Introduction
Colorectal cancer (CRC) is the third most common
type of cancer worldwide (1).
Even though the survival rate has doubled over the past 20 years,
it remains one of the most fatal malignancies worldwide (2–4).
CRC is a heterogeneous disease, and a complex series of molecular
changes have been reported to be involved in the transformation of
the normal colonic mucosa into a potentially invasive tumor over a
long period of time (5). The
development of CRC is generally believed to result from complex
interactions between genetic and environmental factors (6). Molecular biomarkers based on gene
mutations located in oncogenes or tumor suppressors, such as
adenomatous polyposis coli (APC), v-raf murine sarcoma viral
oncogene homolog B1 (BRAF) and p53 have been established for over a
decade (6); however, the clinical
applications of these biomarkers remain limited. For this reason,
the development of novel molecular markers is necessary in order to
improve the diagnosis, prognosis and treatment of CRC.
MicroRNAs (miRNAs or miRs) are a class of small
non-coding RNAs, approximately 22 nucleotides in length, that
post-transcriptionally regulate the expression of up to 1/3 human
genes. miRNAs target mRNAs through imperfect base pairing of the
5′-end of miRNAs to multiple sites in the 3′-untranslated regions
(UTRs) of the target transcript, and this imperfect miR-mRNA hybrid
with central bulges (9–12 nt) recruits a microRNA ribonucleoprotein
complex (miRNP) that enables the translational inhibition or
exonucleolytic mRNA decay (7).
More than half of the annotated human miRNAs have been found to be
located in tumor-related chromosome segments or fragile hot spots
that are predisposed to translocation, insertion or deletion in a
number of human malignancies, including colon cancer (8), and thus, some miRNAs can function
either as oncogenes or tumor suppressors (9–11).
Using microarray technology, we can detect the
expression levels of thousands of genes simultaneously, which makes
it a promising tool with which to discover novel potential
diagnostic or therapeutic biomarkers, and it can assist us in
exploring the underlying molecular mechanisms (12,13). A growing body of evidence
indicates that DNA microarray profiling performed on clinical
specimens may be directly applied to cancer diagnosis, as well as
to the evaluation of prognosis (14,15). Additionally, expression profiling
has been used to predict the response to cancer treatment (16). Expression profiling analysis has
also been used to establish characteristic miRNA signatures that
can predict the clinical outcomes of various malignancies (17).
In order to broaden our understanding of the
molecular mechanisms underlying the pathogenesis of human CRC and
explore novel potential biomarkers for use in clinical practice, in
this study, we examined and compared gene expression profiles in
CRC tissues and adjacent non-cancerous control tissues obtained
from surgical resections, and using functional analysis, we focused
on the differentially expressed miRNAs, their potential target
gene, and their roles in regulating colon cancer cell behavior.
Materials and methods
Tissue sample collection
Paired primary CRC samples and adjacent
histologically normal tissues were collected from a total of 60
patients with CRC in the Department of General Surgery at The Third
Affiliated Hospital of Guangzhou Medical University (Guangdong,
China). Tumor tissues and adjacent non-cancerous tissues that were
at least 2.0 cm distal to the tumor margins were snap-frozen in
liquid nitrogen and then stored at −80°C until use. All samples
were evaluated by two pathologists, according to the World Health
Organization (WHO) guidelines (18). Those who had received chemotherapy
or radiotherapy prior to surgery were excluded from this study.
Informed consent was obtained from all patients, and the study was
approved by the Human Research Ethics Committee of Guangzhou
Medical University.
Gene expression analysis and data
processing
RNA was isolated from each tissue sample and
subjected to gene expression analyses using whole genome expression
microarrays (Funeng Biotech Co., Ltd., Guangzhou, China). Following
array data processing, differentially expressed genes were
identified using the GeneSpring GX software package (Funeng
Biotech), and only those miRNAs with differences >1.6-fold
between the 2 groups (normal and tumor tisues) are listed in
Table I.
 | Table IDifferentially expressed microRNAs
between cancerous tissues and adjacent non-cancerous control
tissues, as identified by microarray analysis. |
Table I
Differentially expressed microRNAs
between cancerous tissues and adjacent non-cancerous control
tissues, as identified by microarray analysis.
| Gene | Mean
| t | P-value |
|---|
| Normal tissues | Tumor tissues |
|---|
| Downregulated |
| hsa-let-7c | 0.98 | −0.43 | 3.809 | 0.001 |
| microRNA-126 | 2.8 | 1.56 | 3.852 | 0.001 |
| microRNA-451 | 1.91 | 0.27 | 4.153 | 0.001 |
| microRNA-23b | 2.7 | 1.14 | 4.394 | <0.001 |
| microRNA-1979 | 1.79 | 0.47 | 4.453 | <0.001 |
| microRNA-27b | 1.3 | −0.19 | 4.543 | <0.001 |
| hsa-let-7b | 1.1 | −0.52 | 5.702 | <0.001 |
| microRNA-125b | 1.43 | −0.37 | 6.202 | <0.001 |
|
microRNA-768-3p | 3.34 | 1.56 | 6.564 | <0.001 |
| microRNA-99a | 3.79 | 1.83 | 6.73 | <0.001 |
| microRNA-4770 | 3.17 | 0.91 | 7.162 | <0.001 |
|
microRNA-125a-5p | 2.66 | 0.84 | 7.195 | <0.001 |
| microRNA-1 | 6.47 | 2.68 | 7.218 | <0.001 |
| microRNA-100 | 3.45 | 1.58 | 7.294 | <0.001 |
| microRNA-143 | 2.3 | −0.5 | 7.389 | <0.001 |
|
microRNA-145* | 8.42 | 4.99 | 7.897 | <0.001 |
| Upregulated |
| hsa-let-21 | 0.96 | −0.49 | 3.901 | 0.001 |
| microRNA-128 | 2.5 | 1.48 | 3.912 | 0.001 |
| microRNA-375 | 2.01 | 0.24 | 4.024 | 0.001 |
| microRNA-26b | 2.81 | 1.09 | 4.283 | <0.001 |
| microRNA-146a | 1.81 | 0.46 | 4.503 | <0.001 |
In silico analysis
The target genes of the miRNA or the regulatory
miRNA of certain genes were identified by searching online miRNA
databases (www.TargetScan.org and www.miRanda.org). The candidate gene list was
shortened based on their pathophysiological function.
RNA isolation and quantitative PCR
(qPCR)
Total RNA was isolated from HCT116 cells and tissues
using the total RNA isolation kit (Invitrogen, Carlsbad, CA, USA).
RNA was reverse transcribed into cDNA using the SuperScript III kit
(Invitrogen). Total RNA concentrations were measured using a
NanoDrop ND-1000 fluorospectrometer (NanoDrop Technologies,
Wilmington, DE, USA). To confirm the differential expression of the
selected miRNAs identified in our microarray analyses, standard
SYBR-Green-based qPCR (Sigma-Aldrich, St. Louis, MO, USA) analyses
were conducted. Specific primer sequences were as follows: miR-1
forward, 5′-aggcaaagagaatagttccccag-3′ and reverse,
5′-cggacttcgtggagatgga-3′; miR-145, forward,
5′-ctggctcttgatacccccct-3′ and reverse, 5′-tcaacactgagacgggctcc-3′;
miR-451, forward, 5′-ggaaaccaggaagcctagca-3′ and reverse,
5′-tgattcagtgccattttgcc-3′. Fluorescence was measured at the final
step of each cycle using a real-time PCR system (ABI-7900; Applied
Biosystems, Foster City, CA, USA). The housekeeping gene, U6, was
used to normalize the expression data for the gene of interest.
Luciferase reporter assay
The human NLR family, apoptosis inhibitory protein
(NAIP) 3′-UTRs were amplified by PCR and cloned into a modified
version of pcDNA3.1(+) that contained a firefly luciferase reporter
gene (Invitrogen), at a position downstream of the luciferase
reporter. The vectors were termed wild-type 3′UTRs, and the primers
for cloning the 3′-UTRs of NAIP were as follows: sense, 5′-GAT GAA
TTC TTA TCC CCT GCC CCT TCC-3′ and antisense, 5′-TAT CTC GAG TGG
GTC CAC CAT GGC TAA GTG A-3′. Site-directed mutagenesis of the
miRNA binding sites in the NAIP 3′UTR was performed using a
Site-Directed Mutagenesis kit (SBS Genetech, Beijing, China) and
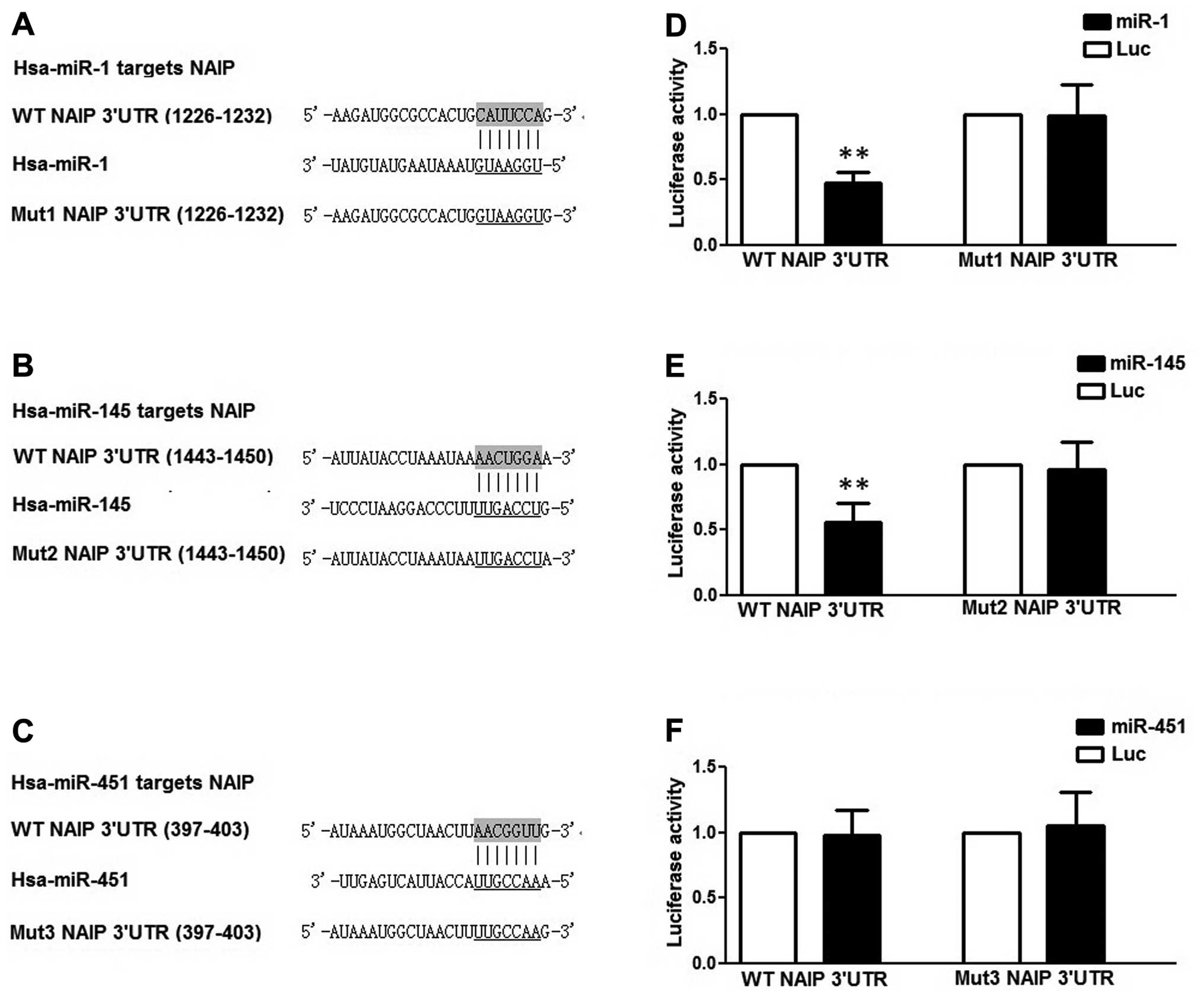
termed mutant-1, -2 and -3 3′UTRs, as shown in Fig. 2. All constructs were confirmed by
DNA sequencing. CRC cells (HCT116) were grown in a 48-well plate
and co-transfected with 400 ng of either individual miR mimics
(miR-1, miR-145 or miR-451) or the control, 40 ng of the firefly
luciferase reporter plasmid including the 3′-UTR of the target
gene, and 4 ng of pRL-TK, a plasmid expressing Renilla
luciferase (Promega, Madison, WI, USA). After 24 h, the cells were
collected and the luciferase activity was determined using a
TD-20/20 luminometer (Turner Biosystems, Sunnyvale, CA, USA).
Cell cycle and apoptosis analysis
At 48 h following transfection, the cells were
collected by trypsinization and washed with phosphate-buffered
saline (PBS). An Annexin-V-FLUOS Staining kit (Roche, Mannheim,
Germany) was used to stain the cells, according to the
manufacturer's instructions. Apoptosis was evaluated through
fluorescence-activated cell sorting (FACS) using CellQuest software
(Becton-Dickinson and Company, Franklin Lakes, NJ, USA), and
Annexin-V-FLUOS-positive cells were regarded as apoptotic cells.
Flow cytometric analysis was performed using a Becton-Dickinson
flowcytometer (BD Biosciences, San Jose, CA, USA).
Western blot analysis
The cells were lysed in RIPA lysis buffer [50 mM
Tris-HCl, pH 8.0, 250 mM NaCl, 1% NP40, 0.5% (w/v) sodium
deoxycholate, 0.1% sodium dodecylsulfate] (from Beyotime, Nanjing,
China). The lysates were then centrifuged at 12,000 × g/min at 4°C
for 15 min, and were subjected to 10% SDS polyacrylamide gel
electrophoresis. The separated proteins were then transferred onto
polyvinylidene difluoride membranes (Millipore, Billerica, MA,
USA), and the membranes were subsequently incubated with primary
antibodies at 4°C overnight and washed 3 times with PBS for 5 min,
followed by incubation with horseradish peroxidase-conjugated
secondary antibodies (1:15,000 dilution; Cat. no. ZB-2301; Beijing
Zhongshan Golden Bridge Biotechnology Co., Ltd., Beijing, China) at
room temperature for 1.5 h and detected with an ECL kit (Applygen,
Beijing, China). The primary anti-NAIP antibody (Cat. no. sc-11064;
Santa Cruz Biotechnology, Santa Cruz, CA, USA), and anti-β-actin
antibody (Cat. no. sc-47778; Santa Cruz Biotechnology) were diluted
at 1:2,000.
Cell culture
The human CRC cell line, HCT116, was purchased from
the American Type Culture Collection (ATCC; Manassas, VA, USA). The
HCT116 cells were cultured in Dulbecco's modified Eagle's medium
(DMEM; Invitrogen) supplemented with 10% heat-inactivated fetal
bovine serum (FBS; Invitrogen), 100 U/ml penicillin and 100 lg/ml
streptomycin (Invitrogen), at 37°C in a 5% CO2
atmosphere.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay
The HCT116 cells were transfected with miR-1 mimics
and/or miR-145 mimics, or miR-1 inhibitors, or negative control
(NC) and incubated for 48 h. MTT soloution (25 µl 0.5%;
Sigma-Aldrich) was added to each well followed by incubation for a
further 3 h. The medium was then replaced with 150 µl
dimethyl sulfoxide (DMSO; Sigma-Aldrich), which was added to the
microplate, and shaken on a rotary platform for 15 min. The cell
survival rate was determined by measuring the optical density (OD)
values at a 492 nm wavelength and comparing these values with the
OD values of the controls.
RNA oligoribonucleotide and
transfection
MicroRNA mimics were purchased from Dharmacon
(Lafayette, CO, USA): miR-145 mimic (MIMAT0000437), miR-1 mimic
(MIMAT0000416) and miR-451 mimic (MIMAT0001631). miRNA inhibitors
were purchased from Guangzhou RiboBio Co., Ltd., Guangzhou, China:
miR-1 inhibitor (miR10005824-1-2) and miR-145 inhibitor
(miR20000437-1-5). The HCT116 cells were cultured to 70%
confluence, and Lipofectamine 2000 (Invitrogen) was used to
transfect the DNA or RNA.
Statistical analysis
Data are expressed as the means ± SD. Statistical
analysis was performed using the SPSS 19.0 software package (IBM
Inc., Chicago, IL, USA). P-values were calculated using the
two-tailed Student's t-test or one-way ANOVA. P-values <0.05
were considered to indicate statistically significant differences,
and P-values <0.01 were considered to indicate highly
significant differences.
Results
Microarray analysis of miRNAs
differentially expressed in cancer and adjacent non-cancerous
tissues from patients with CRC
To identify the miRNAs involved in the regulation of
CRC cell proliferation, invasion and metastasis, we examined and
compared the gene expression profiles of whole CRC tissues and
adjacent non-cancerous mucosa obtained from surgical resections
using an miRNA microarray platform (whole genome expression
microarrays as described in the Materials and methods). Of the
human miRNAs on the array, a variety of miRNAs exhibited a
significant difference in expression between the cancerous and
non-cancerous samples. Among these miRNAs, 5 were upregulated and
16 were downregulated by >1.6-fold, between the 2 groups
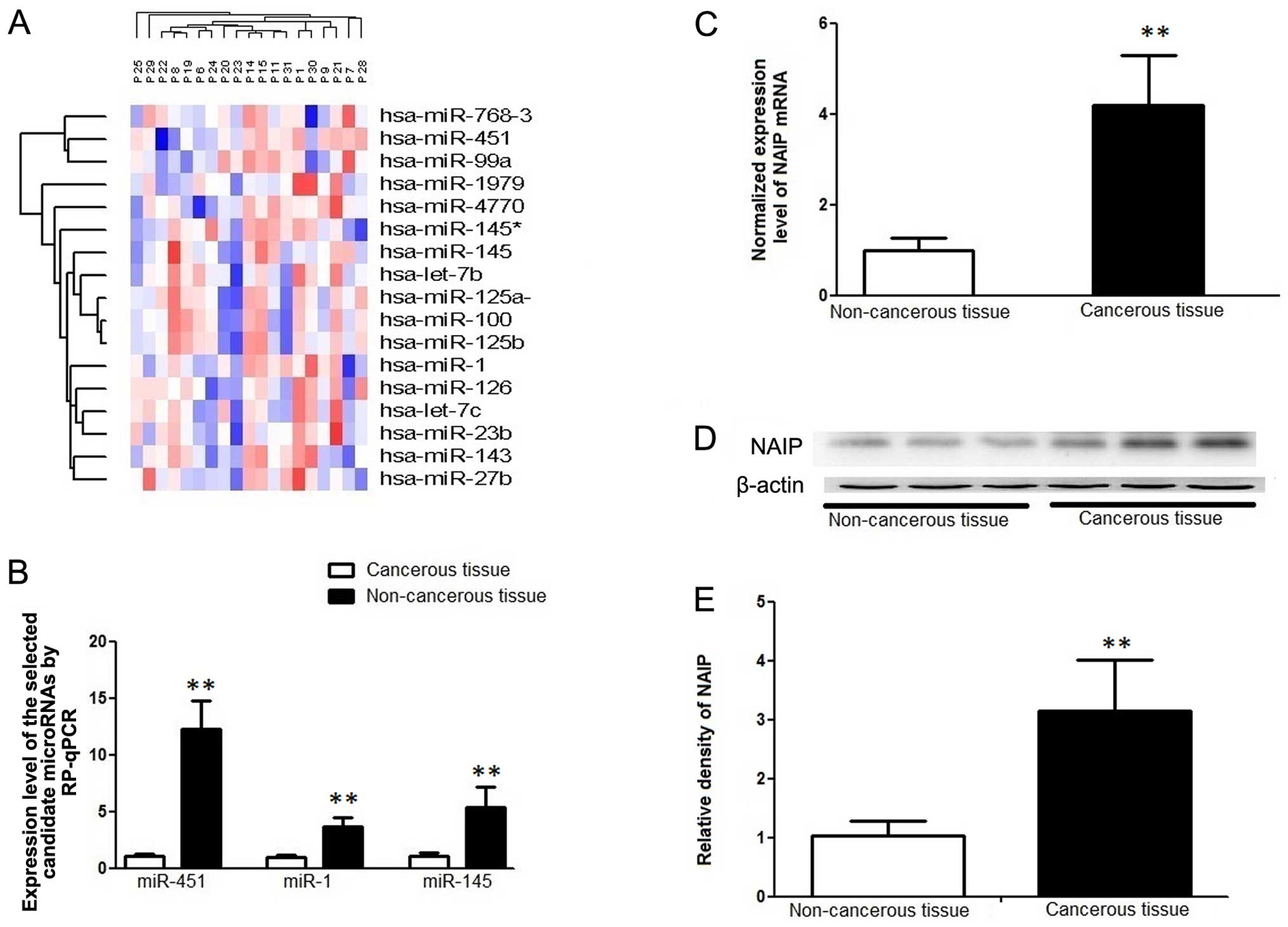
(Fig. 1A and Table I). In particular, miR-145
exhibited the most significant difference in expression, with a
log2 reduction of 3.43 in the cancer tissues compared with the
controls. By using in silico analsyis and searching online
microRNA databases, we identified NAIP as a potential target of
miR-145. Furthermore, we found that NAIP was also a possible shared
target of two other significantly downregulated miRNAs, miR-1 and
miR-451.
Microarray validation by RT-qPCR of
miR-145, miR-1 and miR-451 in CRC tissues
To validate the miRNA microarray data and examine
the expression patterns of the focused miRNAs and the target gene,
we measured the expression of miR-145, miR-1 and miR-451 in an
additional 40 CRC tissue samples by RT-qPCR. These were quantified
using the ΔΔCt method together with the previous 20 samples. One of
the tested samples was selected as the calibrator, and the relative
expression value of this sample was set as 1. As shown in Fig. 1B, we found that the basal
expression levels of miR-145, miR-1 and miR-451 were significantly
decreased in the tumor tissues as compared with the adjacent
non-cancerous control tissues.
Expression patterns of NAIP in cancerous
and non-cancerous tissues
To determine the clinical significance of the target
gene of miR-145 and miR-1, we measured the NAIP mRNA and protein
levels in 40 pairs of matched CRC specimens by qPCR and western
blot analysis. The protein expression of NAIP was significantly
upregulated in the CRC tissues compared with the adjacent
non-cancerous tissues (Fig.
1C–E).
NAIP is a shared target of miR-145 and
miR-1, but not miR-451 in HCT116 cells
The aforementioned findings indicated that miR-145,
miR-1 and miR-451 acted as a cell growth inhibitor in CRC tissues.
Therefore, we searched for potential gene targets of miR-145, miR-1
and miR-451, which may contribute to its prometastatic function. We
performed in silico analysis to search for the potential
gene targets of miR-145, miR-1 and miR-451 using the bioinformatics
algorithms, TargetScan (http://www.targetscan.org/) and miRanda (http://www.microrna.org/microrna/home.do). Both
algorithms identified NAIP as a potential target gene of miR-145,
miR-1 and miR-451 (Fig.
2A–C).
NAIP is a well-known inhibitor of
apoptosis that promotes tumor growth
The above findings suggest that the oncogenic
function of miR-145, miR-1 and miR-451 in CRC may be attributed to
the suppression of NAIP expression. To examine this hypothesis,
luciferase reporter assays were performed using luciferase
reporters carrying the wild-type NAIP 3′-UTR, and we found that
miR-145 and miR-1, but not miR-451, had a significantly decreased
activity compared with the controls in the HCT116 cells (Fig. 2D–F).
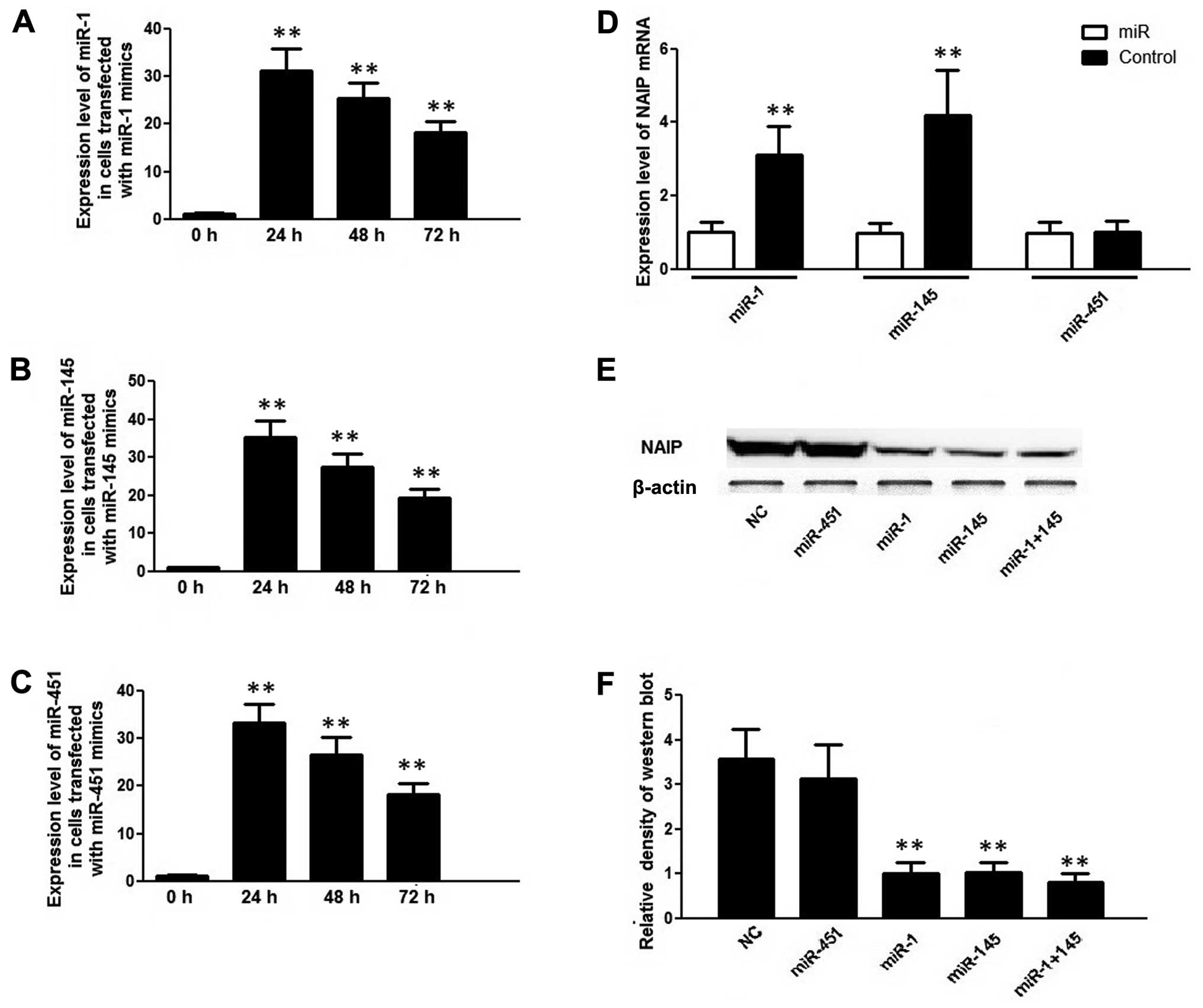
Furthermore, the HCT116 cells transfected with
miR-145, miR-451 and miR-1 mimics exhibited an approximately
30-fold increase in each corresponding miR at 24 h following
transfection compared with the controls (0 h) (Fig. 3A–C); however, only miR-1 and
miR-145 decreased the mRNA and protein expression of NAIP in the
HCT116 cells, and no such effect was observed in the cells
transfected with the miR-451 mimic (Fig. 3D–F). These findings corroborate
the hypothesis that NAIP expression has an inverse correlation with
miR-145 and miR-1 expression, but not with miR-451 expression in
CRC cells.
Additionally, miR-1 and miR-145 inhibitors were
transfected into the HCT116 cells, but only the miR-1 inhibitor
suppressed the expression level of miR-1, and endogenous miR-145
expression was already too low to be further reduced (Fig. 4A and B). As expected, the decrease
in the expression of miR-1, caused by the miR-1 inhibitor, led to
an increase in the protein and mRNA expression of NAIP in the
HCT116 cells (Fig. 4C and D).
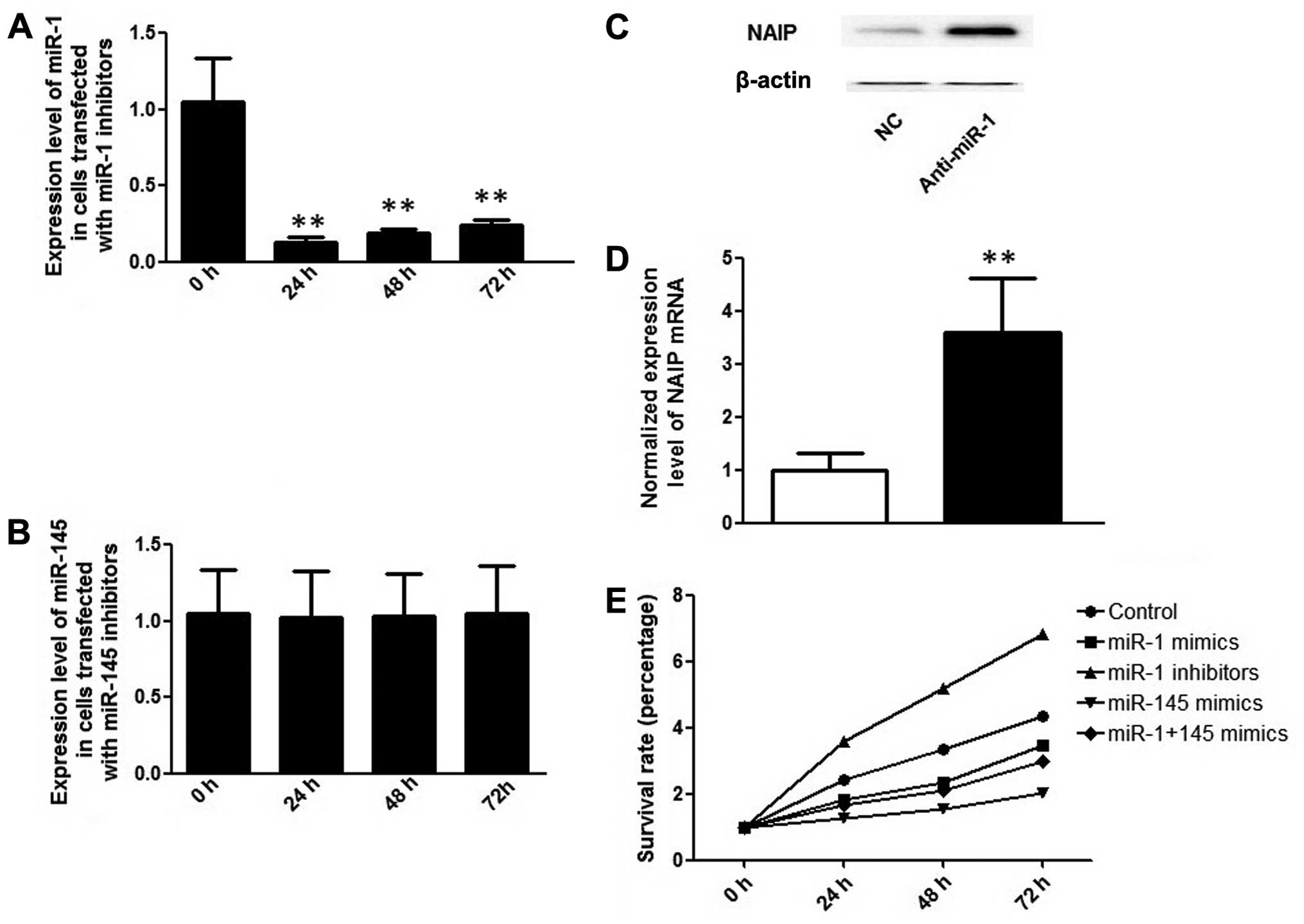 | Figure 4(A) Expression levels of miR-1 24,
48, 72 and 96 h following transfection with miR-1 inhibitors; (B)
expression levels of miR-145 24, 48, 72 and 96 h following
transfection with miR-145 inhibitors; **P<0.01,
compared to the 0 h time point. (C) protein expression level of NLR
family, apoptosis inhibitory protein (NAIP) following transfection
with miR-1 inhibitors; (D) mRNA expression level of NAIP following
transfection with miR-1 inhibitors; **P<0.01,
compared to control. (E) Survival rate analysis of colorectal
cancer (CRC) cells transfected with miR-1 and/or miR-145 mimics, as
determined by MTT assay 24, 48 and 72 h following transfection. NC,
negative control. |
Upregulation of miR-1 and miR-145
inhibits HCT116 cell proliferation in vitro
To further examine whether the differentially
expressed miRNAs alter the proliferative capacity of the HCT116
cells, miR-145 mimic, miR-1 mimic and miR-145 inhibitor were
sequentially transfected into the HCT116 cells, and RT-qPCR was
performed to determine miRNA upregulation or downregulation
(Figs. 3A, 3B and 4A). As shown in Fig. 4E, the overexpression of miR-145
and miR-1 significantly suppressed the proliferation of the HCT116
cells, as evidenced by a decrease in the percentage of surviving
cells compared with the controls. The introduction of the miR-1
inhibitor evidently suppressed the expression of miR-1, causing an
increase in NAIP expression (Fig.
4C) and promoting the proliferation of the cells (Fig. 4E).
Introducing miR-1 and miR-145
significantly induces the apoptosis of HCT116 cells
To elucidate the molecular mechanisms underlying the
observed inhibitory effect on CRC cell proliferation of miR-1 and
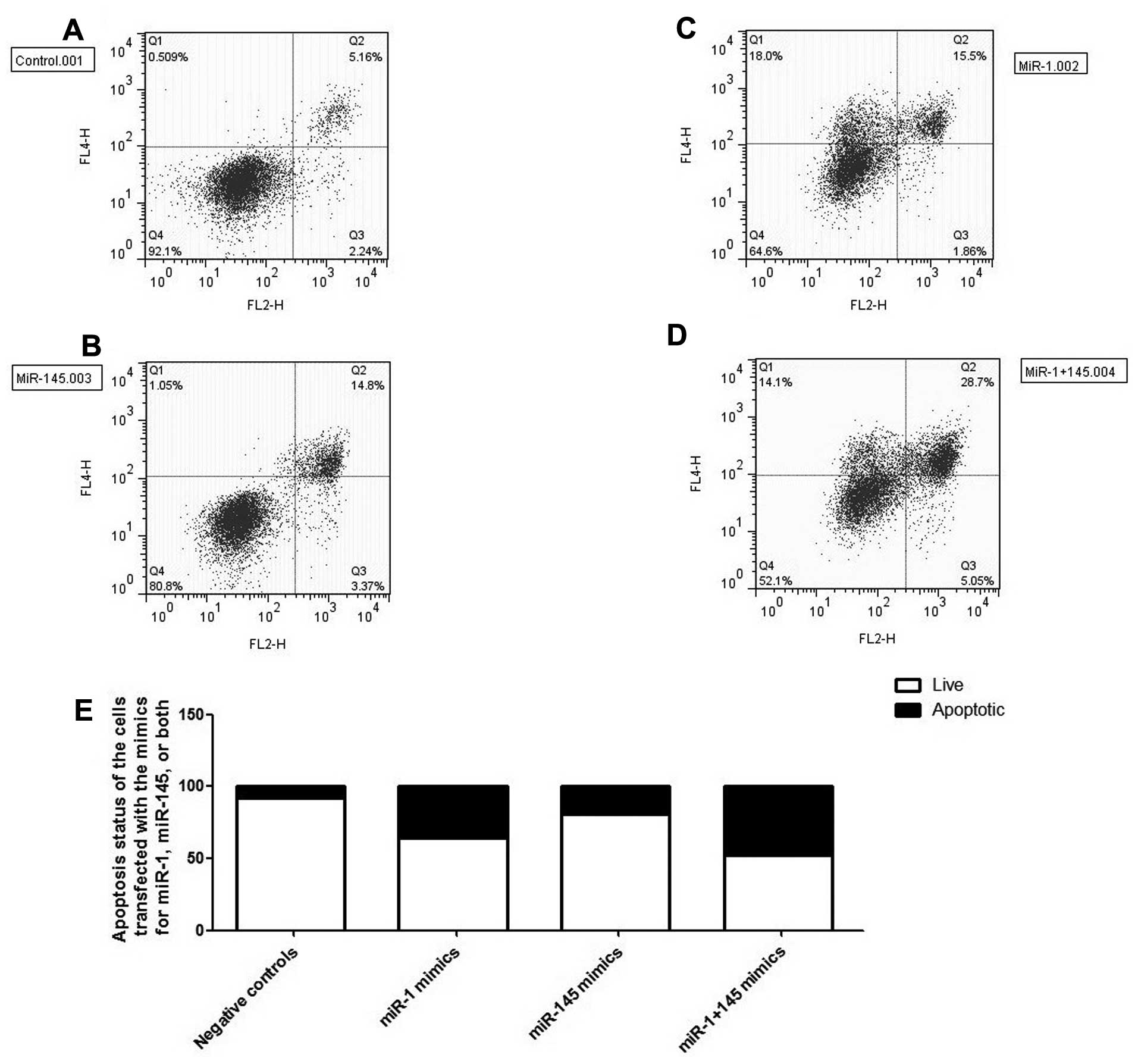
miR-145, we performed flow cytometric analysis using the cells
transfected with miR-1 mimic and/or miR-145 mimic. We found that
the introduction of miR-1 and miR-145 significantly induced
apoptosis both individually and in combination, as shown in
Fig. 5A–D, and the apoptotic
status of the cells is summarized in Fig. 5E. Our results demonstrate that the
inhibitory effect on CRC cell proliferation of miR-1 and miR-145
may be due to their ability to induce the apoptosis of HCT116
cells.
Discussion
CRC is a complex genetic disease which results from
abnormalities in gene structure and/or expression (19). The aberrant expression of
endogenous miRNAs has been noted in multiple types of human cancer,
such as lung and breast cancer, leukemia and CRC (20), and the deregulated expression of
certain miRNAs has been reported to be a marker for early
diagnosis, prognosis, as well as the response to treatment in
patients with CRC, indicating that miR-145 and miR-1 play a
promising role as prognostic or therapeutic biomarkers for patients
with CRC (21–23). The balance of oncogenes and tumor
suppressor genes plays an essential role in regulating cell
proliferation and survival (24,25). Considering the fact that 1/3
protein-coding mRNAs are subjected to miRNA-mediated regulation,
the role of miRNAs in the development of malignancies is critical
(26). In the present study,
using microarray analysis, we identified 21 differentially
expressed miRNAs, 5 upregulated and 16 downregulated mRNAs in the
colon cancer samples as compared with the adjacent non-cancerous
control tissues (Table I).
In non-cancerous tissue, miRNA-mediated control
helps to maintain normal cell activities, including growth,
differentiation and apoptosis. The deregulation of miRNA expression
leads to the abnormal activity of the miRNA target genes, and the
consequent upregulation of oncogene(s) and/or the downregulation of
tumor suppressor gene(s) results in cells with selective
advantages, such as increased proliferation or survival (27). The present study focused on
individual miRNAs and their target genes. By using online target
prediction tools, we identified NAIP as a possible shared target of
three significantly downregulated miRNAs, miR-1, miR-145 and
miR-451, and we subsequently demonstrated that NAIP is a valid
target of miR-1 and miR-145, but not miR-451, using a luciferase
assay (Fig. 2), and this was
further confirmed by the observation that the introduction of miR-1
and miR-145 mimics decreased the expression level of NAIP (Fig. 3). miR-145 and miR-1 have been
reported to be aberrantly expressed in numerous types of cancer,
and their tumor-suppressive roles have been well established. miR-1
is located at 18q11 in the human genome, and is abundantly
expressed in the heart and skeletal muscle tissue (28). It has been demonstrated that miR-1
is among the most frequently downregulated miRs in solid human
tumors. miR-1 gene expression is reduced in human hepatocellular
carcinoma cell lines compared to normal liver cells due to
hypermethylation. When the miR-1 gene is hypomethylated, it
re-expresses miR-1 and can induce apoptosis (29). In mouse myoblast cells, the mutant
MyoD transcription factor downregulates miR-1 expression and
inhibits cell apoptosis (30).
miR-1 expression has bene shown to be downregulated in human lung
cancer tissues and cell lines in comparison to normal human lungs,
and miR-1 influences the response of lung cancer cells to
anticancer drugs (31).
Additionally, miR-1 is downregulated in human bladder cancer
(32) and head-and neck squamous
cell carcinoma (HNSCC) cells (33). It has been demonstrated that miR-1
acts as a tumor suppressor miRNA by directly targeting various
oncogenes, such as prothymosin alpha (PTMA) and transgelin 2
(TAGLN2) (33).
miR-145 is located in a cluster in the 5q32
chromosomal region. Several mRNAs have been demonstrated to be
direct targets of miR-145 (34)
in colon cancer, and the dysregulation of this miRNA is believed to
be responsible for the development of the disease. miR-145, or its
analogue, has also been studied in relation to breast (35), leukemia (36), non-small cell lung (37) and prostate cancer (38). One of the best-characterized
candidate targets of miR-145 is the oncogene cadherin-17 (CDH17),
which is a member of the cadherin superfamily. Cadherins function
as calcium-dependent cell adhesion proteins and their dysregulated
expression is associated with tumor formation and metastasis
(39). It has also been
previously demonstrated that CDH17 is involved in proliferation, as
it activates Wnt signaling (40).
CDH17 was suppressed by >2-fold at the protein and mRNA level
following transfection with miR-145 or its analogue miR-143
(40). Furthermore, it has been
shown that chemically unmodified miR-145 complexed with
polyethylenimine (PEI) can be used as an efficient and
biocompatible strategy for miRNA replacement therapy, which is used
in the treatment of colon cancer (41). In the present study, we confirmed
the tumor suppressive effect of miR-145 and miR-1 by demonstrating
the proliferation-suppressive and apoptosis-inducing effect of the
mimics, as well as the proliferation-promoting effect of the
inhibitors on CRC cells (Figs. 4
and 5). Moreover, we demonstrated
that NAIP, a major anti-apoptotic protein, is an effective target
gene of both miR-1 and miR-145 via a luciferase reporter system,
and this was further confirmed by the observation that transfection
with miR-1 and/or miR-145 suppressed the expression of NAIP.
As its name implies, the inhibitor of apoptosis
(IAP) protein family plays an inhibitory role in the regulation of
apoptosis and therefore, the overexpression of IAP family members
is regarded as an unfavorable feature when diagnosing cancer
(42). IAP family proteins are
characterized by the presence of one or more baculovirus
IAP-repeats (BIR) domains (43),
which are zinc-binding folds approximately 70 amino acids long,
including three conserved cysteine and one conserved histidine
residue (44). The BIR domain is
responsible for its ability to inhibit the function of apoptosis
executioner caspase-3 and -7, as well as initiator caspase-9 by
interacting with IAP-binding motifs (IBMs) in those proteins
(43). In addition to the BIR
domain, IAPs possess other domains, such as really interesting new
gene (RING), caspase activation and recruitment domain (CARD) and
nucleotide-binding oligomerization domain (NOD), the presence of
which endow IAPs with additional abilities to regulate other
accessory biological activities, including cell differentiation,
cell cycle progression, cell signal transduction, and immune
responses (43). The
NAIP/baculo-viral IAP repeat-containing protein 1 (BIRC1) gene
coding region spans 4,212 nucleotides encoding a 1,403-amino acid
protein of the 156 kDa protein, and it contains 3 sequential BIR
domains at the N-terminus, which makes it quite typical among other
IAPs.
In addition to the BIR domain, NAIP harbors a NOD
and a leucine-rich repeat (LRR) (45,46), which makes NAIP function
differently from other IAP proteins (43). Due to the presence of the NOD
domain and LRR motif, NAIP possesses the properties of both the IAP
protein family and the nucleotide-binding domain and leucine-rich
repeat containing (NLR) protein family. The NOD domain is essential
for oligomerization, a key step in signal transduction, and the LRR
domain functions as an intracellular sensor of microbial motifs
(47). NAIP, together with some
other members of NLR, promotes the synthesis and control of the
cytoplasmic multiprotein complex termed 'inflammasome' (47). Considering the significant role
that NAIP plays in regulating the inflammatory repsonse as well as
the fact that miR-145 is substantially downregulated in patients
with ulcerative colitis, it has been postulated that miR-145 may
contribute to the pathogenesis of ulcerative colitis and is
possibly involved in the mechanisms underlying the malignant
transformation of the colonic epithelium into ulcerative colitis
(48).
In summary, our data provide evidence that miR-1 and
miR-145, which were found to be downregulated in colon carcinoma
tissues, are involved in the multistep process of CRC development
through the modulation of NAIP expression, which in turn affects
cell apoptosis. Furthermore, in CRC cells, miR-1 and miR-145 induce
cell death by targeting a major anti-apoptotic protein, NAIP, and
contribute to the development of colon cancer. Therefore, both
miR-1/miR-145 and NAIP can be regarded as having great potential in
colon cancer therapy, and can be targeted by either re-expressing
the miRNAs and/or interfering with NAIP function.
Acknowledgments
The present study was fully sponsored by the
following three programs: the Guangzhou City Science and Technology
and Information Bureau Science and Technology Supporting Program
(grant no. 2010J-E141), the Guangdong Province, the Third Batch of
Major Science and Technology Projects (grant no. 2011A080300002),
and the Academic-Industrial Department of Guangdong Province
Supported Pilot project (grant no. 2011B090400526).
References
|
1
|
Jemal A, Murray T, Samuels A, Ghafoor A,
Ward E and Thun MJ: Cancer statistics, 2003. CA Cancer J Clin.
53:5–26. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pasetto LM, Jirillo A, Iadicicco G, Rossi
E, Paris MK and Monfardini S: FOLFOX versus FOLFIRI: a comparison
of regimens in the treatment of colorectal cancer metastases.
Anticancer Res. 25:563–576. 2005.PubMed/NCBI
|
|
3
|
Goldberg RM and Gill S: Recent phase III
trials of fluorouracil, irinotecan, and oxaliplatin as chemotherapy
for metastatic colorectal cancer. Cancer Chemother Pharmacol.
54(Suppl 1): S57–S64. 2004.PubMed/NCBI
|
|
4
|
Loeve F, van Ballegooijen M, Snel P and
Habbema JD: Colorectal cancer risk after colonoscopic polypectomy:
a population-based study and literature search. Eur J Cancer.
41:416–422. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Koehler A, Bataille F, Schmid C, Ruemmele
P, Waldeck A, Blaszyk H, Hartmann A, Hofstaedter F and Dietmaier W:
Gene expression profiling of colorectal cancer and metastases
divides tumours according to their clinicopathological stage. J
Pathol. 204:65–74. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kolonel LN, Altshuler D and Henderson BE:
The multiethnic cohort study: exploring genes, lifestyle and cancer
risk. Nat Rev Cancer. 4:519–527. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ohtsuka M, Ling H, Doki Y, Mori M and
Calin GA: MicroRNA processing and human cancer. J Clin Med.
4:1651–1667. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang J, Song YX, Ma B, Wang JJ, Sun JX,
Chen XW, Zhao JH, Yang YC and Wang ZN: Regulatory roles of
non-coding RNAs in colorectal cancer. Int J Mol Sci.
16:19886–19919. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Anwar SL and Lehmann U: MicroRNAs:
emerging novel clinical biomarkers for hepatocellular carcinomas. J
Clin Med. 4:1631–1650. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kedmi M, Sas-Chen A and Yarden Y:
MicroRNAs and growth factors: an alliance propelling tumor
progression. J Clin Med. 4:1578–1599. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Han F, He J, Li F, Yang J, Wei J, Cho WC
and Liu X: Emerging roles of nicroRNAs in EGFR-targeted therapies
for lung cancer. Biomed Res Int. 2015:6727592015. View Article : Google Scholar
|
|
12
|
Croner RS, Foertsch T, Brueckl WM,
Guenther K, Siebenhaar R, Stremmel C, Matzel KE, Papadopoulos T,
Kirchner T, Behrens J, et al: Common denominator genes that
distinguish colorectal carcinoma from normal mucosa. Int J
Colorectal Dis. 20:353–362. 2005. View Article : Google Scholar
|
|
13
|
Raetz EA and Moos PJ: Impact of microarray
technology in clinical oncology. Cancer Invest. 22:312–320. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chiu ST, Hsieh FJ, Chen SW, Chen CL, Shu
HF and Li H: Clinicopathologic correlation of up-regulated genes
identified using cDNA microarray and real-time reverse
transcription-PCR in human colorectal cancer. Cancer Epidemiol
Biomarkers Prev. 14:437–443. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nambiar PR, Nakanishi M, Gupta R, Cheung
E, Firouzi A, Ma XJ, Flynn C, Dong M, Guda K, Levine J, et al:
Genetic signatures of high- and low-risk aberrant crypt foci in a
mouse model of sporadic colon cancer. Cancer Res. 64:6394–6401.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mariadason JM, Arango D, Shi Q, Wilson AJ,
Corner GA, Nicholas C, Aranes MJ, Lesser M, Schwartz EL and
Augenlicht LH: Gene expression profiling-based prediction of
response of colon carcinoma cells to 5-fluorouracil and
camptothecin. Cancer Res. 63:8791–8812. 2003.PubMed/NCBI
|
|
17
|
Neviani P and Fabbri M: Exosomic microRNAs
in the tumor microenvironment. Front Med (Lausanne). 2:472015.
|
|
18
|
Eisinger F, Roussel C, Morère JF and
Viguier J: Cancer screening: reaching the limits or terra
incognita? Lessons from the EDIFICE surveys. Eur J Cancer Prev.
20(Suppl 1): S42–S44. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Peters U, Bien S and Zubair N: Genetic
architecture of colorectal cancer. Gut. 64:1623–1636. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ng EK, Chong WW, Jin H, Lam EK, Shin VY,
Yu J, Poon TC, Ng SS and Sung JJ: Differential expression of
microRNAs in plasma of colorectal cancer patients: a potential
marker for colorectal cancer screening. Gut. 58:1375–1381. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu CW, Ng SS, Dong YJ, Ng SC, Leung WW,
Lee CW, Wong YN, Chan FK, Yu J and Sung JJ: Detection of miR-92a
and miR-21 in stool samples as potential screening biomarkers for
colorectal cancer and polyps. Gut. 61:739–745. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schetter AJ, Leung SY, Sohn JJ, Zanetti
KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et
al: MicroRNA expression profiles associated with prognosis and
therapeutic outcome in colon adenocarcinoma. JAMA. 299:425–436.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ruddon R: Cancer Biology. Oxford
University Press; New York, NY: 2007
|
|
25
|
Corvinus FM, Orth C, Moriggl R, Tsareva
SA, Wagner S, Pfitzner EB, Baus D, Kaufmann R, Huber LA, Zatloukal
K, et al: Persistent STAT3 activation in colon cancer is associated
with enhanced cell proliferation and tumor growth. Neoplasia.
7:545–555. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chekulaeva M and Filipowicz W: Mechanisms
of miRNA-mediated post-transcriptional regulation in animal cells.
Curr Opin Cell Biol. 21:452–460. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 4:143–159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li J, Dong X, Wang Z and Wu J: MicroRNA-1
in cardiac Diseases and cancers. Korean J Physiol Pharmacol.
18:359–363. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Datta J, Kutay H, Nasser MW, Nuovo GJ,
Wang B, Majumder S, Liu CG, Volinia S, Croce CM, Schmittgen TD, et
al: Methylation mediated silencing of MicroRNA-1 gene and its role
in hepato-cellular carcinogenesis. Cancer Res. 68:5049–5058. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hirai H, Verma M, Watanabe S, Tastad C,
Asakura Y and Asakura A: MyoD regulates apoptosis of myoblasts
through microRNA-mediated downregulation of Pax3. J Cell Biol.
191:347–365. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nasser MW, Datta J, Nuovo G, Kutay H,
Motiwala T, Majumder S, Wang B, Suster S, Jacob ST and Ghoshal K:
Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression
of tumorigenic property of lung cancer cells and their
sensitization to doxorubicin-induced apoptosis by miR-1. J Biol
Chem. 283:33394–33405. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yoshino H, Chiyomaru T, Enokida H,
Kawakami K, Tatarano S, Nishiyama K, Nohata N, Seki N and Nakagawa
M: The tumour-suppressive function of miR-1 and miR-133a targeting
TAGLN2 in bladder cancer. Br J Cancer. 104:808–818. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nohata N, Sone Y, Hanazawa T, Fuse M,
Kikkawa N, Yoshino H, Chiyomaru T, Kawakami K, Enokida H, Nakagawa
M, et al: miR-1 as a tumor suppressive microRNA targeting TAGLN2 in
head and neck squamous cell carcinoma. Oncotarget. 2:29–42. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xu Q, Liu LZ, Qian X, Chen Q, Jiang Y, Li
D, Lai L and Jiang BH: MiR-145 directly targets p70S6K1 in cancer
cells to inhibit tumor growth and angiogenesis. Nucleic Acids Res.
40:761–774. 2012. View Article : Google Scholar :
|
|
35
|
Götte M, Mohr C, Koo CY, Stock C, Vaske
AK, Viola M, Ibrahim SA, Peddibhotla S, Teng YH, Low JY, et al:
miR-145-dependent targeting of junctional adhesion molecule A and
modulation of fascin expression are associated with reduced breast
cancer cell motility and invasiveness. Oncogene. 29:6569–6580.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Akao Y, Nakagawa Y, Iio A and Naoe T: Role
of microRNA-143 in Fas-mediated apoptosis in human T-cell leukemia
Jurkat cells. Leuk Res. 33:1530–1538. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gao W, Yu Y, Cao H, Shen H, Li X, Pan S
and Shu Y: Deregulated expression of miR-21, miR-143 and miR-181a
in non small cell lung cancer is related to clinicopathologic
characteristics or patient prognosis. Biomed Pharmacother.
64:399–408. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Clapé C, Fritz V, Henriquet C, Apparailly
F, Fernandez PL, Iborra F, Avancès C, Villalba M, Culine S and
Fajas L: miR-143 interferes with ERK5 signaling, and abrogates
prostate cancer progression in mice. PLoS One. 4:e75422009.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lee NP, Poon RT, Shek FH, Ng IO and Luk
JM: Role of cadherin-17 in oncogenesis and potential therapeutic
implications in hepatocellular carcinoma. Biochim Biophys Acta.
1806:138–145. 2010.PubMed/NCBI
|
|
40
|
Liu LX, Lee NP, Chan VW, Xue W, Zender L,
Zhang C, Mao M, Dai H, Wang XL, Xu MZ, et al: Targeting cadherin-17
inactivates Wnt signaling and inhibits tumor growth in liver
carcinoma. Hepatology. 50:1453–1463. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ibrahim AF, Weirauch U, Thomas M,
Grünweller A, Hartmann RK and Aigner A: MicroRNA replacement
therapy for miR-145 and miR-33a is efficacious in a model of colon
carcinoma. Cancer Res. 71:5214–5224. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Grzybowska-Izydorczyk O and Smolewski P:
The role of the inhibitor of apoptosis protein (IAP) family in
hematological malignancies. Postepy Hig Med Dosw (Online).
62:55–63. 2008.In Polish.
|
|
43
|
Herman MD, Moche M, Flodin S, Welin M,
Trésaugues L, Johansson I, Nilsson M, Nordlund P and Nyman T:
Structures of BIR domains from human NAIP and cIAP2. Acta
Crystallogr Sect F Struct Biol Cryst Commun. 65:1091–1096. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Verhagen AM, Coulson EJ and Vaux DL:
Inhibitor of apoptosis proteins and their relatives: IAPs and other
BIRPs. Genome Biol. 2:reviews3009–reviews3009.10. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Davoodi J, Ghahremani MH, Es-Haghi A,
Mohammad-Gholi A and Mackenzie A: Neuronal apoptosis inhibitory
protein, NAIP, is an inhibitor of procaspase-9. Int J Biochem Cell
Biol. 42:958–964. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Dubrez-Daloz L, Dupoux A and Cartier J:
IAPs: more than just inhibitors of apoptosis proteins. Cell Cycle.
7:1036–1046. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Franchi L, Eigenbrod T, Munoz-Planillo R
and Nunez G: The inflammasome: a caspase-1-activation platform that
regulates immune responses and disease pathogenesis. Nat Immunol.
10:241–247. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Pekow JR, Dougherty U, Mustafi R, Zhu H,
Kocherginsky M, Rubin DT, Hanauer SB, Hart J, Chang EB, Fichera A,
et al: miR-143 and miR-145 are downregulated in ulcerative colitis:
putative regulators of inflammation and protooncogenes. Inflamm
Bowel Dis. 18:94–100. 2012. View Article : Google Scholar
|