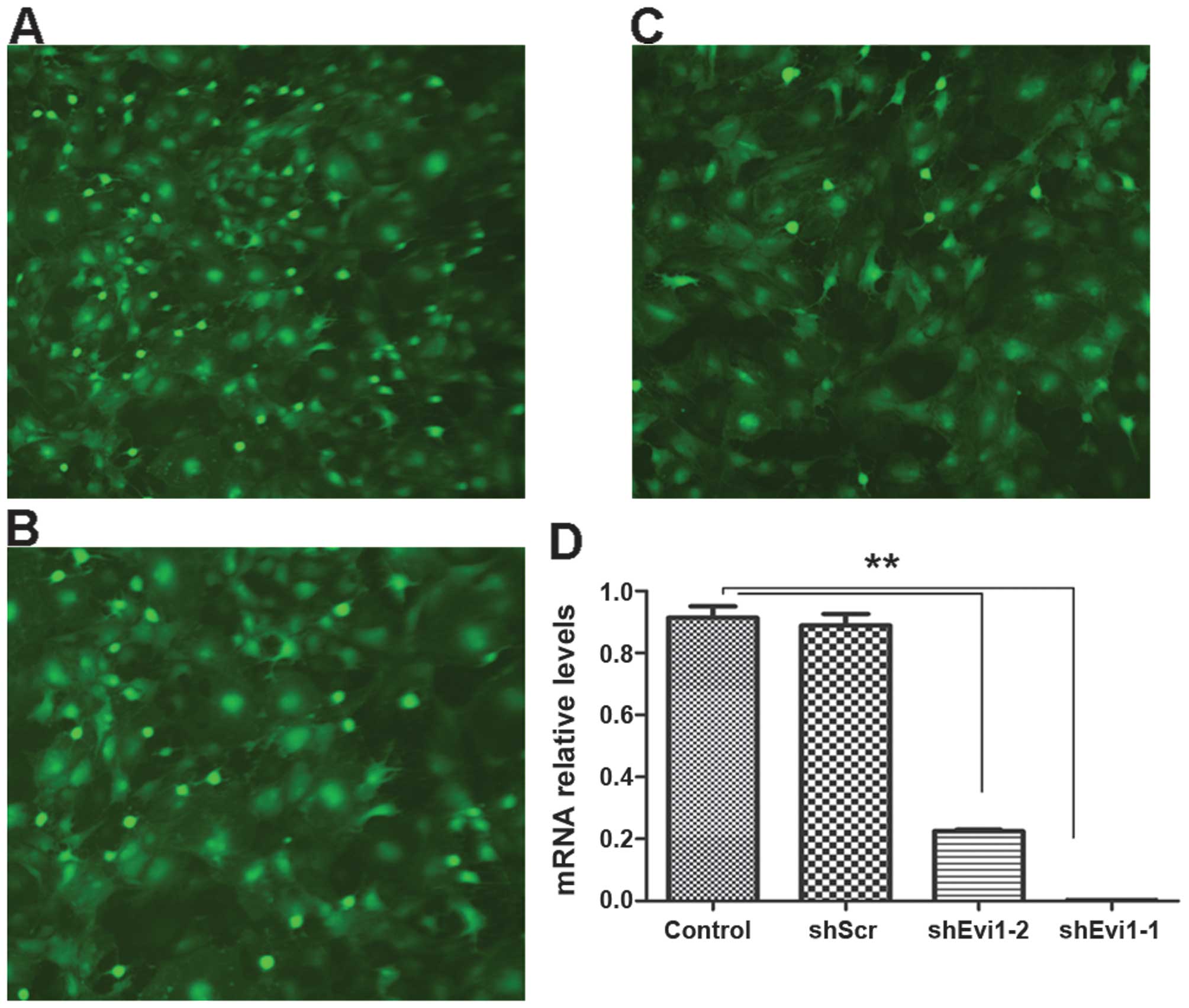
Evi1 knockdown using Ad-shEvi1 expression
vector
We created an adenoviral vector encoding shRNA
against Evi1 (Evi1 shRNA) for expressing Evi1 siRNA to knockdown
the Evi1 gene at the onset of BMSC adipogenesis. We used two
different siRNA sequences to target the rat Evi1 gene, termed
Ad-shEvi1-1 (KD1) (Fig. 2A) and
Ad-shEvi1-2 (KD2) (Fig. 2B). The
cells in the mock group were transfected with an adenoviral vector
expressing scrambled shRNA Ad-shScr (Fig. 2C). KD1 and KD2 markedly reduced
Evi1 expression at the mRNA level (Fig. 2). The inhibition rate of KD1 and
KD2 was 90.9 and 72.3%, respectively, in the BMSCs undergoing
adipogenic differentiation compared to the mock (scrambled) and
control (untransfected) groups at the mRNA level (Fig. 2D).
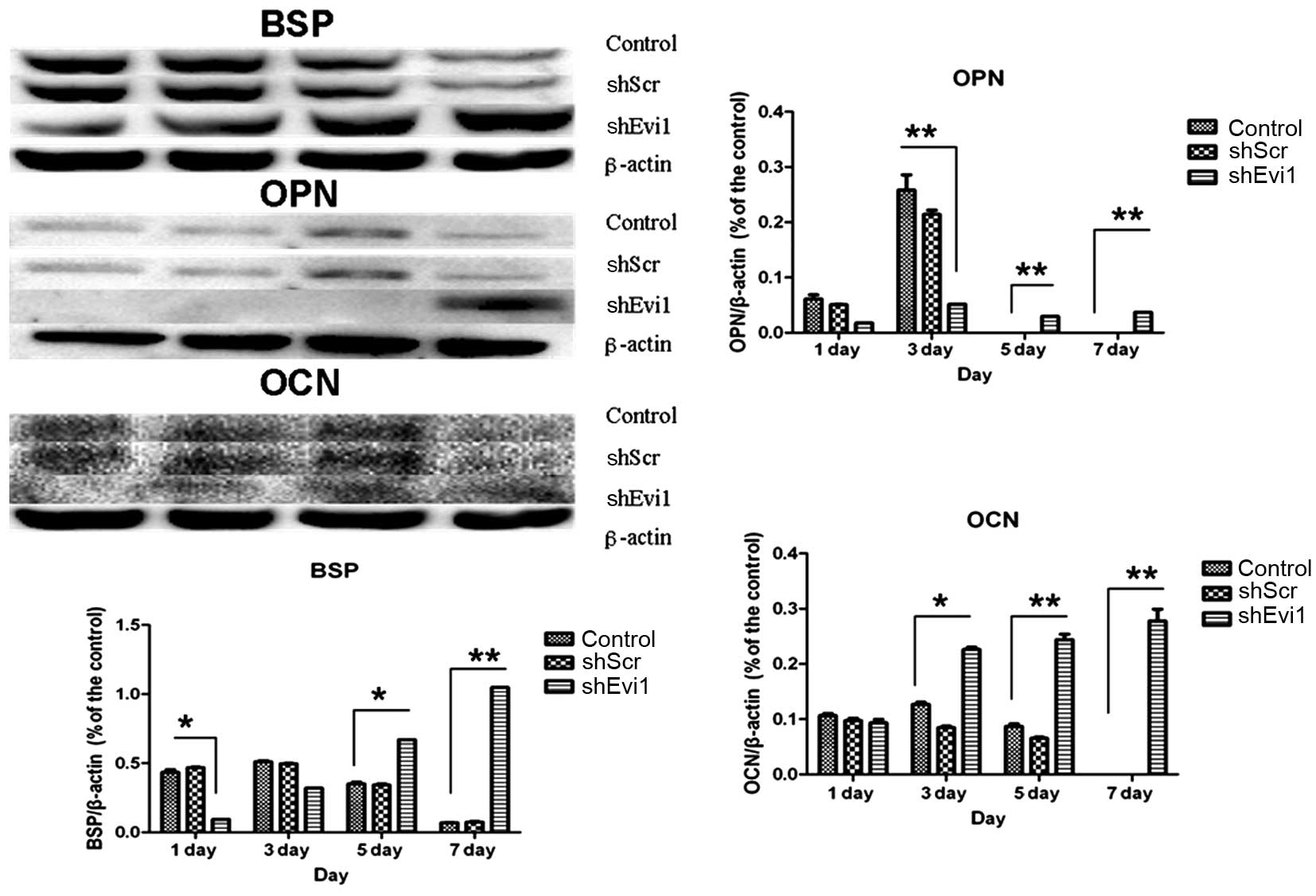
Effects of Evi1 shRNA on adipogenic and
osteogenic differentiation of BMSCs
As mentioned above, in two representative subclones,
infection with Evi1 shRNA-1 caused a more profound reduction in the
expression of the Evi1 gene than infection with Evi1 shRNA-2.
Hereafter, we mainly used the Ad-shEvi1-1 subline for further
analysis. The expression levels of specific osteogenic and
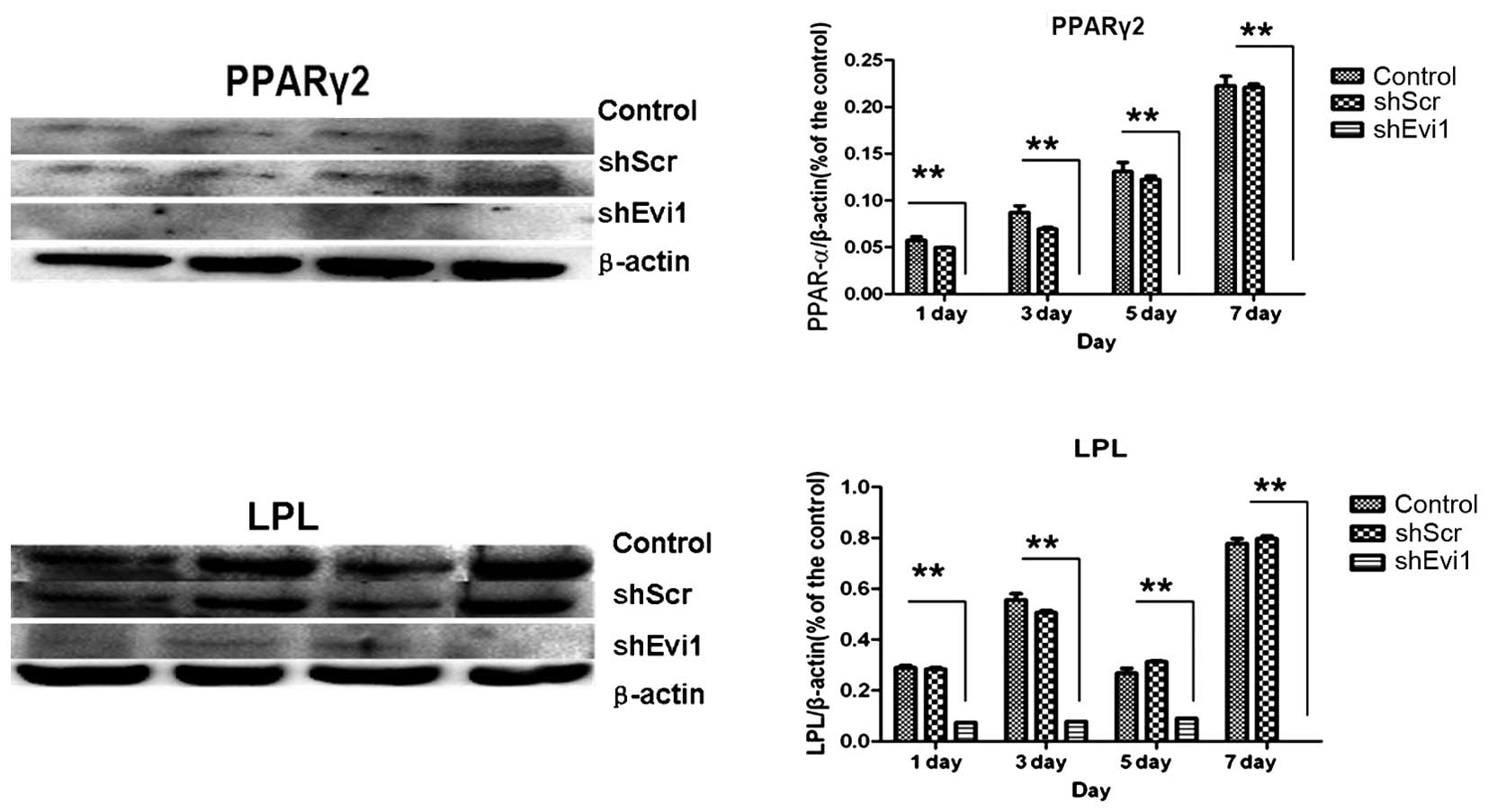
adipogenic genes were measured by RT-qPCR (Fig. 4) and western blot analysis
(Figs. 5 and 6) on days 1, 3, 5 and 7
post-transduction. Our results revealed that the specific
osteogenic markers, BSP, OPN and OCN, were downregulated during the
adipogenesis of rat BMSCs and were upregulated in the
Ad-shEvi1-transduced BMSCs at both the mRNA and protein level,
particularly on day 7 (Figs. 4
and 5). The specific adipogenic
markers, PPARγ2 and LPL, were downregulated in the
Ad-shEvi1-transduced BMSCs in comparison to the BMSCs undergoing
adipogenic differentiation at both the mRNA and the protein level
(Figs. 4 and 6). Statistical analysis indicating that
knocking down the expression of Evi1 by Ad-shEvi1 significantly
inhibited the adipogenic differentiation of BMSCs and
simultaneously enhanced the osteogenic differentiation of
BMSCs.
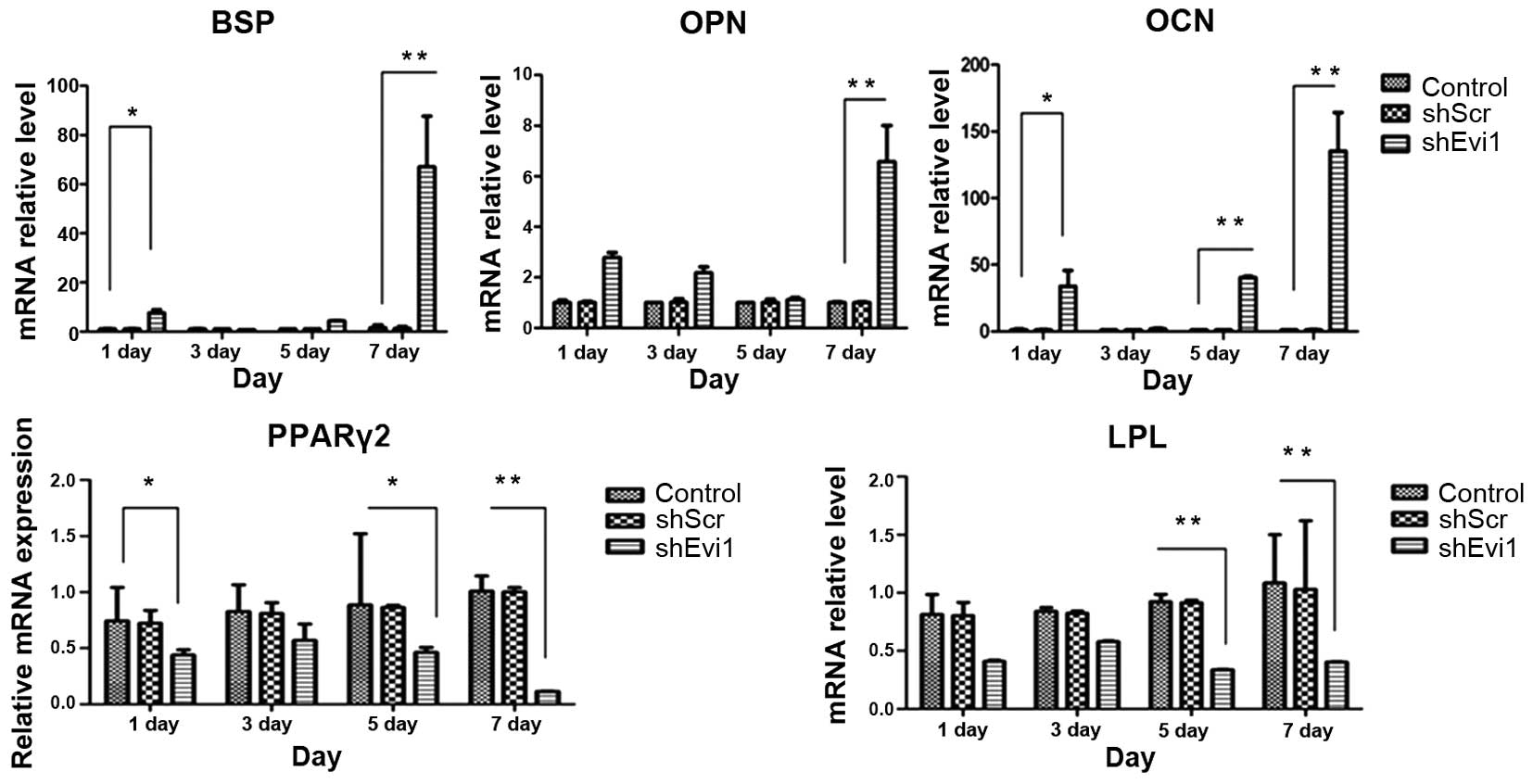 | Figure 4The mRNA levels of specific osteogenic
and adipogenic genes in each group. mRNA expression was evaluated
by RT-qPCR on days 1, 3, 5, and 7 post-transduction. Adipocyte
differentiation of the bone mesenchymal stem cell (BMSC) group was
used as a control. The specific osteogenic markers, markers
sialoprotein (BSP), osteopontin (OPN) and osteocalcin (OCN), were
upregulated in the Ad-shEvi1-transduced BMSCs, particularly on day
7. The specific adipogenic markers, peroxisome
proliferator-activated receptor γ2 (PPARγ2) and lipoprotein lipase
(LPL), were downregulated in the Ad-shEvi1-transduced BMSCs (mean ±
SEM, n=3 experiments). *P<0.05;
**p<0.01. |
Discussion
In this study, we substantiated that Evi1 was highly
expressed during the of adipogenesis of rat BMSCs, and that the
Evi1 expression level markedly increased on day 3 of adipogenic
differentiation following the addition of adipogenic inducers. The
experimental data further verified that the silencing of Evi1 by
RNAi in the BMSCs inhibited adipogenic differentiation, while it
promoted osteogenic differentiation. The inhibition of Evi1 may
thus represent a therapeutic strategy for promoting bone
formation.
Primary OP is a polygenetic disease characterized by
low bone mineral density and microarchitectural deteriorations,
leading to an increased risk of fragility fractures of the
vertebrae, the femoral neck and other typical localizations of
lower incidence (19). As OP is
characterized by pathologically enhanced osteoclast development and
function, researchers over the past decaces have mainly focused on
the imbalance of bone resorption and bone formation (20). Therefore, anti-resorptive
treatment targeting mature osteoclasts and osteoclastogenesis to
promote the RANK/RANKL pathway, has evolved as a standard therapy
over the past decades (4,19,21). However, a considerable number of
clinical trials on OP have demonstrated that anti-resorptive
treatments lead to a reduction in bone remodeling and turnover and
are assoicated with a series of adverse events (22–24). Research on presumptive
deficiencies in bone anabolism has been neglected. The importance
of the balance between bone formation and bone resorption in OP
prompted us to focus on bone formation rather than bone
resorption.
A theoretical inverse association exists between
osteogenic and adipogenic lineage commitment and differentiation,
indicating that differentiation towards an osteoblast phenotype
occurs at the expense of an adipocytic phenotype (25,26). Previous studies have revealed
enhanced BMSC differentiation into adipocytes in the BM of
post-menopausal women. All the aforementioned research results led
us to hypothesize that the inhibition of adipogenesis in stem cells
may alleviate OP using the simultaneous stimulation of
osteogenesis, which was confirmed by the results of the present
study. In addition, our data validated the results of Gimble and
Nuttall, as well as those of Beresford et al, who
demonstrated in vitro that the balance between BMSC
adipogenic and osteogenic processes exhibits an inverse or
reciprocal association (27,28).
The Evi1 isoform of MECOM is a member of the PR
(PRDI-BF1 and RIZ homology) domain-containing family of zinc finger
transcriptional regulatory proteins and is closely related in
sequence and structure to Prdm16 (15). Previous studies have revealed that
Evi1 is not expressed or is expressed at low levels in normal BM
(28). In this study, we found
that Evi1 expression increased at the onset of adipogenesis of rat
BMSCs, and that the mRNA level of Evi1 was low in normal growing
BMSCs. Taken together, these results indicate that the increased
expression of Evi1 in early differentiating BMSCs is critical to
the adipogenic process.
Our data validated Evi1 gene expression in the
process of BMSC adipogenic differentiation using mature adipogenic
induction medium at the cellular level. Our results revealed that
the Evi1 gene was not only expressed in the first 5 days after the
addition of adipogenic inducers, but that its mRNA expression
reached peak levels on day 3. We also measured the protein
expression level of Evi1, but we were unable to produce a result.
In order to determine whether the high expression of Evi1
determines the adiopogenic lineage commitment, we further measyred
the RNA and protein levels of adipogenic and osteogenic
markers.
We observed a lower expression of the adipogenic
markers, PPARγ2 and LPL, in the RNAi group in comparison to the
adipocyte differentiation group on days 1, 3, 5, and 7 of
differentiation. Furthermore, our results revealed a higher
expression of the osteogenic markers, BSP, OPN and OCN, in the RNAi
group in comparison to the adipocyte differentiation group on days
1, 3, 5, and 7 of differentiation. Notably, the levels of the
markers, BSP, OPN, OCN, PPARγ2 and LPL, varied markedly on day 7.
This phenomenon can be explained by the fact that the expression of
the Evi1 gene increased in the first 5 days of BMSC adipogenesis
(Fig. 3) and that the
corresponding osteogenic and adipogenic markers as the Evi1
downstream genes possess hysteresis. The mechanisms involved remain
to be elucidated. It will now be important to examine the role of
Evi1 in estrogen deficiency-induced OP using gain-
and-loss-of-function studies on animals.
Evi1 is highly expressed in certain cytogenetic
subsets of adult acute myeloid leukaemia (AML), and has been
associated with inferior survival (29). Consequently, we presume that the
side-effects caused by knocking down the Evi1 gene are relatively
limited.
In conclusion, in this study, we identified Evi1 as
a key competency factor that allows rat BMSCs to undergo
adipogenesis. While animals are not exactly the same as humans, it
is logical to make the inference that an effect observed in rats
may also apply to humans. These findings indicate that the
suppression of the Evi1 gene prevents estrogen deficiency-induced
bone loss by the simultaneous stimulation of osteogenesis and the
inhibition of adipogenesis.
Abbreviations:
|
BMSCs
|
bone marrow-derived mesenchymal stem
cells
|
|
Evi1
|
ectopic viral integration site-1
|
|
BSP
|
bone sialoprotein
|
|
OPN
|
osteopontin
|
|
OCN
|
osteocalcin
|
|
PPARγ
|
peroxisome proliferator- activated
receptor γ
|
|
LPL
|
lipoprotein lipase
|
References
|
1
|
Weinstein RS, Jilka RL, Parfitt AM and
Manolagas SC: Inhibition of osteoblastogenesis and promotion of
apoptosis of osteoblasts and osteocytes by glucocorticoids.
Potential mechanisms of their deleterious effects on bone. J Clin
Invest. 102:274–282. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Morrison NA, Qi JC, Tokita A, Kelly PJ,
Crofts L, Nguyen TV, Sambrook PN and Eisman JA: Prediction of bone
density from vitamin D receptor alleles. Nature. 367:284–287. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Seeman E: Bone quality: The material and
structural basis of bone strength. J Bone Miner Metab. 26:1–8.
2008. View Article : Google Scholar
|
|
4
|
Pietschmann P, Rauner M, Sipos W and
Kerschan-Schindl K: Osteoporosis: An age-related and
gender-specific disease - a mini-review. Gerontology. 55:3–12.
2009. View Article : Google Scholar
|
|
5
|
Kassem M and Abdallah BM: Human
bone-marrow-derived mesenchymal stem cells: Biological
characteristics and potential role in therapy of degenerative
diseases. Cell Tissue Res. 331:157–163. 2008. View Article : Google Scholar
|
|
6
|
Egermann M, Heil P, Tami A, Ito K, Janicki
P, Von Rechenberg B, Hofstetter W and Richards PJ: Influence of
defective bone marrow osteogenesis on fracture repair in an
experimental model of senile osteoporosis. J Orthop Res.
28:798–804. 2010.
|
|
7
|
Rodriguez JP, Astudillo P, Rios S and Pino
AM: Involvement of adipogenic potential of human bone marrow
mesenchymal stem cells (MSCs) in osteoporosis. Curr Stem Cell Res
Ther. 3:208–218. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pittenger MF, Mackay AM, Beck SC, Jaiswal
RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S and
Marshak DR: Multilineage potential of adult human mesenchymal stem
cells. Science. 284:143–147. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Uejima S, Okada K, Kagami H, Taguchi A and
Ueda M: Bone marrow stromal cell therapy improves femoral bone
mineral density and mechanical strength in ovariectomized rats.
Cytotherapy. 10:479–489. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ocarino Nde M, Boeloni JN, Jorgetti V,
Gomes DA, Goes AM and Serakides R: Intra-bone marrow injection of
mesenchymal stem cells improves the femur bone mass of osteoporotic
female rats. Connect Tissue Res. 51:426–433. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lien CY, Chih-Yuan Ho K, Lee OK, Blunn GW
and Su Y: Restoration of bone mass and strength in
glucocorticoid-treated mice by systemic transplantation of CXCR4
and cbfa-1 co-expressing mesenchymal stem cells. J Bone Miner Res.
24:837–848. 2009. View Article : Google Scholar
|
|
12
|
Park S-H, Sim WY, Min B-H, Yang SS,
Khademhosseini A and Kaplan DL: Chip-based comparison of the
osteogenesis of human bone marrow- and adipose tissue-derived
mesenchymal stem cells under mechanical stimulation. PLoS One.
7:e466892012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mimeault M and Batra SK: Recent progress
on tissue-resident adult stem cell biology and their therapeutic
implications. Stem Cell Rev. 4:27–49. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rodríguez JP, Garat S, Gajardo H, Pino AM
and Seitz G: Abnormal osteogenesis in osteoporotic patients is
reflected by altered mesenchymal stem cells dynamics. J Cell
Biochem. 75:414–423. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ishibashi J, Firtina Z, Rajakumari S, Wood
KH, Conroe HM, Steger DJ and Seale P: An Evi1-C/EBPβ complex
controls peroxisome proliferator-activated receptor γ2 gene
expression to initiate white fat cell differentiation. Mol Cell
Biol. 32:2289–2299. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yue B, Lu B, Dai KR, Zhang XL, Yu CF, Lou
JR and Tang TT: BMP2 gene therapy on the repair of bone defects of
aged rats. Calcif Tissue Int. 77:395–403. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fu L, Tang T, Miao Y, Zhang S, Qu Z and
Dai K: Stimulation of osteogenic differentiation and inhibition of
adipogenic differentiation in bone marrow stromal cells by
alendronate via ERK and JNK activation. Bone. 43:40–47. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liang J, Liu C, Qiao A, Cui Y, Zhang H,
Cui A, Zhang S, Yang Y, Xiao X, Chen Y, et al: MicroRNA-29a-c
decrease fasting blood glucose levels by negatively regulating
hepatic gluconeogenesis. J Hepatol. 58:535–542. 2013. View Article : Google Scholar
|
|
19
|
Rachner TD, Khosla S and Hofbauer LC:
Osteoporosis: Now and the future. Lancet. 377:1276–1287. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Manolagas SC: Birth and death of bone
cells: Basic regulatory mechanisms and implications for the
pathogenesis and treatment of osteoporosis. Endocr Rev. 21:115–137.
2000.PubMed/NCBI
|
|
21
|
Khosla S: Minireview: The OPG/RANKL/RANK
system. Endocrinology. 142:5050–5055. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schwartzman J and Yazici Y: Denosumab in
postmenopausal women with low bone mineral density. N Engl J Med.
354:2390–2391; author reply 2390–2391. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Watts NB, Roux C, Modlin JF, Brown JP,
Daniels A, Jackson S, Smith S, Zack DJ, Zhou L, Grauer A, et al:
Infections in postmenopausal women with osteoporosis treated with
denosumab or placebo: Coincidence or causal association? Osteoporos
Int. 23:327–337. 2012. View Article : Google Scholar :
|
|
24
|
Sobacchi C, Frattini A, Guerrini MM,
Abinun M, Pangrazio A, Susani L, Bredius R, Mancini G, Cant A,
Bishop N, et al: Osteoclast-poor human osteopetrosis due to
mutations in the gene encoding RANKL. Nat Genet. 39:960–962. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
James AW, Pang S, Askarinam A, Corselli M,
Zara JN, Goyal R, Chang L, Pan A, Shen J, Yuan W, et al: Additive
effects of sonic hedgehog and Nell-1 signaling in osteogenic versus
adipogenic differentiation of human adipose-derived stromal cells.
Stem Cells Dev. 21:2170–2178. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pei L and Tontonoz P: Fat's loss is bone's
gain. J Clin Invest. 113:805–806. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gimble JM and Nuttall ME: The relationship
between adipose tissue and bone metabolism. Clin Biochem.
45:874–879. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Beresford JN, Bennett JH, Devlin C, Leboy
PS and Owen ME: Evidence for an inverse relationship between the
differentiation of adipocytic and osteogenic cells in rat marrow
stromal cell cultures. J Cell Sci. 102:341–351. 1992.PubMed/NCBI
|
|
29
|
Ho PA, Alonzo TA, Gerbing RB, Pollard JA,
Hirsch B, Raimondi SC, Cooper T, Gamis AS and Meshinchi S: High
EVI1 expression is associated with MLL rearrangements and predicts
decreased survival in paediatric acute myeloid leukaemia: A report
from the children's oncology group. Br J Haematol. 162:670–677.
2013. View Article : Google Scholar : PubMed/NCBI
|