Introduction
One of the challenges currently facing gene therapy
researchers is finding a means of efficiently delivering exogenous
genes into cells. There are many factors which affect the
efficiency of this process; the gene therapy vector construct plays
a key role. Gene delivery systems are broadly classified into viral
vectors or non-viral vectors. Non-viral vectors include liposomes,
polymeric micelles and other nanoparticles. Compared with viral
vectors, non-viral vectors possess several advantages, including
low toxicity, biodegradability, ease of synthesis and low immune
responsiveness, and therefore non-viral vectors are attracting more
research interest (1,2). In particular, cationic liposomes
(CLs) composed of N-[1-(2,3-dioleoyloxy)
propyl]-N,N,N-trimethylammonium chloride (DOTAP) and cholesterol
(chol), or DOTAP/chol, have been classified as one of the most
efficient vectors for plasmid DNA (pDNA) transfection into cells
(3,4). However, the molar ratio of
DOTAP/chol has an impact on the transfection efficiency. Previous
research has indicated that DOTAP/chol liposomes with a molar ratio
of 1:1 or 2:1 had a high transfection efficiency (3,5).
To achieve a higher transfection efficiency, on the one hand, it is
necessary to identify the optimal molar ratio of DOTAP/chol for use
in the preparation of liposomes. On the other hand, the hydration
medium has an effect on the liposome properties, such as entrapment
efficiency and stability (6).
According to previous research, hydration media commonly used in
the preparation of CLs include 0.9% NaCl, 5% glucose and Tris-HCl
(1,4,7).
However, a comprehensive study of the effects of different
hydration media on CL-based gene delivery systems has not yet been
reported, to the best of our knowledge. In addition, previous
studies have demonstrated that CL-mediated gene transfer may be
enhanced by the addition of natural polycations which act as
DNA-condensing agents and form liposome-polycation-DNA complexes.
Natural polycations, such as protamine sulfate (8,9),
chitosan (10) and poly-L-lysine
(11), have been shown to
condense DNA and form nanoparticles which are mechanically stable,
of uniform particle size and of controllable morphology. Protamine,
which is non-toxic and is composed of natural cationic peptides,
may provide unique membrane-translocating and nuclear-localizing
activities owing to its abundant amino acid sequence (12).
RNA interference (RNAi) is a process which involves
the post-transcriptional silencing of gene expression in molecular
biology, and has become more popular as a gene therapy technique
and has been applied in the treatment of various diseases. RNAi
tools include synthetic short interfering RNA and endogenous
microRNA (miRNA or miR) (13). As
small non-coding RNAs, miRNAs regulate the expression of multiple
proteins at the translation level. miR-145 has been reported to
exert potent antitumor effects by targeting multiple genes which
are associated with tumor growth, metastasis and invasion. It has
been reported that miR-145 is expressed at low levels in breast
cancer, colon cancer, lung cancer and other tumor tissues compared
with normal tissues (14). As
previously demonstrated, by transfecting synthetic miR-145
oligonucleotides into neuroblastoma cells, endometrial stem cells,
and bladder cancer cells, the expression of genes, such as
insulin-like growth factor receptor I (IGF-IR) and
hypoxia-inducible factor 2α (HIF-2α) was downregulated, which
resulted in the inhibition of cancer cell growth and invasion
(15–17).
In the present study, we evaluated the effects of
various factors on the transfection efficiency of DOTAP/chol
liposomes, including the hydration medium used for the preparation
of the liposomes, the molar ratio of DOTAP/chol, the mass of
DOTAP/DNA and CLs combined with protamine, in order to create a
non-viral vector with a higher transfection efficiency.
Additionally, we found that miR-145 was expressed at low levels in
HepG2 cells compared with other cancer cell lines; therefore, we
further explored the role of miR-145 in HepG2 cells and its related
downstream genes.
Materials and methods
Materials
The plasmid, pCDH-miR-145-GFP, was constructed in
our own laboratory. DOTAP was purchased from Corden Pharma
(Liestal, Switzerland), and chol and protamine sulfate salt were
purchased from Sigma (St. Louis, MO, USA); and Lipofectamine™ 2000
(Lipo 2000) and TRIzol reagent were purchased from Invitrogen
(Carlsbad, CA, USA). The miRcute miRNA isolation kit was obtained
from Tiangen Biotech Co., Ltd. (Beijing, China). The primers were
synthesized by Sangon Biotech (Shanghai, China).
Cell culture
The 293T cells, hepatoma cells (HepG2), cervical
cancer cells (HeLa), breast adenocarcinoma cells (MCF-7), lung
adenocarcinoma cells (A549), human gastric cancer (BGC-823) and
human colorectal cancer cells (LoVo) were all purchased from
Shanghai Institutes for Biological Sciences (Shanghai, China).
These cells were cultured in Dulbecco's modified Eagle's medium
(DMEM) or RPMI-1640 containing 10% fetal bovine serum (FBS) at 37°C
with 5% CO2.
Construction of gene carriers
Preparation of CLs and CL/DNA
complexes
The CLs were prepared using the thin-film dispersion
method. Different molar ratios of DOTAP and cholesterol (1:1 to
7:1) were mixed to a total amount of 10 µmol, and the
above-mentioned lipids were dissolved in chloroform, and the
solvent was then evaporated under a vacuum by rotary evaporation
for 30 min at 45°C in order to form a thin lipid film. The correct
amount of hydration medium was then added to the film for 45 min at
50°C. The multicellular liposomes were obtained following
sufficient hydration. Using a handheld extruder, the liposomes were
extruded 10 times repeatedly, each through 2 stacks of
polycarbonate membranes with progressively decreasing pore sizes of
400, 200 and 100 nm (ME-25S). The resulting CLs were extruded
through a 0.22-µm filter for sterilization and stored at
4°C. The liposomes were diluted with deionized water and the
appropriate amount of DNA (pCDH-miR-145-GFP) by mixing in equal
volumes and incubated for 20 min at room temperature to form the
CL/DNA complexes.
Preparation of CL-protamine-DNA (LPD)
complexes
Protamine/DNA nanoparticles were prepared by mixing
equal volumes of protamine and DNA solution (pCDH-miR-145-GFP) and
incubating the resulting mixture for 10 min at room temperature.
LPD complexes were prepared by mixing equal volumes of CLs and
protamine/DNA nanoparticles and incubating for 20 min at room
temperature.
Single factor tests
Firstly, to examine the the effects of the hydration
medium on the CLs, we used DOTAP/chol (at a molar ratio of 1:1) to
prepare the CLs, and hydration medium (deionized water, 0.9% NaCl,
5% glucose, 5% sucrose, 10 mM Tris buffer and 10 mM phosphate
buffer). A weight ratio of DOTAP/DNA of 4:1 was used for
transfection. Secondly, varying amounts of DNA (2, 4, 6, 8, 10 and
15 µg) were used to compare the relative transfection
efficiency, and we selected the DOTAP/chol liposomes (at a molar
ratio of 1:1) and the weight ratio of DOTAP/DNA at 4:1 to determine
the optimal mass of DNA. Thirdly, to identify the optimal molar
ratio of DOTAP/chol, 5% glucose was used as the hydration medium,
the molar ratio of DOTAP/chol was 1, 2, 3, 4, 5 and 6, and the mass
ratio of DOTAP/DNA was 4:1, the weight of the DNA was 8 µg;
under these conditions, we examined the transfection efficiency.
Fourthly, we examined the weight ratio of DOTAP/DNA (1, 2, 3, 4, 5,
6, 8 and 10) and used DOTAP/chol liposomes (at a molar ratio of
3:1) to determine the transfection efficiency. Lastly, to examine
the weight of protamine in the LPD complexes, we set a weight ratio
of DOTAP/protamine/DNA at 3:0.2:1, 3:0.5:1, 3:0.8:1, 3:1:1,
3:1.5:1, 3:2:1 and 3:3:1 for transfection.
Measurement of particle size and
ζ-potential
The particle size and ζ-potential of the liposomes,
lipoplexes and LPD complexes were measured using a Zetasizer Nano
ZS90 (Malvern Instruments, Malvern, UK) after being dispersed in
deionized water.
Agarose gel retardation assay
The DNA binding ability of the DOTAP/chol liposomes
was evaluated by a gel retardation assay. The lipoplex included 0.5
µg at different molar ratios of DOTAP/chol and a different
mass ratio of DOTAP/DNA which was loaded into individual wells on a
1% agarose gel, and electrophoresis was performed in
Tris/Borate/EDTA (TBE) buffer at 120 V for 30 min, followed by
staining with ethidium bromide.
In vitro transfection
The cells were seeded in a 6-cm dish at a density of
approximately 1×106 cells per dish and incubated
overnight prior to transfection. The lipoplexes and LPD complexes
were transfected into the cells for 6 h with minimum essential
medium (MEM) medium in the absence of FBS. The complexes were then
removed and added to 10% FBS medium. After 48 h, the GFP expression
levels in the transfected cells were analyzed under a fluorescence
microscope (Zeiss, Oberkochen, Germany) and using a flow cytometer
(BD Biosciences, Franklin Lakes, NJ, USA). Lipo 2000 was used as a
positive control according to the manufacturer's instructions.
DNase assay
Naked DNA, lipoplexes and LPD complexes were added
to DNase solution at 37°C for 30 min and EDTA was used to terminate
the reaction. Triton X-100 (1%) was added to disrupt the lipid
membranes and subseqently, heparin was added to release the DNA
from the protamine/DNA complexes. The resulting products were
analyzed by electrophoresis on a 1% agarose gel at 120 V for 30
min.
Analysis of endogenous miR-145
expression levels in different cancer cell lines by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
According to the instructions of the manufacturer
(provider of TRIzol reagent), we extracted the total RNA from the
A549, BGC-823, HepG2, HeLa, LoVo and MCF-7 cells. miR-145 was
reverse transcribed into cDNA using an miRcute miRNA cDNA kit and
miRcute miRNA qPCR detection kits, to measure the expression levels
of miR-145. The PCR cycles were as follows: denaturation at 94°C
for 2 min, followed by 40 cycles at 94°C for 20 sec and 60°C for 34
sec and snRU6 was used as an internal normalization control as
previously described (16). The
relative changes in miRNA expression were calculated using the
2−ΔΔCT method.
Cell proliferation assay
Cell proliferation was measured using by
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay as previously described (16). Following transfection for 48 h,
10,000 HepG2 cells were seeded in 96-well plates and cultured for
24 h. Briefly, MTT solution was added and followed by further
incubation for 4 h; DMSO was then added to dissolve the formazan
crystals and the optical density was measured at 570 and 630 nm
using a microplate reader (Multiskan MK3, Thermo Fisher Scientific
Inc.,Waltham, MA, USA).
Analysis of the downsteam genes
regulated by miR-145 by RT-qPCR
TRIzol reagent was used to extract all RNA which was
then reverse transcribed into cDNA using a ReverTra Ace®
qPCR RT kit (Toyobo Co., Ltd., Osaka, Japan). qPCR was performed
using SYBR-Green Real-Time PCR Master Mix (Toyobo). We also
searched the TargetScan and miRanda databases in order to predict
the binding sites of miR-145 based on different algorithms; genes
found on both databases which may affect cell proliferation were
selected, including CDK6, cyclinD1, c-Myc and Sp1. The primer
sequences of cyclin-dependent kinase 6 (CDK6), cyclinD1, c-Myc, Sp1
transcription factor (Sp1) and β-actin are listed in Table I.
 | Table ISequences of the primers used for
RT-qPCR. |
Table I
Sequences of the primers used for
RT-qPCR.
| Gene | Forward sequence
(5′→3′) | Reverse sequence
(3′→5′) |
|---|
| CDK6 |
CGAATGCGTGGCGGAGATC |
CCACTGAGGTTAGAGCCATC |
| CyclinD1 |
GGATGCTGGAGGTCTGCGAGGAAC |
GAGAGGAAGCGTGTGAGGCGGTAG |
| c-Myc |
CTTCTCTCCGTCCTCGGATTCT |
GAAGGTGATCCAGACTCTGACCTT |
| Sp1 |
GCCTCCAGACCATTAACCTCAGT |
GCTCCATGATCACCTGGGGCAT |
| β-actin |
GGGACCTGACTGACTACCTC |
TCATACTCCTGCTTGCTGAT |
| miR-145 |
GTCCAGTTTTCCCAGGAATCCCT | |
| U6 |
CTCGCTTCGGCAGCACA | |
Statistical analysis
All experimental results are presented as the means
± standard deviation (SD). Statistical analyses were performed
using the Student's t-test. A P-value <0.05 was considered to
indicate a statistically significant difference.
Results
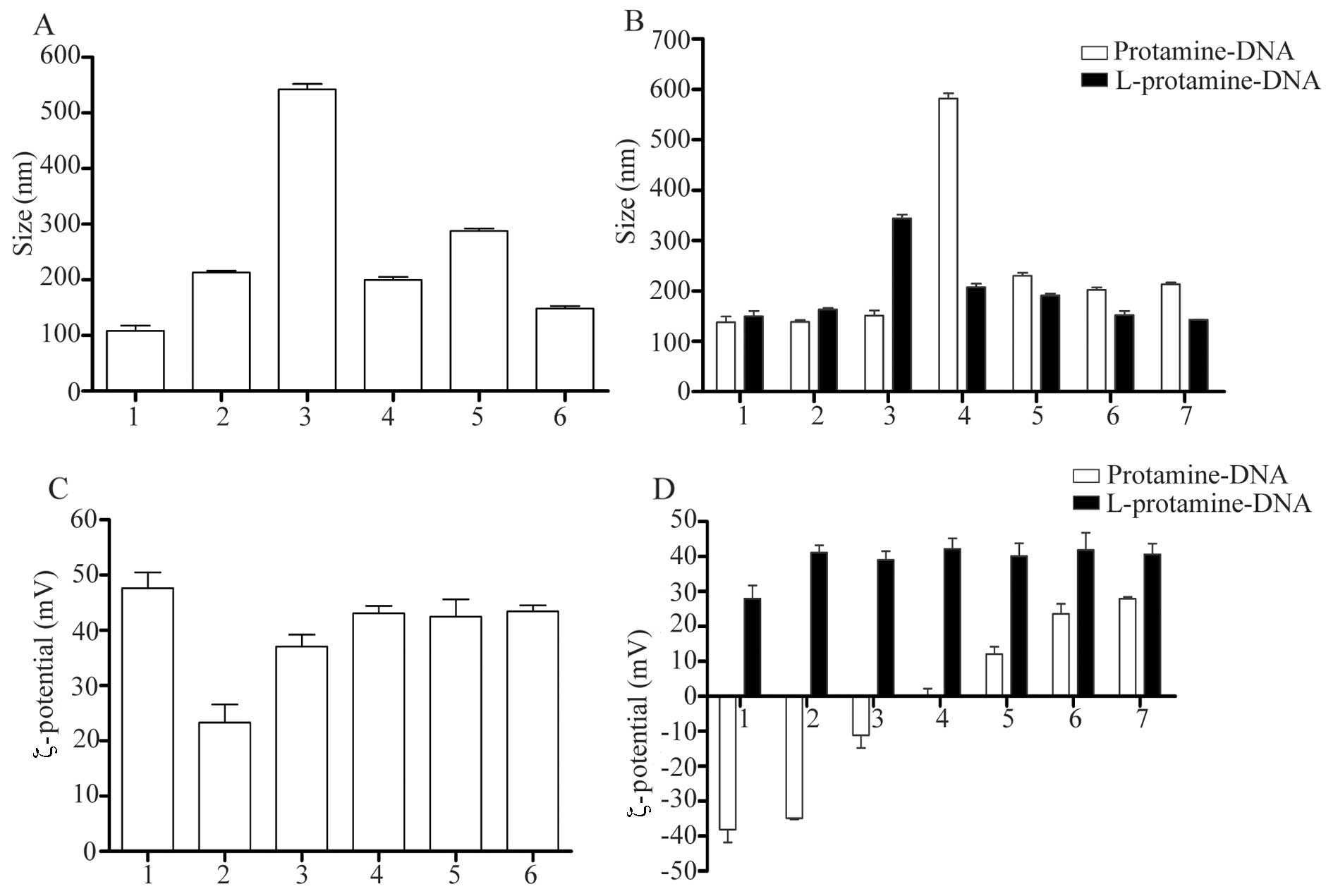
Particle sizes and ζ-potentials of CLs,
lipoplexes and LPD complexes
Particle size and ζ-potential are important
physicochemical parameters of nanoparticles, which significantly
affect the transfection efficiency in vitro or in
vivo (10). The size and
ζ-potential of the lipoplexes and LPD complexes were determined and
compared with those of the corresponding lipoplexes. As shown in
Fig. 1, differences were observed
in the particle size between the lipoplexes and the LPD complexes.
Following the addition of protamine, the mean particle size of the
complexes decreased; however, the ζ-potential of the complexes
remained unaltered. This indicated that protamine had an effect on
nanoparticle size by condensing the DNA, whereas it had a limited
effect on the ζ-potential of the nanoparticles.
DNA binding affinity of CLs
Gel retardation assays are widely used to measure
the DNA binding affinity of CLs (1). Our results indicated that with the
different weight ratios of DOTAP/DNA ranging from 1 to 7, the
migration path of the DNA in the lipoplexes changed (Fig. 2). When the weight ratio was ≥2 in
formulation, minimal free DNA was observed in the gel. These
results suggested that when the weight ratio of DOTAP/DNA was ≥2,
the liposomes and DNA were completely bound and also indicated that
the DNA binding affinity of the CLs was unaffected by the hydration
medium.
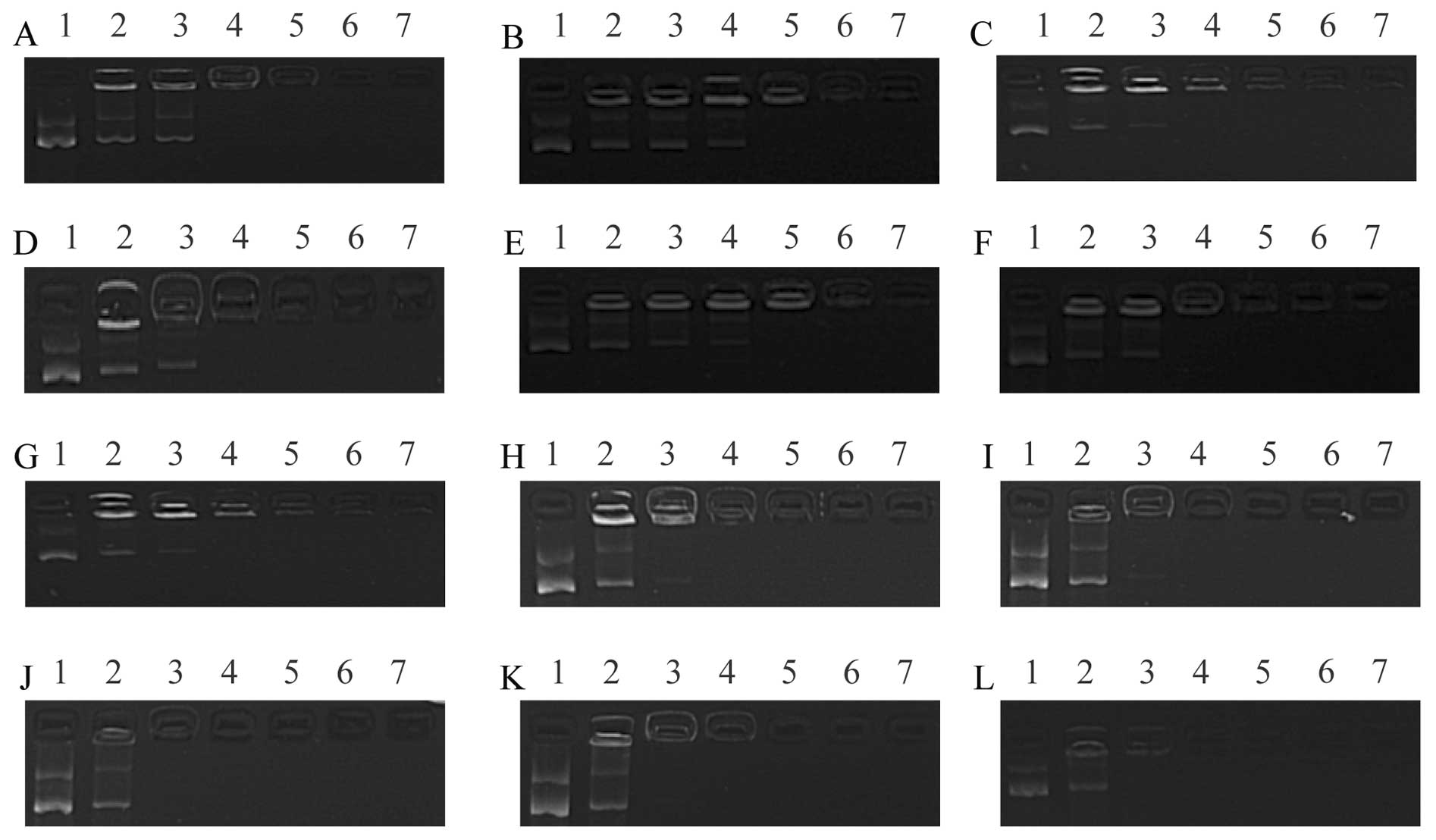 | Figure 2Gel retardation assays. The mass ratio
of DOTAP/DNA was 1, 2, 3, 4, 5 and 7 (lanes 2, 3, 4, 5, 6 and 7,
respectively). Lanes 1, 0.5 µg plasmid. (A–E) Different
hydration medium [DOTAP/cholesterol (chol), 1:1)]; (A) deionized
water; (B) 0.9% NaCl; (C) 5% glucose; (D) 10 mM Tris-HCl; (E) 10 mM
phosphate buffer; (F) 5% sucrose. (G–L) Different liposomes were
complexed with DNA; (G) DOTAP/chol, 1:1; (H) DOTAP/chol, 2:1; (I)
DOTAP/chol, 3:1; (J) DOTAP/chol, 4:1; (K) DOTAP/chol, 5:1; (L)
DOTAP/chol, 7:1. |
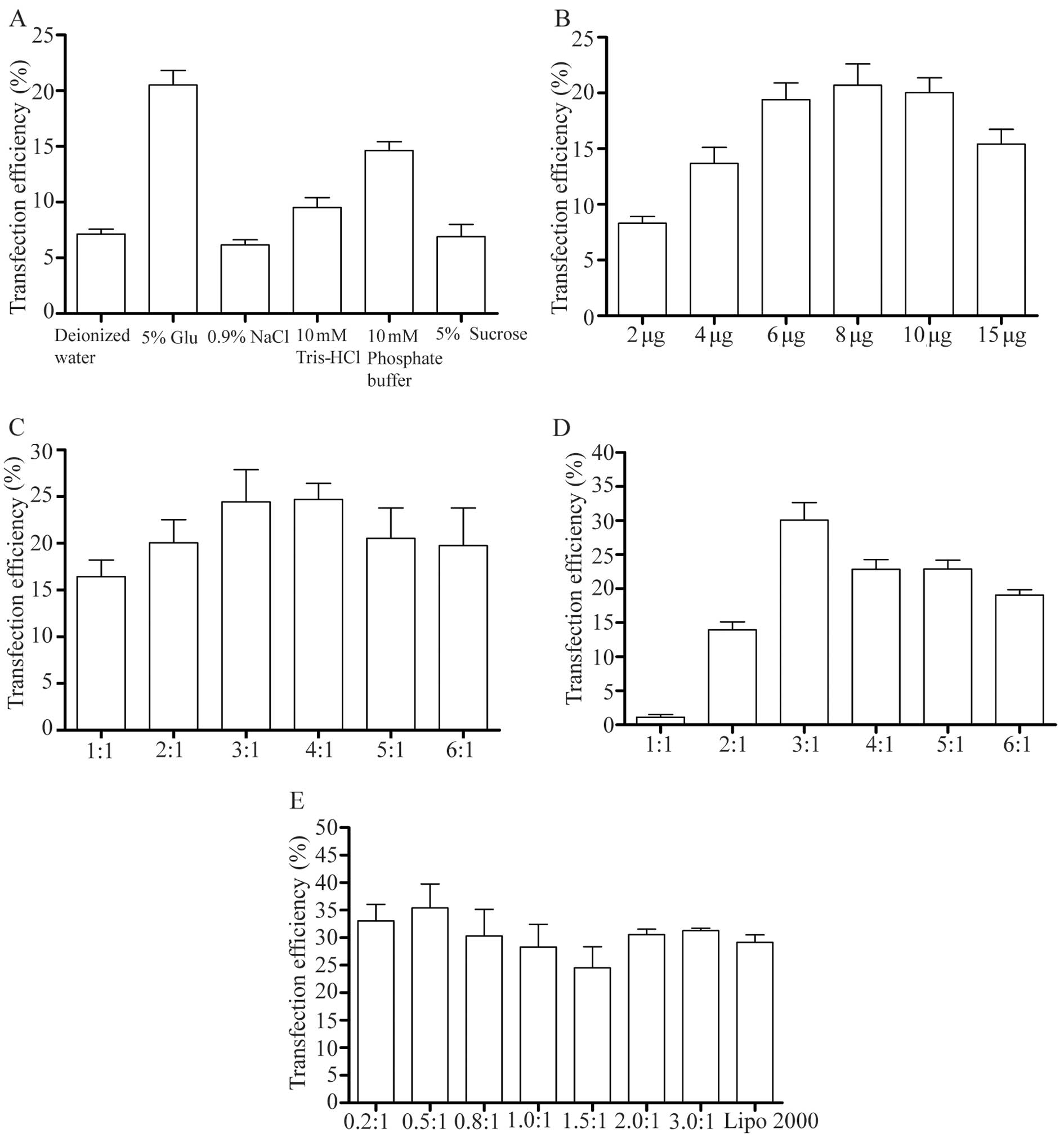
Transfection efficiency of CL/DNA and LPD
complexes
Previous research has indicated that the
transfection efficiency of CLs is affected by a number of factors,
including the molar ratio of DOTAP/chol and the charge ratio of
DOTAP/DNA, and these must be taken into account when determining
the optimal transfection protocol in vitro (1,23).
The present study examined these factors (hydration medium, the
amount of DNA, the molar ratio of DOTAP/chol, the weight ratio of
DOTAP/DNA and the weight of protamine/DNA) in detail. By examining
these factors, we determined the optimal conditions to prepare
DOTAP/chol liposomes for use in a transfection protocol. To
determine the effects of the hydration medium on transfection,
water, 5% glucose, 0.9% NaCl, 10 mM Tris-HCl and 10 mM phosphate
buffer and 5% sucrose were used. We found that the optimal
hydration medium for DOTAP/chol liposomes was 5% glucose (Figs. 3A and 4A). When the hydration medium was
changed from deionized water to 5% glucose, the transfection
efficiency increased from 6.2 to 20.5%, respectively (Fig. 4A). Furthermore, we investigated
the optimal amount of DNA for transfection in a 6-cm dish, by
testing different amounts of DNA (2, 4, 6, 8, 10 and 15 µg).
The results revealed that when the amount of DNA increased from 2
to 8 µg, the transfection efficiency increased from 8.2 to
19.8%, respectively. However, with further increases in the amount
of DNA, the transfection efficiency decreased (Figs. 3B and 4B). To examine the optimal molar ratio
of DOTAP/chol, we tested different molar ratios of DOTAP/chol (1,
2, 3, 4, 5 and 6) to determine the effects on transfection
efficiency. The results revealed that for DNA delivery, the optimal
molar ratio of DOTAP/chol was 3:1 (Figs. 3C and 4C). When the ratio of DOTAP/chol was
changed from 3:1 to 6:1, the effects of DNA transfection
progressively diminished. The effect of changing the mass ratio of
DOTAP/DNA (1, 2, 3, 4, 5 and 6) on the transfection efficiency was
also examined. The results revealed that the transfection
efficiency was highest at a DOTAP/DNA weight ratio of 3 (Figs. 3D and 4D), which also indicated that there is
not always a positive correlation between the transfection
efficiency and the mass ratio of DOTAP/DNA. In order to further
increase the transfection efficiency, we also considered the DNA
condensing agent and the protamine added to the formulation. The
weight ratio of the protamine/DNA ranged from 0.2 to 3 (0.2, 0.5,
0.8, 1.0, 1.5, 2 and 3) and these nanoparticles were combined with
CLs to form LPD complexes for transfection. The results indicated
that the addition of a certain amount of protamine to form the LPD
complexes improved the effects of DNA delivery. The highest
transfection efficiency was observed with a weight ratio of
protamine/DNA of 0.5 (Figs. 3E
and 4E). Compared to transfection
only with Lipo 2000, the LPD complexes had better transfection
efficiency.
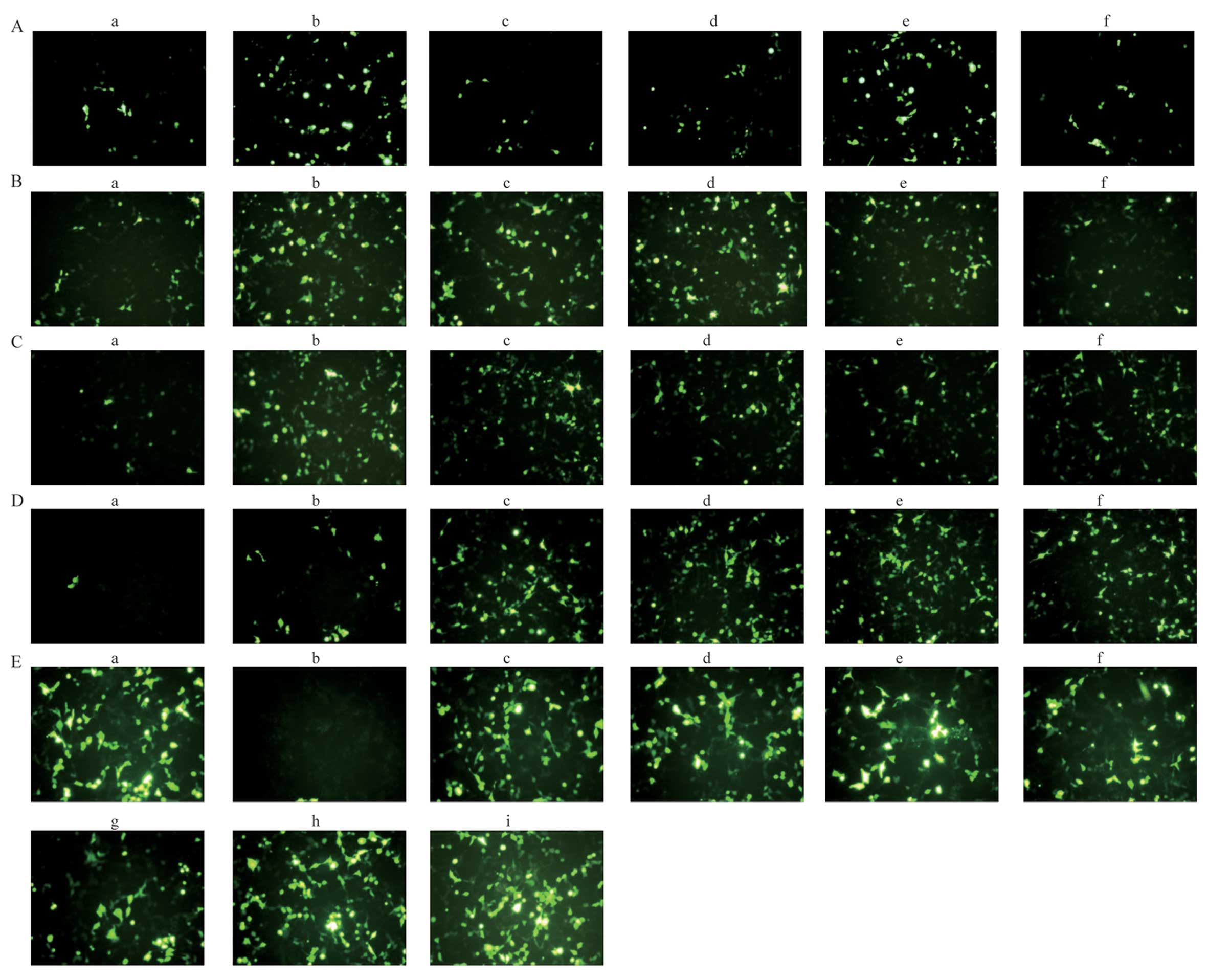 | Figure 3Representative GFP fluorescence
images of transfection experiments. The fluorescence intensity of
GFP was detected using a fluorescence microscope. (A)
DOTAP/cholesterol (chol) liposome (molar ratio of 1:1 and fixed DNA
of 8 µg) in different hydration media [(panels a–f) water,
5% glucose (Glu), 0.9% NaCl, 10 mM Tris-HCl, 10 mM phosphate buffer
and 5% sucrose, respectively]; (B) DOTAP/chol liposome (molar ratio
of 1:1 and 5% Glu as the hydration medium) and different amounts of
DNA [(panels a–f) 2, 4, 6, 8, 10 and 15 µg, respectively];
(C) DOTAP/chol liposome (fixed DNA of 8 µg and 5% Glu as the
hydration medium) at different molar ratios of DOTAP and chol
[(panels a–f) the molar ratio of DOTAP/chol with 1, 2, 3, 4, 5 and
6, respectively]; (D) DOTAP-chol liposome (molar ratio of 3:1, pDNA
fixed of 8 µg) were complexed with DNA at various weight
ratios (panels a–f) the weight ratio of DOTAP/DNA with 1, 2, 3, 4,
5 and 6, respectively; (E) protamine combined with liposome at
different weight ratios of protamine and DNA (panel a)
Lipofectamine™ 2000 (Lipo 2000), (panel b) protamines and DNA
complex, (panels c–i) the LPD with different weight ratio of
protamines/DNA (0.2, 0.5, 0.8, 1.0, 1.5, 2.0 and 3.0, respectively)
and DNA. |
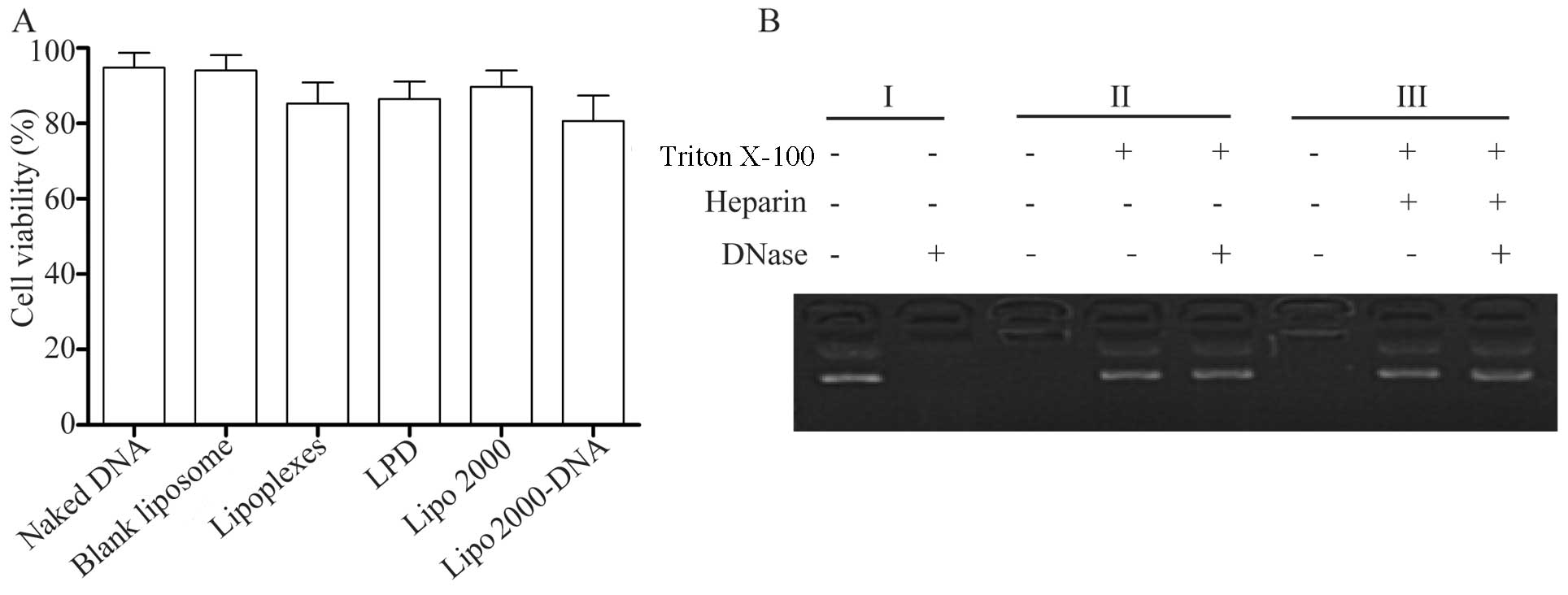
Toxic effects of the complex on 293T cell
proliferation
To determine the toxicity of the liposomes, an MTT
assay was performed to evaluate the effects of different gene
carriers on 293T cell proliferation. Compared to transfection with
Lipo 2000 and the CLs, the lipoplexes and LPD complexes did not
exhibit any significant toxic effects (Fig. 5A). The results suggested that
these preparations had a good compatibility with the cells.
Compared with the cells transfected with the CLs, the viability of
the cells transfected with the complexes containing DNA was
relatively low; this may perhaps be due to the presence of miR-145
expression, which inhibited cell proliferation.
DNase assay
DNA is easily degraded in vivo and is a
factor which restricts the effectiveness of gene therapy. Thus, we
also used DNase to confirm that the liposomes and the LPD complexes
encapsulated the DNA in order to prevent DNA degradation. Naked DNA
was easily degraded by DNase; however, in the lipoplexes or the LPD
complexes, DNA degradation occurred to a lesser extent (Fig. 5B). These results indicated that
the liposomes and protamine had a better encapsulation efficiency
and protected the DNA from degradation by DNase.
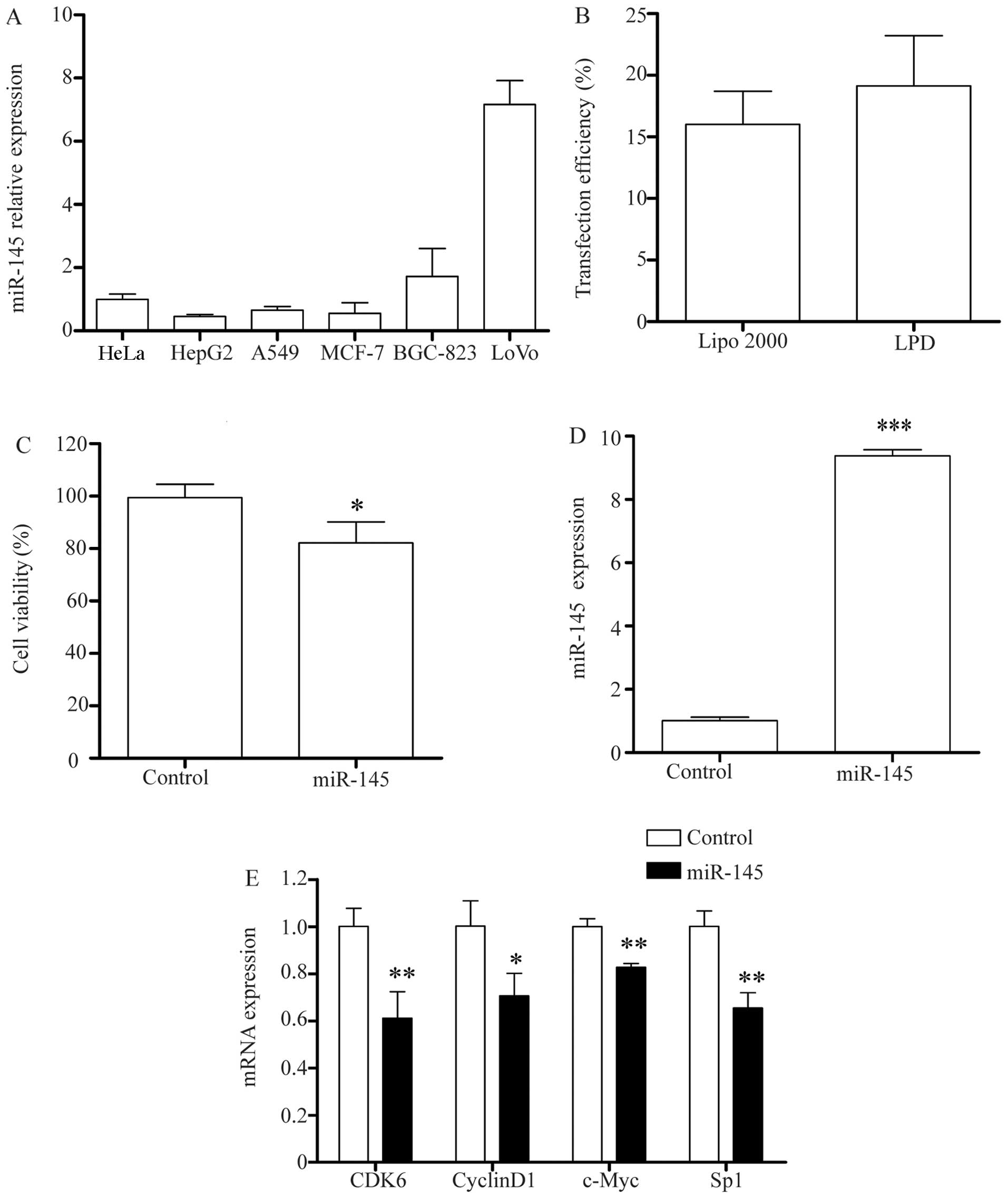
Endogenous miR-145 expression levels
In order to determine the basal expression of
miR-145 in different cancer cell lines, we first detected the
expression of miR-145 in the A549, BGC-823, HepG2, HeLa, LoVo and
MCF-7 cell lines. As shown in Fig.
6A, the majority of the cell lines exhibited a low expression
level of miR-145, of which the HepG2 cells had the lowest. Thus,
the HepG2 cells were selected for use in the subsequent
experiments.
miR-145 inhibits HepG2 cell proliferation
by regulating cell cycle-related genes
In order to investigate the potential mechanisms
responsible for the inhibitory effects of miR-145 in HepG2 cells,
we used LPD complexes to deliver miR-145 to the HepG2 cells, with a
higher transfection efficiency than Lipo 2000 (Fig. 6B). The results of MTT assay
indicated that the delivery of miR-145 significantly inhibited the
proliferation of the HepG2 cells (Fig. 6C, P<0.05). Following
transfection, miR-145 expression was significantly increased in the
HepG2 cells by approximately 9-fold compared to the endogenous
level of miR-145. RT-qPCR confirmed the overexpression of miR-145
in the HepG2 cells (Fig. 6D).
Previous studies have demonstrated that miR-145 regulates certain
genes and consequently affects cancer cell proliferation, migration
and invasion. We used the TargetScan and miRanda websites to
predict the target genes of miR-145. We found that miR-145 combines
with the mRNA 3′UTR of the target gene, and found several genes
associated with the cell cycle, including CDK6, cyclinD1, the
transcription factor, Sp1, and the proto-oncogene, c-Myc. The
results of RT-qPCR indicated that the overexpression of miR-145 in
the HepG2 cells had a significant effect on the expression levels
of these genes (Fig. 6E). RT-qPCR
revealed an approximate 39% decrease in the mRNA expression of CDK6
with the overexpression of miR-145 (P<0.01), and an approximate
30% decrease in the mRNA expression of cyclinD1. c-Myc mRNA
expression was downregulated by approximately 18% with the
overexpression of miR-145 (P<0.05). Sp1 mRNA expression was
downregulated by approximately 35% (P<0.01). These data indicate
that the inhibitory effects induced by miR-145 may occur through
the regulation of the expression of cell cycle-associated
genes.
Discussion
Non-viral gene vectors have been widely used in
research and gene therapy as they are considered safe to use, are
easily prepared and provide a good targeting efficiency. However,
their use as a liposome-mediated gene delivery system for the
delivery of DNA non-viral gene vectors is often associated with two
main concerns: difficulty in escaping from endosomes and difficulty
in entering the nucleus (18).
DOTAP/chol liposomes are an efficient gene delivery vector and
effectively overcome the above two concerns; however, there is
still potential to improve aspects of the design of DOTAP/chol CLs,
including optimization of the hydration medium, the mass of DNA and
the molar ratio of DOTAP/chol (19). The screening and comparison of
different hydration media, with the aim of improving the
transfection efficiency, has been rarely reported in the
literature, to the best of our knowledge. Water, 5% dextrose, 0.9%
NaCl and 10 mM Tris-HCl have been used as hydration media in
liposome preparation. In the present study, we compared different
hydration media in order to determine whether the selection of
hydration media affects the transfection efficiency. The results of
the gel retardation assay demonstrated that there were no
significant changes to the migration path of the DNA in the
presence of the different hydration media, which may suggest that
the hydration medium has minimal effect on the DNA binding affinity
of CLs. However, the transfection experiments revealed a difference
in transfection efficiency among the liposomes prepared with
different hydration media; there was a 4-fold increase in the
transfection efficiency of CLs prepared with 5% glucose compared
with those prepared with either water or 0.9% NaCl (Figs. 3A and 4A).
These findings confirm that the selection of
hydration medium has an effect on CL-mediated gene delivery, and
suggest that it should be considered as a key step in liposome
preparation. The mechanism responsible for 5% glucose enhancing
gene delivery remains unclear; however, we presumed that it not
only acted as an isosmotic solution, but may also meet the glucose
addiction of cell lines cultured in vitro, which may
effectively enhance cellular uptake or improve endosomal
escape.
In previous research,
dioleoylphosphatidylethanolamine (DOPE) or chol were often used as
helper lipids. DOPE which may destabilize cell membranes, has been
shown to be beneficial for lipid membrane fusion with the cell and
endosomal membrane, which enables escape from the endosomes
(20). However, this material has
great defects in the presence of serum, as liposomes obtained from
DOPE would present obstacles as regards to fusion with the cell
membrane, thus reducing the transfection efficiency. It has been
found that chol does not have a prominent effect on promoting cell
fusion; however, chol does not have ionizable groups and does not
greatly affect surface charge. In the presence of serum, the
carrier holds more positive charges and also maintains its
integrity. Moreover, chol promotes phase alteration to form
separated domains (21,22). Thus, for in vivo
transfection or transfection under approximate physiological
conditions, chol is a much better helper lipid. The molar ratio of
DOTAP/chol in CLs plays a crucial role in transfection. In this
study, the results of the gel retardation assay demonstrated that
when the weight ratio of DOTAP/DNA exceeded 2, the DNA was totally
encapsulated within the CLs which were composed of different molar
ratios of DOTAP/chol (Fig. 2G–L).
The results also illustrated that chol had a minimal effect on the
surface charge of carriers. When the molar ratio of DOTAP/DNA was
3:1, the expression of GFP and the transfection efficiency
(measured by flow cytometry), exhibited optimal results (Figs. 3C and 4C). By increasing the mass ratio of
DOTAP/DNA, the transfection efficiency increased and then decreased
(Figs. 3D and 4D). Thus, we hypothesized that at a low
weight ratio of DOTAP/DNA and the surface charge of the lipoplexes
were less (Fig. 1C) which may
induce the complexes to enter the cells or the that the escape from
endosomes was less. While the high weight ratio of DOTAP/DNA was
probably the reason for the DNA to bind firmly with the CLs and
enter the cells, the lipoplexes only released small amounts of DNA,
resulting in a reduced transfection efficiency. Furthermore, it is
possible that the cytotoxicity of lipoplexes increased with the
increasing weight ratio of DOTAP/DNA, and this may prove harmful to
transfection (23).
Liposome-polycation-DNA complexes are based on the
CL/DNA lipoplexes and have been developed as a novel non-viral
vector in recent years. Compared with CLs, LPD complexes are more
efficient at condensing DNA which significantly improves
transfection efficiency, and therefore they are promising clinical
gene therapy vectors (24).
Protamine is a natural polymer and has received FDA approval for
use in clinical treatments. Protamine is a basic protein which is
positively charged and arginine-rich, and it was extracted from
marine fish sperm. Moreover, protamine has several advantages, such
as low antigenicity and a good safety profile (25,26). As a cationic polymer, protamine
may be used to condense a long chain of DNA to improve resistance
to the radical changes of external forces. Furthermore, it has been
found that protamine has a specific nuclear localization signal,
since it is rich in amino acids, and therefore it is more
beneficial in terms of enabling the complex to enter nuclear
translocation (12). Our study
proved that protamine improved the transfection efficiency of a
CL-based gene delivery system (Figs.
3E and 4E). The mechanism of
action of LPD may that a core complex is formed with protamine and
DNA, and this complex in conjunction with CLs then forms a LPD
nanoparticle with a shell-core structure (9). Moreover, in our study, the size of
the nanoparticles decreased following the addition of protamine to
form the LPD complexes (Fig. 1).
Particle size also plays an important role in gene delivery. The
complexes of different sizes enter cells through different
endocytic pathways, which consequently affects the transfection
efficiency. DNA easily undergoes enzymatic degradation in
vivo, which is a restriction of gene therapy (27); thus ways of preventing DNA
degradation by DNase must be considered when designing systemic
gene therapy. In our study, we found that the shell-core
construction of LPD complexes not only protected the DNA from
enzymatic degradation (Fig. 5B),
but that the lipid membrane restricted DNase from touching the
encapsulated DNA.
In previous studies, it has been suggested that
miR-145 has an anti-tumor ability and targets multiple tumor genes
which are involved in cancer cell growth, metastasis and invasion
(16,28,29). It has been shown that miR-145 is
expressed at low levels in a different types of tumor tissue, such
as colon, breast, liver and lung cancer tissue. We selected several
cancer cell lines and then observed the endogenous expression of
miR-145 (Fig. 6A). It was found
that miR-145 had the lowest expression level in the HepG2 cells
compared with the other cancer cells. miR-145 as a tumor
suppressor, has been shown to inhibit cancer cell growth by
targeting c-Myc and p53 in colon and breast cancers (30), IGF-IR in human bladder cancer
(17) and HIF-2α in neuroblastoma
(15). In our study, our results
indicated that miR-145 reduced the mRNA expression of CDK6,
cyclinD1, c-Myc and Sp1 in the HepG2 cells, thus indicating that
the inhibitory effects of miR-145 on HepG2 cell proliferation may
occur through the regulation of the cell cycle-associated genes,
which results in anticancer effects.
In conclusion, in this study, we systematically
evaluated the various factors which affect the preparation of
DOTAP/chol liposomes in order to improve the transfection
efficiency. Furthermore, we clarified that protamine improved the
transfection efficiency of the CLs. Finally, the present study
demonstrated that miR-145 is a potential novel target for
anticancer therapeutics.
Acknowledgments
The present study was funded by the National Natural
Science Foundation of China (no. 81201350).
References
|
1
|
Yang SY, Zheng Y, Chen JY, Zhang QY, Zhao
D, Han DE and Chen XJ: Comprehensive study of cationic liposomes
composed of DC-Chol and cholesterol with different mole ratios for
gene transfection. Colloids Surf B Biointerfaces. 101:6–13. 2013.
View Article : Google Scholar
|
|
2
|
Masotti A, Mossa G, Cametti C, Ortaggi G,
Bianco A, Grosso ND, Malizia D and Esposito C: Comparison of
different commercially available cationic liposome-DNA lipoplexes:
parameters influencing toxicity and transfection efficiency.
Colloids Surf B Biointerfaces. 68:136–144. 2009. View Article : Google Scholar
|
|
3
|
Templeton NS, Lasic DD, Frederik PM, Strey
HH, Roberts DD and Pavlakis GN: Improved DNA: liposome complexes
for increased systemic delivery and gene expression. Nat
Biotechnol. 15:647–652. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim SY, Lee SJ and Lim SJ: Formulation and
in vitro and in vivo evaluation of a cationic emulsion as a vehicle
for improving adenoviral gene transfer. Int J Pharm. 475:49–59.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Roberts R, Al-Jamal WT, Whelband M, Thomas
P, Jefferson M, van den Bossche J, Powell PP, Kostarelos K and
Wileman T: Autophagy and formation of tubulovesicular
autophagosomes provide a barrier against nonviral gene delivery.
Autophagy. 9:667–682. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fritze A, Hens F, Kimpfler A, Schubert R
and Peschka-Süss R: Remote loading of doxorubicin into liposomes
driven by a transmembrane phosphate gradient. Biochim Biophys Acta.
1758:1633–1640. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
de la Torre LG, Rosada RS, Trombone APF,
Frantz FG, Coelho-Castelo AA, Silva CL and Santana MH: The synergy
between structural stability and DNA-binding controls the antibody
production in EPC/DOTAP/DOPE liposomes and DOTAP/DOPE lipoplexes.
Colloids Surf B Biointerfaces. 73:175–184. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen J, Yu Z, Chen H, Gao J and Liang W:
Transfection efficiency and intracellular fate of polycation
liposomes combined with protamine. Biomaterials. 32:1412–1418.
2011. View Article : Google Scholar
|
|
9
|
Pozzi D, Marchini C, Cardarelli F,
Salomone F, Coppola S, Montani M, Zabaleta ME, Digman MA, Gratton
E, Colapicchioni V and Caracciolo G: Mechanistic evaluation of the
transfection barriers involved in lipid-mediated gene delivery:
interplay between nanostructure and composition. Biochim Biophys
Acta. 1838:957–967. 2014. View Article : Google Scholar :
|
|
10
|
Liu R, Gan L, Yang X and Xu H: Chitosan as
a condensing agent induces high gene transfection efficiency and
low cytotoxicity of liposome. J Biosci Bioeng. 111:98–103. 2011.
View Article : Google Scholar
|
|
11
|
Hartono SB, Gu W, Kleitz F, Liu J, He L,
Middelberg AP, Yu C, Lu GQ and Qiao SZ: Poly-L-lysine
functionalized large pore cubic mesostructured silica nanoparticles
as biocompatible carriers for gene delivery. ACS Nano. 6:2104–2117.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Reynolds F, Weissleder R and Josephson L:
Protamine as an efficient membrane-translocating peptide. Bioconjug
Chem. 16:1240–1245. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kanasty R, Dorkin JR, Vegas A and Anderson
D: Delivery materials for siRNA therapeutics. Nat Mater.
12:967–977. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sachdeva M and Mo YY: miR-145-mediated
suppression of cell growth, invasion and metastasis. Am J Transl
Res. 2:170–180. 2010.PubMed/NCBI
|
|
15
|
Zhang H, Pu J, Qi T, Qi M, Yang C, Li S,
Huang K, Zheng L and Tong Q: MicroRNA-145 inhibits the growth,
invasion, metastasis and angiogenesis of neuroblastoma cells
through targeting hypoxia-inducible factor 2 alpha. Oncogene.
33:387–397. 2014. View Article : Google Scholar
|
|
16
|
Adammek M, Greve B, Kässens N, Schneider
C, Brüggemann K, Schüring AN, Starzinski-Powitz A, Kiesel L and
Götte M: MicroRNA miR-145 inhibits proliferation, invasiveness, and
stem cell phenotype of an in vitro endometriosis model by targeting
multiple cytoskeletal elements and pluripotency factors. Fertil
Steril. 99:1346–1355.e5. 2013. View Article : Google Scholar
|
|
17
|
Zhu Z, Xu T, Wang L, Wang X, Zhong S, Xu C
and Shen Z: MicroRNA-145 directly targets the insulin-like growth
factor receptor I in human bladder cancer cells. FEBS Lett.
588:3180–3185. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yao J, Fan Y, Li Y and Huang L: Strategies
on the nuclear-targeted delivery of genes. J Drug Target.
21:926–939. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Samadikhah HR, Majidi A, Nikkhah M and
Hosseinkhani S: Preparation, characterization, and efficient
transfection of cationic liposomes and nanomagnetic cationic
liposomes. Int J Nanomedicine. 6:2275–2283. 2011.PubMed/NCBI
|
|
20
|
Morille M, Passirani C, Vonarbourg A,
Clavreul A and Benoit JP: Progress in developing cationic vectors
for non-viral systemic gene therapy against cancer. Biomaterials.
29:3477–3496. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hirsch-Lerner D, Zhang M, Eliyahu H,
Ferrari ME, Wheeler CJ and Barenholz Y: Effect of 'helper lipid' on
lipoplex electrostatics. Biochim Biophys Acta. 1714:71–84. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zuhorn IS, Visser WH, Bakowsky U, Engberts
JBFN and Hoekstra D: Interference of serum with lipoplex-cell
interaction: modulation of intracellular processing. Biochim
Biophys Acta. 1560:25–36. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang Y, Li H, Sun J, Gao J, Liu W, Li B,
Guo Y and Chen J: DC-Chol/DOPE cationic liposomes: a comparative
study of the influence factors on plasmid pDNA and siRNA gene
delivery. Int J Pharm. 390:198–207. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang XP and Cui WH: Comparison of
liposome-polycation-DNA(LPD) and monophosphoryl lipid A(MPL)
adjuvant formulations in BALB/c mice models. Immunol Invest.
41:356–366. 2012. View Article : Google Scholar
|
|
25
|
Ma K, Wang DD, Lin Y, Wang J, Petrenko V
and Mao C: Synergetic targeted delivery of sleeping-beauty
transposon system to mesenchymal stem cells using LPD nanoparticles
modified with a phage-displayed targeting peptide. Adv Funct Mater.
23:1172–1181. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jalali SA, Sankian M, Tavakkol-Afshari J
and Jaafari MR: Induction of tumor-specific immunity by
multi-epitope rat HER2/neu-derived peptides encapsulated in LPD
nanoparticles. Nanomedicine. 8:692–701. 2012.
|
|
27
|
Obata Y, Saito S, Takeda N and Takeoka S:
Plasmid DNA-encapsulating liposomes: effect of a spacer between the
cationic head group and hydrophobic moieties of the lipids on gene
expression efficiency. Biochim Biophys Acta. 1788:1148–1158. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shi B, Sepp-Lorenzino L, Prisco M, Linsley
P, deAngelis T and Baserga R: Micro RNA 145 targets the insulin
receptor substrate-1 and inhibits the growth of colon cancer cells.
J Biol Chem. 282:32582–32590. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Suh SO, Chen Y, Zaman MS, Hirata H,
Yamamura S, Shahryari V, Liu J, Tabatabai ZL, Kakar S, Deng G, et
al: MicroRNA-145 is regulated by DNA methylation and p53 gene
mutation in prostate cancer. Carcinogenesis. 32:772–778. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sachdeva M, Zhu S, Wu F, Wu H, Walia V,
Kumar S, Elble R, Watabe K and Mo YY: p53 represses c-Myc through
induction of the tumor suppressor miR-145. Proc Natl Acad Sci USA.
106:3207–3212. 2009. View Article : Google Scholar : PubMed/NCBI
|