Introduction
Gliomas are the most common primary brain tumors in
humans. Malignant gliomas are highly aggressive. Although the
standard treatment for malignant gliomas has evolved over the past
30 years, the average life expectancy for patients with malignant
gliomas is only 14 months (1).
With the development of molecular biology, previous research has
suggested gene therapy strategies for the treatment of gliomas
(2–6).
The receptor for activated C kinase 1 (RACK1) is a
versatile scaffold protein involved in the regulation of a variety
of cellular processes, including cell growth, migration, survival
and metastasis (7–9). Previous studies have indicated that
RACK1 is abnormally expressed in various malignant tumors, and its
dysregulation may contribute to tumorigenesis (10–13). Certain pathways have been
demonstrated to be activated by RACK1, such as the Src/Akt
signaling pathway, mitogen-activated protein kinase (MAPK)
signaling pathway, and the sonic hedgehog signaling pathway
(14–16). RACK1 is widely expressed in the
central nervous system, and it has been suggested that RACK1 is
associated with the development of gliomas (14).
Contactin-2 (also known as CNTN2 or axonal
glycoprotein TAG-1) protein is a member of the immunoglobulin
super-family. It functions as a cell adhesion molecule and is
involved in the development of a variety of tumors (17,18). Rickman et al reported that
CNTN2 is overexpressed in gliomas (19). CNTN2 is the hub node in the
protein-protein interaction network of oligodendroglioma, and it
may serve an essential function in the pathogenesis of
oligodendroglioma (20). It is
thus of interest to identify the proteins that interact with CNTN2
in order to mediate its role in tumor development.
RACK1 is a scaffold protein that physically or
functionally interacts with numerous proteins (8,21–23), and CNTN2 has been linked to the
highest degree in the oligodendroglioma protein-protein interaction
network (20). However, to the
best of our knowledge, the interaction between RACK1 and CNTN2 has
not been reported to date. In the present study, we firstly
examined the association between RACK1 and CNTN2 in glioma cells.
Certain studies have demonstrated that the receptor tyrosine kinase
(RTK)/Ras/extracellular signal-regulated kinase (ERK) MAPK
signaling pathway is associated with the growth and progression of
gliomas (24,25). Furthermore, we examined the
possibility that RACK1/CNTN2/RTK/Ras/MAPK axis plays a role in
glioma cell growth and differentiation.
Materials and methods
Clinical specimens
All experimental procedures were approved by the
Ethics Committee of the Second Xiangya Hospital of Central South
University (ID of ethics document: 2014-S021). Written informed
consent was obtained from each patient prior to enrollment. The
malignant glioma tissues were obtained from a total of 48 patients
with gliomas, including 24 males and 24 females, who underwent
surgery at the Second Xiangya Hospital of Central South University
(Changsha, China). The mean patient age was 40.5 years (range,
26–71 years). At the time of diagnosis, 23 patients had low-grade
(grades I–II) glioma, whereas the other 25 patients had high-grade
(grades III–IV) glioma. Adjacent normal brain tissues were obtained
from 14 of these patients, including 7 males and 7 females, and the
mean patient age was 39.2 years (range, 28–68 years).
Cell culture
Human glioma cell lines (U-118 MG, LN-18, SW1783 and
SW1088) were obtained from the American Type Culture Collection
(ATCC; Baltimore, MD, USA), and normal human astrocytes (NHA) were
obtained from Lonza (Allendale, NJ, USA). All the cells were
cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented
with 10% fetal bovine serum (FBS) (both from Invitrogen Life
Technologies, Grand Island, NY, USA), and maintained at 37°C with
5% CO2.
Cell transfection
Cell transfection was performed using Lipofectamine
2000 (Invitrogen Life Technologies) according to the manufacturer's
instructions. The pcDNA3.1 vector and the siRNA were obtained from
Zhonghong Boyuan Biological Technology Co., Ltd (Shenzhen, China).
RACK1-pcDNA3.1 and/or CNTN2 siRNA were transfected into the SW1783
cells, and RACK1 siRNA was transfected into the U-118 MG cells.
Briefly, the cells were seeded onto 6-well plates 1 day prior to
transfection. The pcDNA3.1 vector, RACK1-pcDNA3.1, scrambled siRNA,
RACK1 siRNA, CNTN2 siRNA and Lipofectamine 2000 were diluted in
250-µl volumes of Opti-MEM® I reduced-serum
medium (Invitrogen Life Technologies) without serum. Subsequently,
the plasmid DNA or siRNA were mixed with Lipofectamine 2000 and
incubated at room temperature for 20 min to allow the complexes to
form. The complexes were then added to each well.
Cell proliferation assay
For the cell proliferation assay, the cells were
seeded onto 96-well plates at 3×103 cells/well. The
following day, the cells were transfected with the pcDNA3.1 vector,
RACK1-pcDNA3.1, scrambled siRNA and CNTN2 siRNA. At 24, 48 and 72 h
following transfection, cell proliferation was assessed using an
MTT cell proliferation and cytotoxicity assay kit (Beyotime,
Shanghai, China) according to the manufacturer's instructions.
Briefly, 10 µl MTT solution (5 mg/ml) was added to each well
and incubated at 37°C for 4 h. Subsequently, 100 µl
dissolving buffer was added to dissolve the formazan crystals. The
absorbance at 570 nm was measured using a microplate reader (BioTek
Instruments, Inc., Winooski, VT, USA).
Western blot analysis
Total proteins were extracted from the tissues and
cells using ice-cold lysis buffer [100 mM Tris-HCl (pH 7.4), 150 mM
NaCl, 2 mM DTT, 1% Triton X-100, 100 mM sodium orthovanadate and 1
mM PMSF]. The protein concentration was determined using a Bradford
Protein assay kit (Solarbio, Beijing, China). Equal amounts of
protein lysates (50 µg) were separated by SDS-PAGE, and the
proteins were transferred onto PVDF membranes (Millipore,
Billerica, MA, USA). After blocking with 5% bovine serum albumin
(Amresco LLC, Solon, OH, USA), the membranes were washed with TBST
3 times, and incubated with primary antibodies, namely rabbit
polyclonal antibody to RACK1 (1:800; ab62735), rabbit polyclonal
antibody to CNTN2 (1:400; ab68994), rabbit polyclonal antibody to
epidermal growth factor receptor (EGFR; 1:800; ab2430), rabbit
polyclonal antibody to platelet-derived growth factor receptor, α
polypeptide (PDGFRα; 1:500; ab65258), and mouse monoclonal antibody
to β-actin (1:2,000; ab6276) (all from Abcam, Cambridge, MA, USA),
mouse monoclonal antibody to Ras (1:500; sc-166691), rabbit
polyclonal antibody to glial fibrillary acidic protein (GFAP)
(1:400; sc-9065) (all from Santa Cruz Biotechnology, Inc., Santa
Cruz, CA, USA), rabbit polyclonal antibody to CD133 (1:400;
EAP0023; Elabscience, Hubei, China), rabbit monoclonal antibody to
ERK1/2 (1:400; #4695), rabbit monoclonal antibody to p-ERK1/2
(Thr202/Tyr204; 1:400; #4377) (Cell Signaling Technology, Inc.,
Beverly, MA, USA) at 4°C overnight, followed by incubation with
HRP-conjugated secondary antibodies, namely goat anti-rabbit IgG
HRP (1:2,000; ab6721) and rabbit anti-mouse IgG HRP (1:2,000;
ab6728) (both from Abcam) at 37°C for 1 h. The immunocomplexes were
visualized by enhanced chemiluminescence (Western Blotting Luminol
reagent; Santa Cruz Biotechnology, Inc.).
Co-immunoprecipitation assay
For co-immunoprecipitation (co-IP) assay, the cells
were extracted using immunopre cipitation lysis buffer (Beyotime),
and the supernatants were incubated with 2 µg anti-CNTN2
antibody or normal IgG at 4°C overnight. A total of 20 µl
recombinant fusion protein A and protein G agarose (Sigma, St.
Louis, MO, USA) was then added followed by incubation at 4°C for 3
h. Following centrifugation at 1,000 g for 5 min and washing with
phosphate-buffered saline (PBS), the immunoprecipitates were
collected and subjected to SDS-PAGE.
Statistical analysis
Quantitative data are expressed as the means ±
standard deviation. The two-tailed Student's t-test was performed
to analyze the difference between 2 groups using SPSS 19.0 software
(SPPS, Inc., Chicago, IL, USA). A P-value <0.05 was considered
to indicate a statistically significant difference.
Results
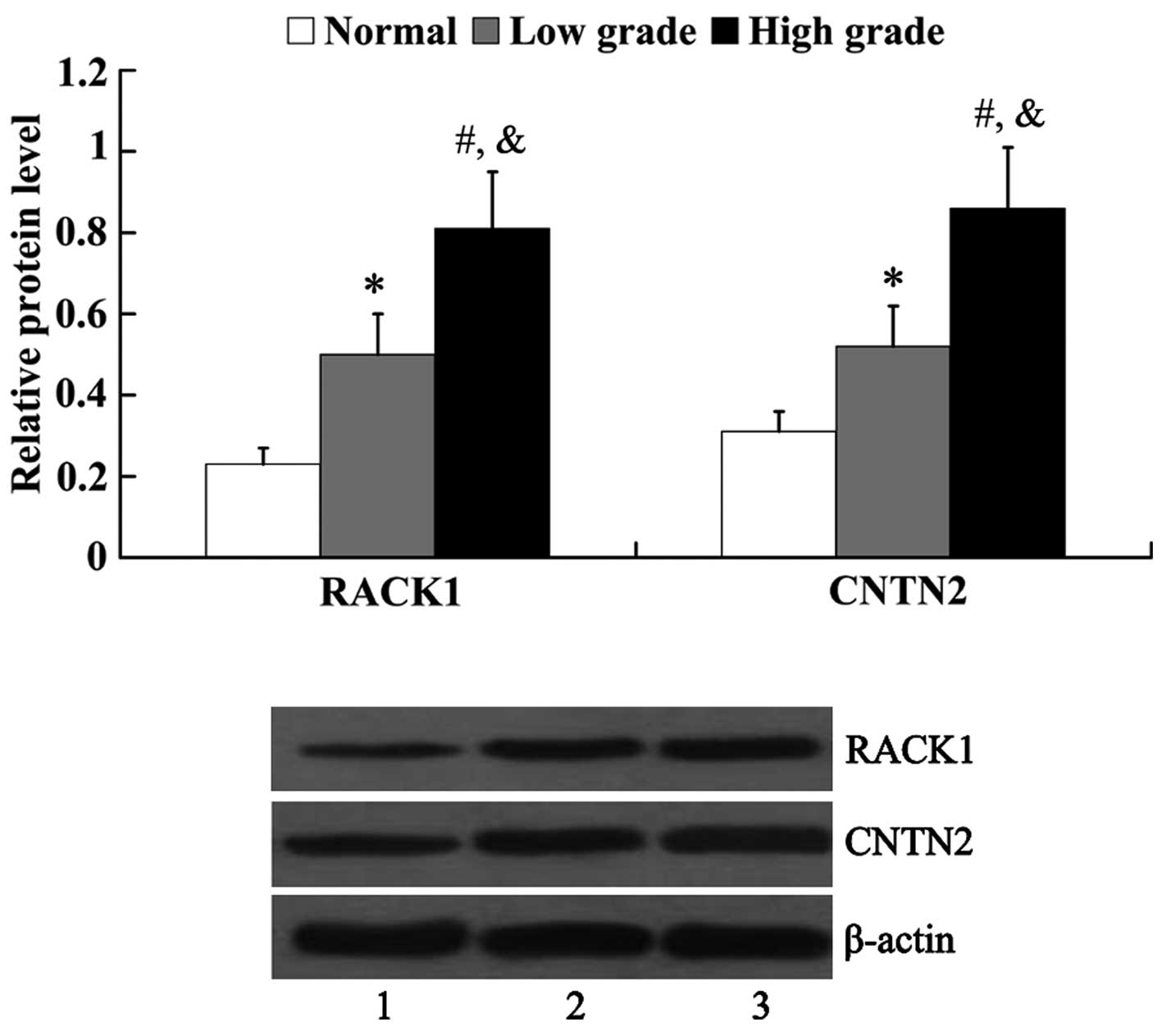
Expression of RACK1 and CNTN2 in human
glioma tissues
The expression of RACK1 and CNTN2 in normal brain
tissues and glioma tissues was examined by western blot analysis.
As shown in Fig. 1, the protein
expression of both RACK1 and CNTN2 was significantly higher in the
glioma tissues [RACK1, 0.5±0.11 (low grade) and 0.81±0.14 (high
grade); CNTN2, 0.52±0.10 (low grade) and 0.86±0.15 (high grade)]
than that in the normal brain tissues (RACK1, 0.23±0.04; CNTN2,
0.31±0.05). Furthermore, it was revealed that the protein
expression of RACK1 and CNTN2 in the high-grade glioma tissues was
significantly higher than that in the low-grade glioma tissues
(P<0.05).
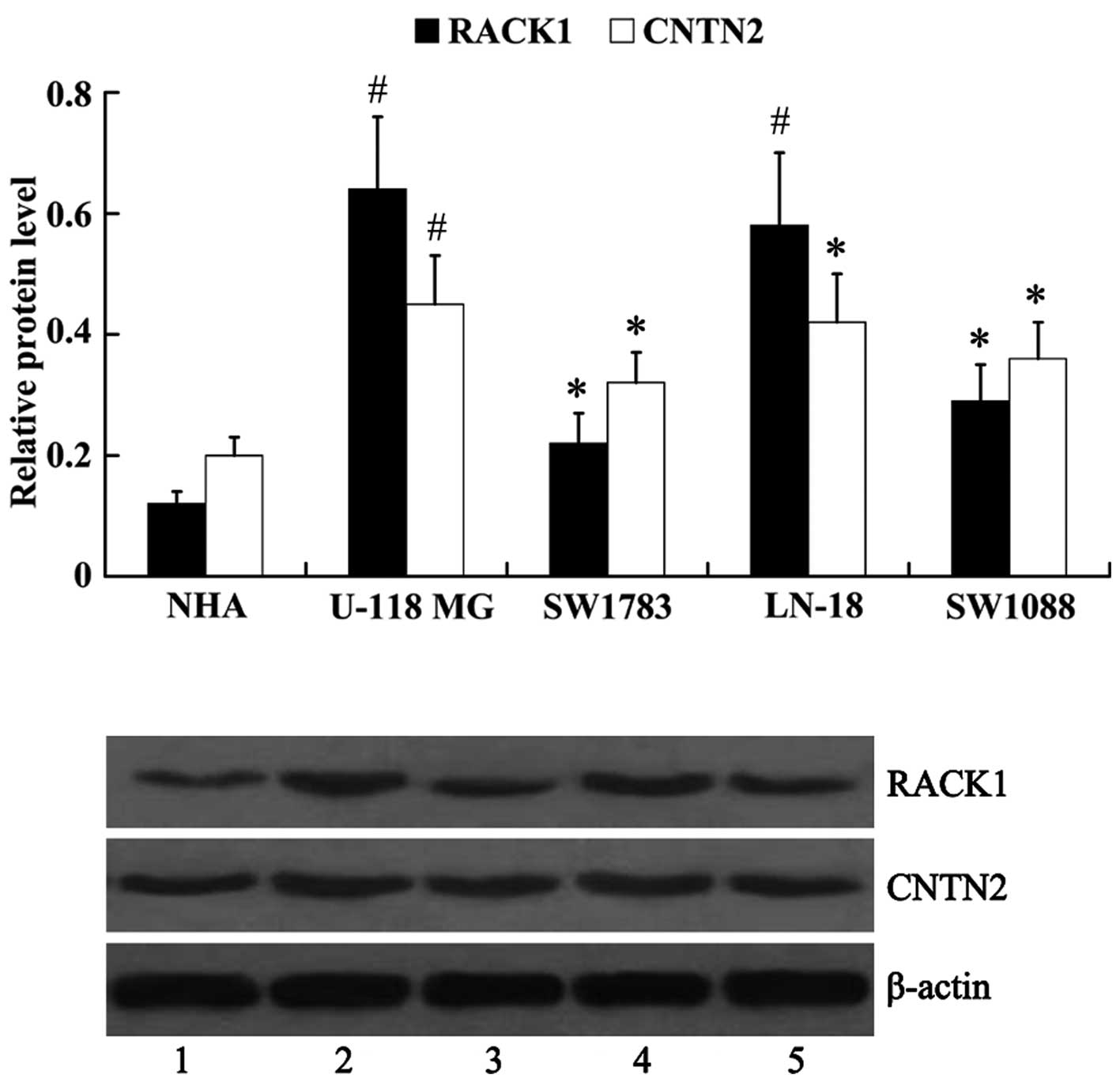
Expression of RACK1 and CNTN2 in human
glioma cell lines
The protein expression of RACK1 and CNTN2 was
examined in human glioma cell lines (U-118 MG, LN-18, SW1783 and
SW1088). Normal human astrocytes (NHA) were used as controls. It
was noted that, compared with the NHA cells (RACK1, 0.12±0.02;
CNTN2, 0.20±0.03), the protein expression levels of RACK1 and CNTN2
were upregulated in human glioma cells [RACK1: 0.64±0.12 (U-118
MG), 0.22±0.05 (SW1783), 0.58±0.12 (LN-18), 0.29±0.06 (SW1088);
CNTN2: 0.45±0.08 (U-118 MG), 0.32±0.05 (SW1783), 0.42±0.08 (LN-18),
0.36±0.06 (SW1088)], with the highest expression noted in the U-118
MG cells, and lowest in the SW1783 cells (Fig. 2).
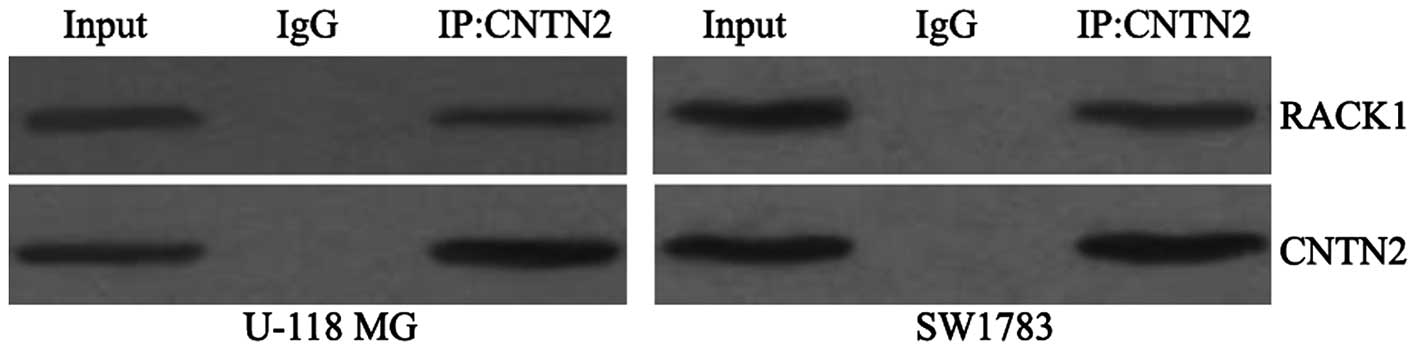
RACK1 interacts with and regulates the
expression of CNTN2
To determine whether RACK1 interacts with CNTN2, we
examined RACK1 expression after cell lysates were
immunoprecipitated with an anti-CNTN2 antibody. As shown in
Fig. 3, RACK1 protein was
detected after immunoprecipitation of endogenous CNTN2 from cell
lysates.
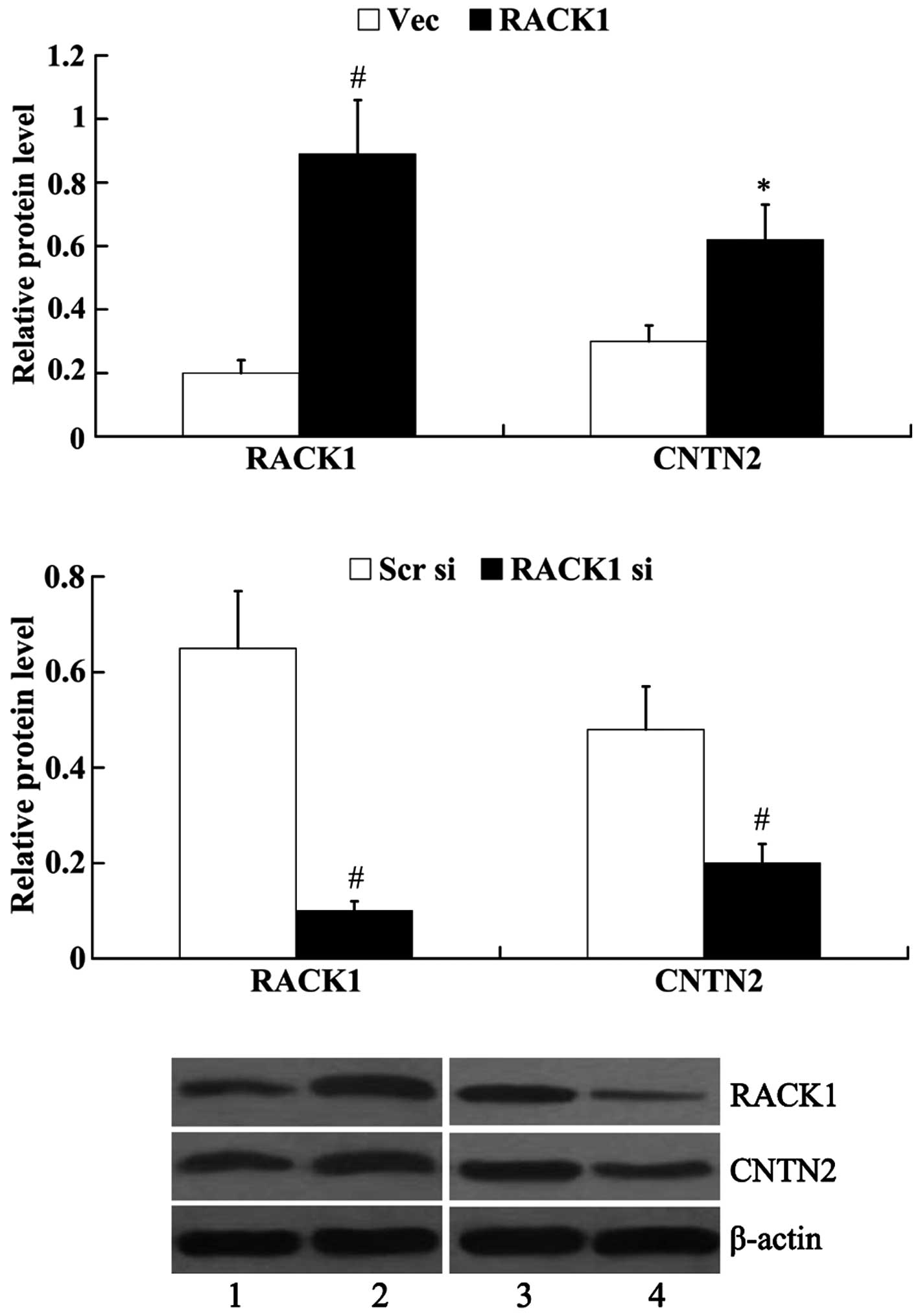
The SW1783 cells demonstrated the lowest protein
expression of RACK1; thus, we transfected RACK1-pcDNA3.1 into the
SW1783 cells to induce the overexpression of RACK1. RACK1 siRNA was
transfected into the U-118 MG cells to knockdown endogenous RACK1,
as the U-118 MG cells exhibited the highest expression of RACK1.
Subsequently, the protein expression of RACK1 and CNTN2 was
examined. The results from western blot analysis revealed that
RACK1 protein expression was significantly increased in the SW1783
cells following transfection with RACK1-pcDNA3.1 (0.89±0.17 vs.
0.20±0.04, P<0.01); however, RACK1 protein expression was
decreased in the U-118 MG cells following transfection with RACK1
siRNA (0.10±0.02 vs. 0.65±0.12, P<0.01). The overexpression of
RACK1 led to the increased expression of CNTN2 (0.62±0.11 vs.
0.30±0.05, P<0.05), whereas the knockdown of RACK1 exerted an
inhibitory effect on the expression of CNTN2 (0.21±0.04 vs.
0.48±0.09, P<0.01) (Fig.
4).
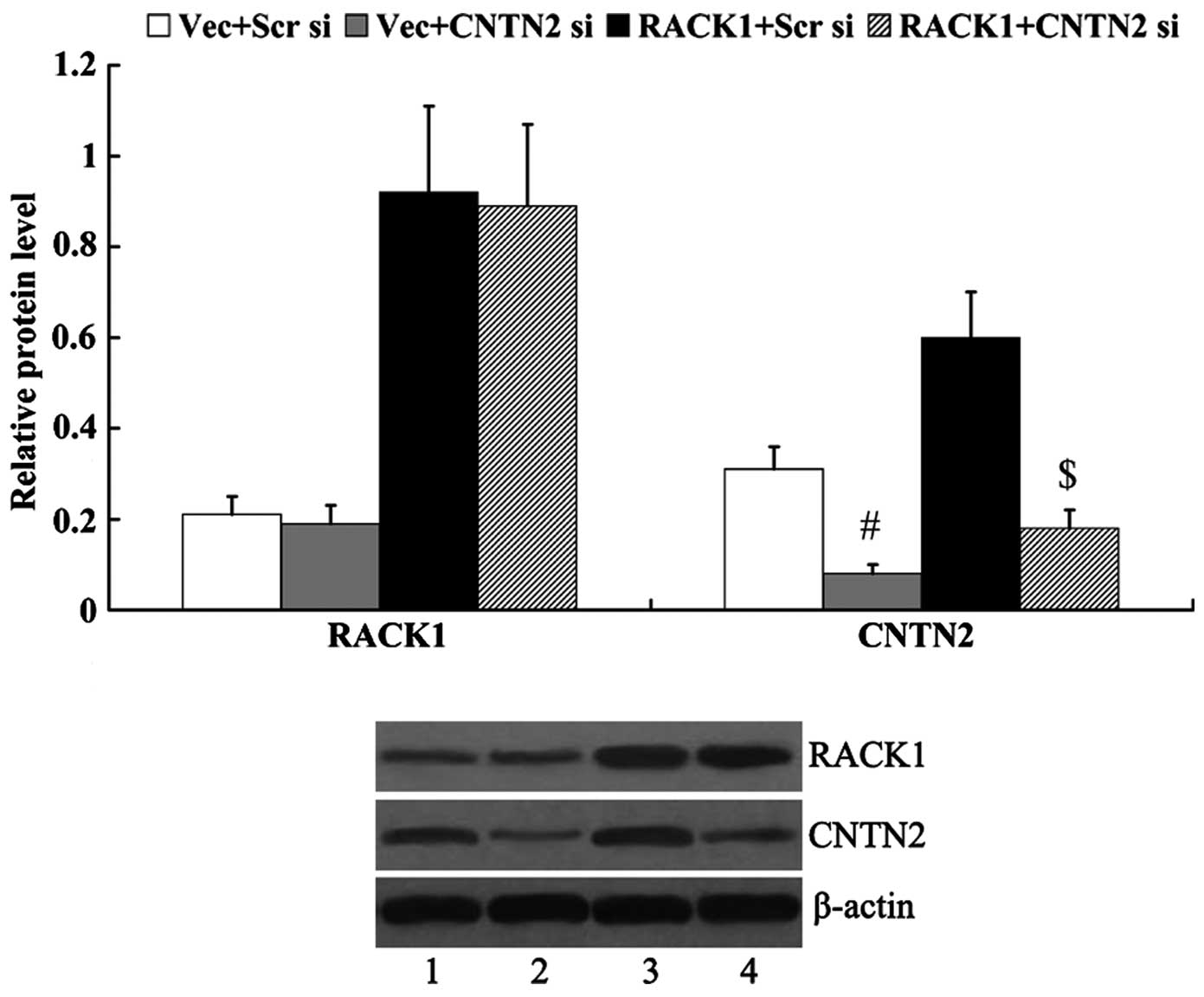
CNTN2 mediates the effects of RACK1 on
cell proliferation
RACK1-pcDNA3.1 and/or CNTN2 siRNA were transfected
into the SW1783 cells, and the protein expression levels of RACK1
and CNTN2 were then examined. As shown in Fig. 5, the expression of CNTN2 was
significantly decreased in the cells following transfection with
CNTN2 siRNA (0.31±0.05 vs. 0.08±0.02, P<0.01; 0.60±0.11 vs.
0.18±0.04, P<0.01). However, the expression of RACK1 was not
markedly altered by transfection with CNTN2 siRNA (0.21±0.04 vs.
0.19±0.04, P>0.05; 0.92±0.19 vs. 0.89±0.18, P>0.05).
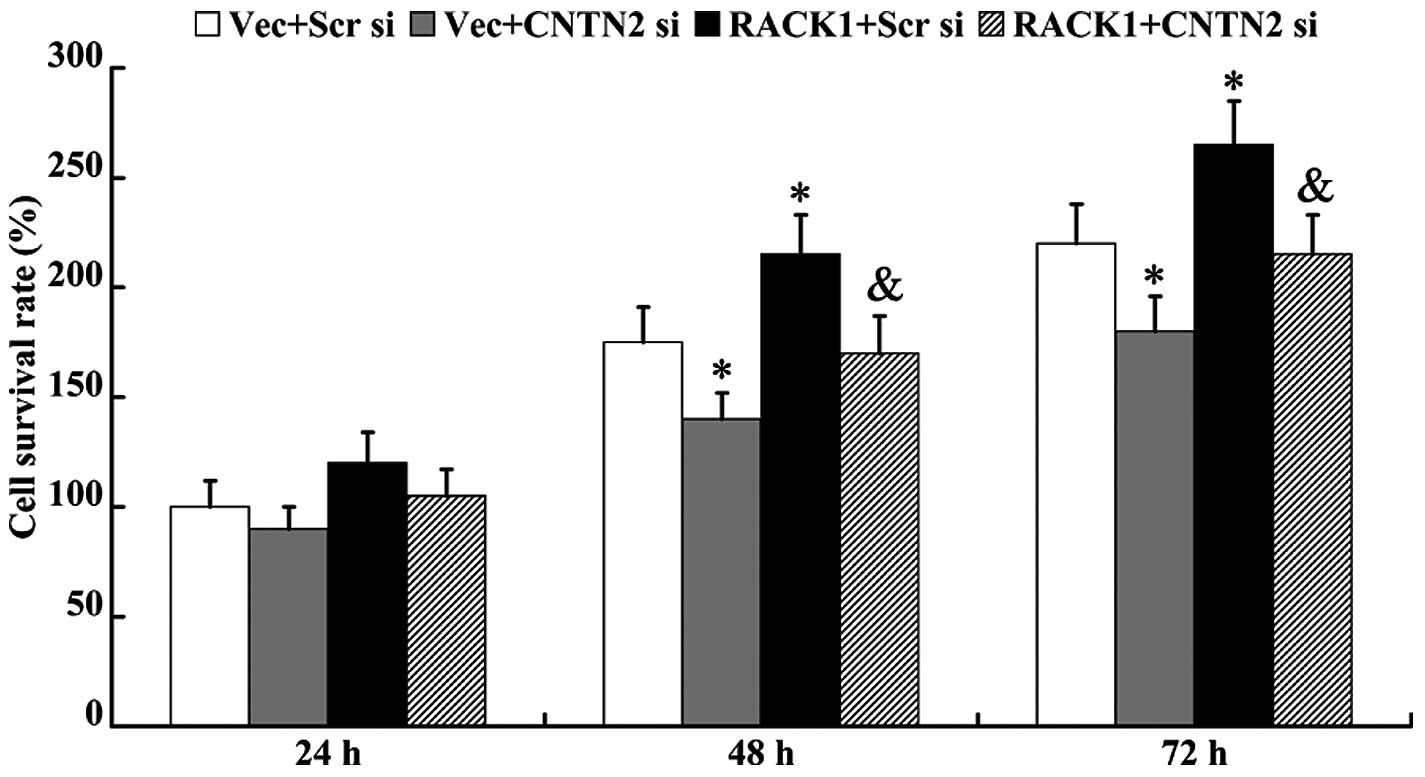
An MTT assay was performed to determine cell
proliferation ability. As shown in Fig. 6, cell viability was inhibited in
the cells following transfection with CNTN2 siRNA for 48 and 72 h
(P<0.05). RACK1-pcDNA3.1 promoted cell proliferation (48 h,
175±16% vs. 216±18%, P<0.05; 72 h, 219±18% vs. 266±20%,
P<0.05); however, this effect was abolished by the knockdown of
CNTN2 (48 h, 216±18% vs. 170±17%, P<0.05; 72 h, 266±20% vs.
213±18%, P<0.05).
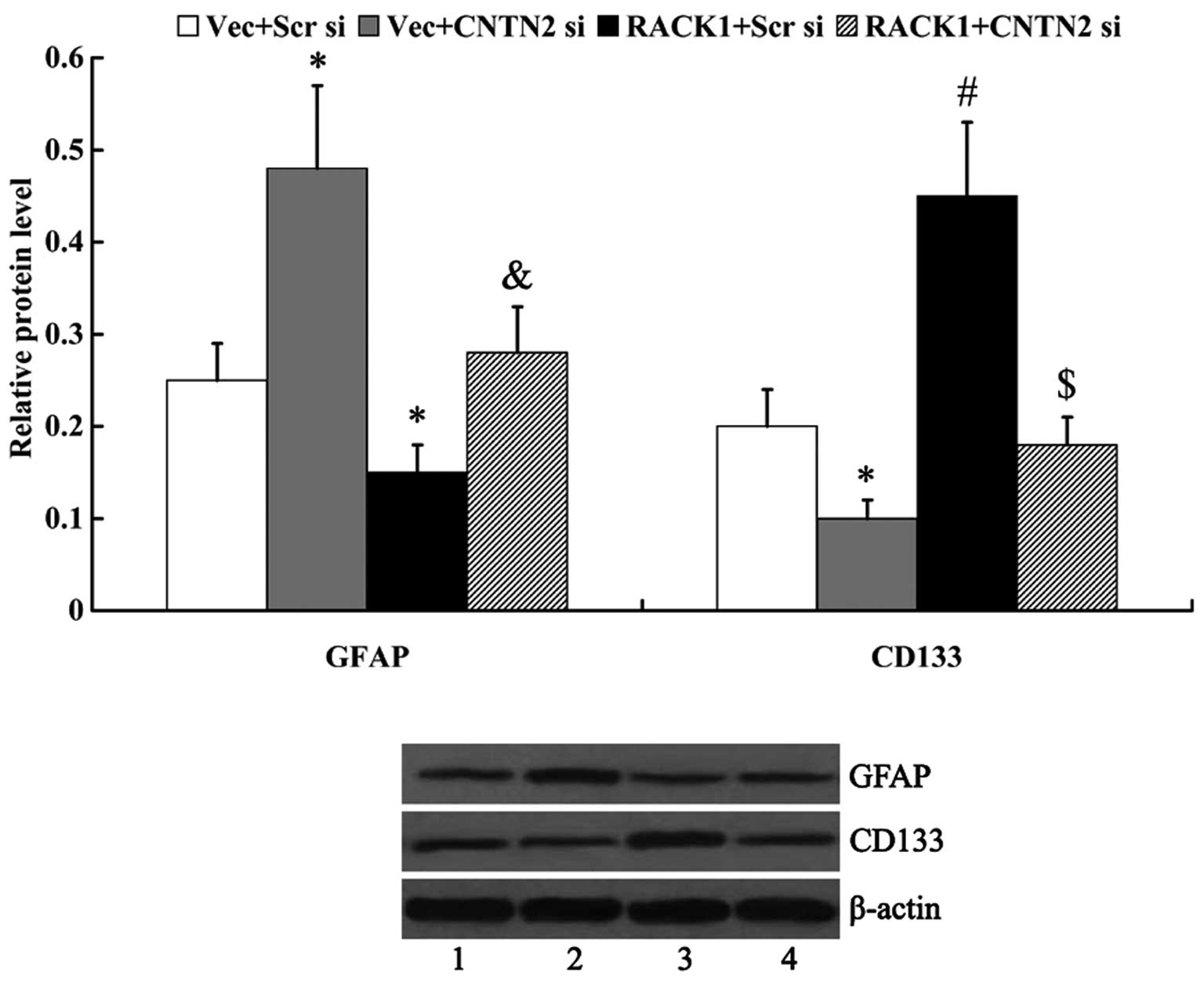
CNTN2 mediates the effect of RACK1 on
cell differentiation
The expression of GFAP and CD133 was examined by
western blot analysis to determine cell differentiation. We found
that RACK1 inhibited cell differentiation, as evidenced by the
markedly decreased expression of GFAP and the increased expression
of CD133 in the cells following transfection with RACK1-pcDNA3.1
(GFAP, 0.25±0.04 vs. 0.15±0.03, P<0.05; CD133, 0.20±0.04 vs.
0.45±0.08, P<0.01). The knockdown of CNTN2 attenuated the
effects of RACK1 on cell differentiation, as the expression of GFAP
was significantly upregulated and CD133 was significantly
downregulated following transfection with CNTN2 siRNA (GFAP,
0.15±0.03 vs. 0.28±0.05, P<0.05; CD133, 0.45±0.08 vs. 0.18±0.03,
P<0.01) (Fig. 7).
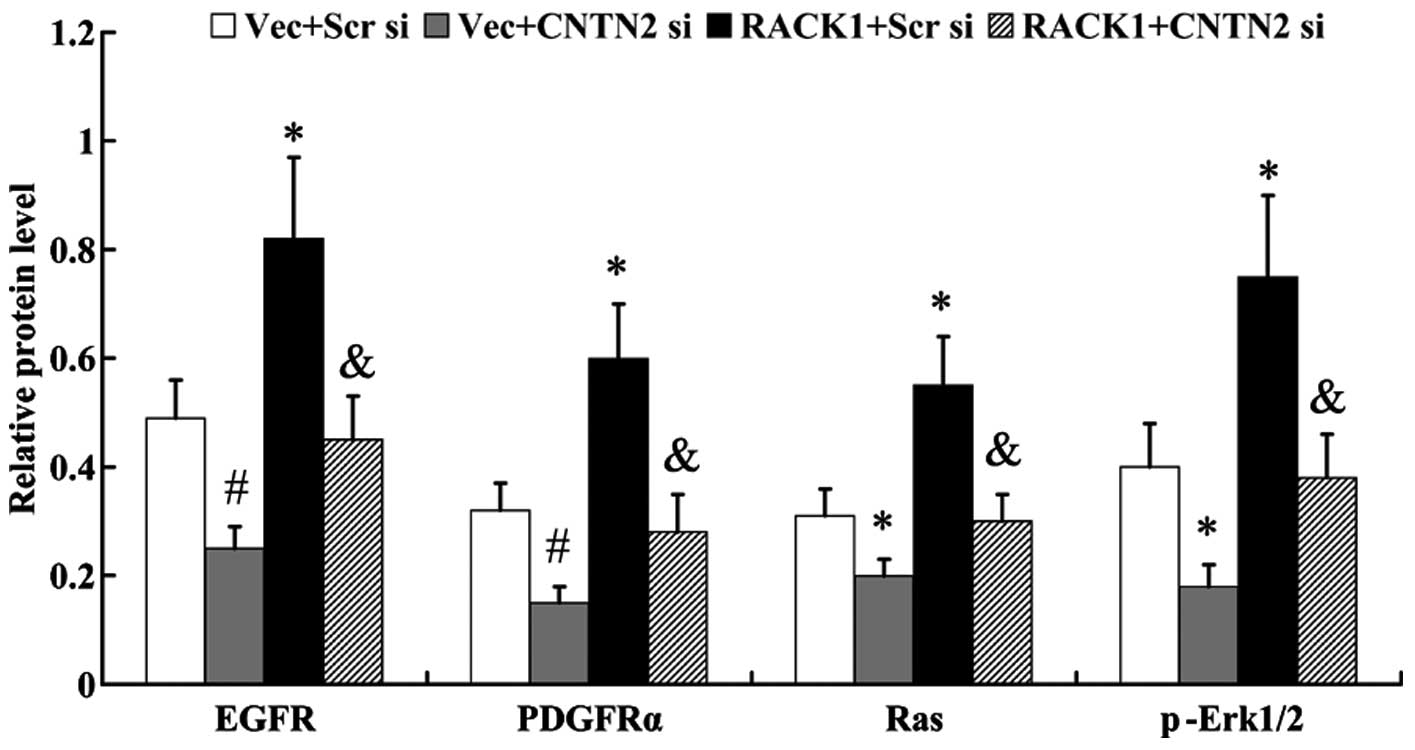
CNTN2 mediates the effect of RACK1 on
RTK/Ras/MAPK signaling
In order to examine the activation of RTK/Ras/MAPK
signaling, the expression of molecules in this signaling pathway,
including EGFR, PDGFRα, Ras and phosphorylated (p-)ERK1/2 were
examined by western blot analysis. As shown in Fig. 8, the RTK/Ras/MAPK signaling
pathway was activated by RACK1, accompanied by the increased
expression of EGFR, PDGFRα, Ras and p-ERK1/2 in the cells
overexpressing RACK1 (EGFR, 0.49±0.07 vs. 0.82±0.15, P<0.05;
PDGFRα, 0.32±0.05 vs. 0.60±0.10, P<0.05; Ras, 0.31±0.05 vs.
0.55±0.09, P<0.05; p-ERK1/2, 0.40±0.08 vs. 0.75±0.15,
P<0.05). However, the activation of the RTK/Ras/MAPK signaling
pathway was abolished by the knockdown of CNTN2 (EGFR, 0.82±0.15
vs. 0.45±0.08, P<0.05; PDGFRα, 0.60±0.10 vs. 0.28±0.07,
P<0.05; Ras, 0.55±0.09 vs. 0.30±0.05, P<0.05; p-Erk1/2,
0.75±0.15 vs. 0.38±0.08, P<0.05).
Discussion
In the present study, we found that the protein
expression levels of RACK1 and CNTN2 were higher in high-grade
glioma tissues and cells, and lower in low-grade glioma tissues and
cells. RACK1 is a scaffold protein that physically or functionally
interacts with numerous proteins (8,21–23). CNTN2 is a cell adhesion protein
that is expressed mainly in axons or regenerating neurons (26). It is involved in the
differentiation of cerebellar neurons (27), and is linked to the highest degree
in the oligodendroglioma protein-protein interaction network
(20). However, to the best of
our knowledge, this is the first study to examine whether RACK1
interacts with CNTN2. To examine the association between RACK1 and
CNTN2, a co-immunoprecipitation assay was performed, and the
results revealed that RACK1 interacts with CNTN2. Furthermore, the
results from western blot analysis demonstrated that RACK1
upregulated the expression of CNTN2. However, the knockdown of
CNTN2 did not have a marked effect on the expression of RACK1.
These data suggest that CNTN2 is a downstream effector of
RACK1.
It has been suggested in previous studies that the
dysregulation of RACK1 is involved in a variety of cancers, such as
esophageal squamous cell carcinoma, lung cancer and ovarian cancer
(10–13). In the study by Peng et al,
it was demonstrated that the enforced downregulation of RACK1
inhibited tumor xenograft growth in nude mice, and inhibited the
proliferation and invasion of human glioma cells in vitro
(14). In the present study, we
examined the effects of RACK1 on glioma cell differentiation. We
found that RACK1 had an inhibitory effect on the differentiation of
glioma cells, and we noted the decreased expression of the
differentiation marker, GFAP, and the increased expression of the
glioma stem cell self-renewal marker, CD133, in the cells
overexpressing RACK1. Similar to the effects of RACK1, CNTN2 also
inhibited glioma cell differentiation, and the effects of RACK1 on
glioma cell differentiation were abolished by the knockdown of
CNTN2.
RTK signaling pathways control a number of
biological processes, including cell proliferation, survival,
differentiation and morphogenesis (28,29). EGFR and PDGFR are two important
RTKs. The increase in RTK activity initiates signaling through the
conserved Ras/MAPK cassette, thus regulating the development of
glioblastoma multiforme and other cancers (30,31). In the present study, we found that
the RTK/Ras/MAPK signaling pathway was activated by RACK1 and CNTN2
in glioma cells. In addition, the knockdown of CNTN2 attenuated the
effects of RACK1 on the activation of the RTK/Ras/MAPK signaling
pathway.
Taken together, our findings firstly demonstrated
that RACK1 interacts with and regulates CNTN2, and the effects of
RACK1 on glioma cell growth and differentiation were mediated by
CNTN2. Thus, we suggest that the RACK1/CNTN2/RTK/Ras/MAPK axis
exists in glioma cells and this axis may be a potential target in
the treatment of gliomas.
References
|
1
|
Katsetos CD, Dráberová E, Smejkalová B,
Reddy G, Bertrand L, de Chadarévian JP, Legido A, Nissanov J, Baas
PW and Dráber P: Class III beta-tubulin and gamma-tubulin are
co-expressed and form complexes in human glioblastoma cells.
Neurochem Res. 32:1387–1398. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Arko L, Katsyv I, Park GE, Luan WP and
Park JK: Experimental approaches for the treatment of malignant
gliomas. Pharmacol Ther. 128:1–36. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yacoub A, Hamed HA, Allegood J, Mitchell
C, Spiegel S, Lesniak MS, Ogretmen B, Dash R, Sarkar D, Broaddus
WC, et al: PERK-dependent regulation of ceramide synthase 6 and
thioredoxin play a key role in mda-7/IL-24-induced killing of
primary human glioblastoma multiforme cells. Cancer Res.
70:1120–1129. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hamed HA, Yacoub A, Park MA, Eulitt PJ,
Dash R, Sarkar D, Dmitriev IP, Lesniak MS, Shah K, Grant S, et al:
Inhibition of multiple protective signaling pathways and Ad.5/3
delivery enhances mda-7/IL-24 therapy of malignant glioma. Mol
Ther. 18:1130–1142. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yu H, Park J, Lee J, Choi K and Choi C:
Constitutive expression of MAP kinase phosphatase-1 confers
multi-drug resistance in human glioblastoma cells. Cancer Res
Treat. 44:195–201. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xing WJ, Zou Y, Han QL, Dong YC, Deng ZL,
Lv XH, Jiang T and Ren H: Effects of epidermal growth factor
receptor and phosphatase and tensin homologue gene expression on
the inhibition of U87MG glioblastoma cell proliferation induced by
protein kinase inhibitors. Clin Exp Pharmacol Physiol. 40:13–21.
2013. View Article : Google Scholar
|
|
7
|
Gandin V, Senft D, Topisirovic I and Ronai
ZA: RACK1 Function in Cell Motility and Protein Synthesis. Genes
Cancer. 4:369–377. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Adams DR, Ron D and Kiely PA: RACK1, a
multifaceted scaffolding protein: structure and function. Cell
Commun Signal. 9:222011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
McCahill A, Warwicker J, Bolger GB,
Houslay MD and Yarwood SJ: The RACK1 scaffold protein: a dynamic
cog in cell response mechanisms. Mol Pharmacol. 62:1261–1273. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li JJ and Xie D: RACK1, a versatile hub in
cancer. Oncogene. 34:1890–1898. 2015. View Article : Google Scholar
|
|
11
|
Wang N, Liu F, Cao F, Jia Y, Wang J, Ma W,
Tan B, Wang K, Song Q and Cheng Y: RACK1 predicts poor prognosis
and regulates progression of esophageal squamous cell carcinoma
through its epithelial-mesenchymal transition. Cancer Biol Ther.
16:528–540. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Choi YY, Lee SY, Lee WK, Jeon HS, Lee EB,
Lee HC, Choi JE, Kang HG, Lee EJ, Bae EY, et al: RACK1 is a
candidate gene associated with the prognosis of patients with early
stage non-small cell lung cancer. Oncotarget. 6:4451–4466. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lin Y, Cui M, Teng H, Wang F, Yu W and Xu
T: Silencing the receptor of activated C-kinase 1 (RACK1)
suppresses tumorigenicity in epithelial ovarian cancer in vitro and
in vivo. Int J Oncol. 44:1252–1258. 2014.PubMed/NCBI
|
|
14
|
Peng R, Jiang B, Ma J, Ma Z, Wan X, Liu H,
Chen Z, Cheng Q and Chen R: Forced downregulation of RACK1 inhibits
glioma development by suppressing Src/Akt signaling activity. Oncol
Rep. 30:2195–2202. 2013.PubMed/NCBI
|
|
15
|
Su J, Xu J and Zhang S: RACK1, scaffolding
a heterotrimeric G protein and a MAPK cascade. Trends Plant Sci.
20:405–407. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shi S, Deng YZ, Zhao JS, Ji XD, Shi J,
Feng YX, Li G, Li JJ, Zhu D, Koeffler HP, et al: RACK1 promotes
non-small-cell lung cancer tumorigenicity through activating sonic
hedgehog signaling pathway. J Biol Chem. 287:7845–7858. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen Y, Wang L, Xu H, Liu X and Zhao Y:
Exome capture sequencing reveals new insights into hepatitis B
virus-induced hepatocellular carcinoma at the early stage of
tumorigenesis. Oncol Rep. 30:1906–1912. 2013.PubMed/NCBI
|
|
18
|
Adair SJ, Carr TM, Fink MJ, Slingluff CL
Jr and Hogan KT: The TAG family of cancer/testis antigens is widely
expressed in a variety of malignancies and gives rise to
HLA-A2-restricted epitopes. J Immunother. 31:7–17. 2008. View Article : Google Scholar
|
|
19
|
Rickman DS, Tyagi R, Zhu XX, Bobek MP,
Song S, Blaivas M, Misek DE, Israel MA, Kurnit DM, Ross DA, et al:
The gene for the axonal cell adhesion molecule TAX-1 is amplified
and aberrantly expressed in malignant gliomas. Cancer Res.
61:2162–2168. 2001.PubMed/NCBI
|
|
20
|
Yu F and Fu WM: Identification of
differential splicing genes in gliomas using exon expression
profiling. Mol Med Rep. 11:843–850. 2015.
|
|
21
|
Birikh KR, Sklan EH, Shoham S and Soreq H:
Interaction of 'readthrough' acetylcholinesterase with RACK1 and
PKCbeta II correlates with intensified fear-induced conflict
behavior. Proc Natl Acad Sci USA. 100:283–288. 2003. View Article : Google Scholar
|
|
22
|
Wang CC, Lo HF, Lin SY and Chen H: RACK1
(receptor for activated C-kinase 1) interacts with FBW2 (F-box and
WD-repeat domain-containing 2) to up-regulate GCM1 (glial cell
missing 1) stability and placental cell migration and invasion.
Biochem J. 453:201–208. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kundu N, Dozier U, Deslandes L, Somssich
IE and Ullah H: Arabidopsis scaffold protein RACK1A interacts with
diverse environmental stress and photosynthesis related proteins.
Plant Signal Behav. 8:e240122013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wee B, Charles N and Holland EC: Animal
models to study cancer-initiating cells from glioblastoma. Front
Biosci (Landmark Ed). 16:2243–2258. 2011. View Article : Google Scholar
|
|
25
|
Akhavan D, Cloughesy TF and Mischel PS:
mTOR signaling in glioblastoma: lessons learned from bench to
bedside. Neuro Oncol. 12:882–889. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Brümmendorf T and Rathjen FG: Axonal
glycoproteins with immunoglobulin- and fibronectin type III-related
domains in vertebrates: structural features, binding activities,
and signal transduction. J Neurochem. 61:1207–1219. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bizzoca A, Virgintino D, Lorusso L,
Buttiglione M, Yoshida L, Polizzi A, Tattoli M, Cagiano R, Rossi F,
Kozlov S, et al: Transgenic mice expressing F3/contactin from the
TAG-1 promoter exhibit developmentally regulated changes in the
differentiation of cerebellar neurons. Development. 130:29–43.
2003. View Article : Google Scholar
|
|
28
|
Schlessinger J: Cell signaling by receptor
tyrosine kinases. Cell. 103:211–225. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Simon MA: Receptor tyrosine kinases:
specific outcomes from general signals. Cell. 103:13–15. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Carracedo A, Ma L, Teruya-Feldstein J,
Rojo F, Salmena L, Alimonti A, Egia A, Sasaki AT, Thomas G, Kozma
SC, et al: Inhibition of mTORC1 leads to MAPK pathway activation
through a PI3K-dependent feedback loop in human cancer. J Clin
Invest. 118:3065–3074. 2008.PubMed/NCBI
|
|
31
|
Wan X, Harkavy B, Shen N, Grohar P and
Helman LJ: Rapamycin induces feedback activation of Akt signaling
through an IGF-1R-dependent mechanism. Oncogene. 26:1932–1940.
2007. View Article : Google Scholar
|