Introduction
Macrophages are a heterogeneous and plastic cell
population, which play crucial roles in the innate and adaptive
immune response. They can undergo a phenotypically dynamic switch
in response to different microenvironments (1). In general, two major macrophage
subsets, including classically activated (M1) and alternatively
activated (M2) macrophages, have long been recognized (2–4).
M1 macrophages are classically induced by T helper type 1 (Th1)
cytokines, such as interferon (IFN)-γ, and bacterial
lipopolysaccharide (LPS). They express high levels of CD86, as well
as a profile of pro-inflammatory cytokines, such as interleukin
(IL)-12, IL-23, IL-6 and tumor necrosis factor (TNF)-α, but low
levels of CD206, and the anti-inflammatory cytokines, IL-10 and
transforming growth factor (TGF)-β. By contrast, M2 macrophages are
induced by T helper type 2 (Th2) cytokines, such as IL-4. They are
characterized by a high expression of CD206, IL-10 and TGF-β, and a
low expression of CD86 and a set of pro-inflammatory cytokines
(5–10).
Macrophages play different roles in diseases,
depending on their distinct phenotypes (2,11,12). In tumor immunity, there is
accumulating evidence to indicate that M1 macrophages are
tumoricidal. It has been demonstrated that M1 macrophages are the
dominant subset in colon carcinomas, which was related to
diminished metastasis and increased survival rate (14). However, M2 macrophages facilitate
tumor progression by promoting migration, angiogenesis and invasion
(13). It has been reported that
a high M1/M2 ratio is associated with an improved survival in solid
tumors, and the presence of M2 macrophages is considered
responsible for a poor prognosis and enhanced disease progression
in breast cancer (14,15). Collectively, macrophage
polarization may have promising applications in the field of tumor
immune therapy.
It has been well established that IFN-γ and LPS are
two key stimuli which induce the M1 polarization of macrophages
(2,16). Although IFN-γ and LPS play
significant roles in the activation of M1 macrophages, they mediate
distinct pathways. IFN-γ exerts its biological effects primarily by
activating the Janus kinase 1 (JAK1)/signal transducer and
activator of transcription (STAT)1 signaling pathway (17). Interferon regulatory factor
(IRF)1, a transcriptional regulator, is only weakly expressed in
resting macrophages, but can be strongly upregulated by IFN-γ
stimulation (18,19). In macrophages, IRF1 takes part in
the regulation of IL-12 and inducible nitric oxide synthase (iNOS)
(20,21). On the other hand, LPS is
recognized by Toll-like receptor 4 (TRL4) and activates
MyD88-dependent or TRIF-dependent pathways (22). IFN-β is one of the significant
molecules involved in the TRIF-dependent pathway (23). It has also been demonstrated that
IFN-β plays a role in the regulation of IL-12p70 production in
granulocyte-macrophage colony-stimulating factor (GM-CSF)-induced
bone marrow-derived macrophages (GM-BMM) (24); however, the mechanisms involved
remain unclear. It should be noted that IRF1 was originally
discovered as a transcriptional activator of IFN-β in virus
infected fibroblasts (25).
Moreover, IRF1 can bind to the IFN-stimulated responsive element
(ISRE)/IRF-E site induced by IFN-β (26). Furthermore, it has been clearly
demonstrated that IRF1 nuclear expression in human monocytes is
principally induced by a combination of IFN-γ and LPS, rather than
by either stimuli alone, which differs from that in mouse
peritoneal macrophages or RAW 264.7 cells (27). Therefore, we could envisage the
possible synergistic action of IRF1 and IFN-β, which are involved
in the two independent, but complementary pathways induced by IFN-γ
and LPS in the M1 polarization of macrophages.
In addition, several recent studies have identified
a dual function of IRF5 in activating M1 genes (IL-12p35, IL-12p40,
IL-23p19, IL-6 and TNF-α), while suppressing M2 genes (IL-10 and
TGF-β) (28–30). Human IRF5 presents multiple
alternatively spliced isoforms (V1–V9), which are cell
type-specific. It has been shown that V1 and V3 possess different
transcription start sites and are modulated by two distinct
promoters. The V1 promoter (P-V1) contains the IRF-E consensus
binding site, and the V3 promoter (P-V3) contains an ISRE-binding
site (31). Interestingly, both
IRF-E and ISRE can be recognized by IRF1 (32) or by the transcripts complex
induced by IFN-β (33,34). However, the involvement of the
interaction of IRF1, IFN-β and IRF5 in the M1 polarization of
macrophages not yet fully understood.
Based on above-mentioned data, we could reasonably
hypothesize that there may exist a certain association between
IRF1, IFN-β and IRF5, and the M1 polarization of macrophages, or
that the three may interact with each other to promote the M1
polarization of macrophages. Therefore, in this study, we examined
the interactions of IRF1 and IFN-β, particularly the regulation of
IRF5, and their role in the M1 polarization of macrophages and M1
macrophage-mediated antitumor effects on hepatocellular carcinoma
(HCC) cell lines.
Materials and methods
Cell culture
The monocyte cell line, U937, was obtained from the
American Type Culture Collection (ATCC; Manassas, VA, USA). The
cells were maintained in RPMI-1640 medium (HyClone, Logan, UT,
USA), supplemented with 10% fetal bovine serum (FBS; Biological
Industries, Kibbutz Beit-Haemek, Israel) and 1% penicillin and
streptomycin. According to previous studies (10,35,36), U937 monocytes were differentiated
into unpolarized macrophages (M0) by 5 ng/ml phorbol 12-myristate
13-acetate (PMA) (S1819; Beyotime Biotechnology, Jiangsu, China)
for 48 h. To establish the M1 polarization of macrophages, the M0
macrophages were stimulated with 20 ng/ml IFN-γ (no. 300-02;
Peprotech, Rocky Hill, NJ, USA) and 100 ng/ml LPS (no. LZ880;
Sigma-Aldrich, St. Louis, MO, USA) for an additional 24 h.
SMMC-7721 HCC cells were obtained from the Shanghai
Institutes for Biological Sciences (Shanghai, China). HepG2 HCC
cells were obtained from ATCC. The cells were both maintained in
DMEM (HyClone), supplemented with 10% FBS (Biological Industries)
and 1% penicillin and streptomycin.
Analysis of macrophage surface marker
expression
Phenotypic analysis of the macrophages was performed
using flow cytometry. In brief, the cells were collected and washed
3 times with ice-cold PBS. Firstly, the cells were incubated with
ice-cold PBS containing 5% mice serum at 4°C to avoid non-specific
binding. The cells were then stained for anti-human-CD86-PE (no.
305405) or anti-human-CD206-PE (no. 321105) antibodies (both from
BioLegend, San Diego, CA, USA) for 30 min. After immunostaining,
the cells were washed twice with PBS and analyzed using the BD
Influx™ cell sorter flow cytometer (BD Biosciences, San Jose, CA,
USA). Isotype control cells used for non-specific background
staining were stained with PE-labeled mouse IgG1K iso control PE
(E11418-1634; eBioscience, Inc., San Diego, CA, USA).
Enzyme-linked immunosorbent assay
(ELISA)
After the U937-M0 cells were stimulated with IFN-γ
(20 ng/ml) and LPS (100 ng/ml) for 24 h, the supernatant was
collected and centrifuged at 1,800 × g at 4°C for 10 min. The
IL-12p70 and IL-10 secretion levels were measured using ELISA MAX™
Deluxe Sets (nos. 431706 and 430607; BioLegend). The IFN-β levels
were measured using an ELISA kit for human IFN-β (SEA222Hu;
Cloud-Clone Corp., Houston, TX, USA) in accordance with the
manufacturer's instructions. Each experiment was repeated 3 times.
The final outcomes were pooled as the average concentration of
cytokines.
Western blot analysis
According to the above-mentioned cell culture, the
M1 macrophages were collected and total protein was extracted using
radio immunoprecipitation assay (RIPA) lysis buffer (Roche
Diagnostics, Basel, Switzerland) and phenyl-methanesulfonyl
fluoride (PMSF; Beyotime Biotechnology) at 100:1. The protein
concentration was quantified by BCA assay. The supernatant
containing 40 µg total protein was extracted for
electrophoresis on a 12% sodium dodecyl sulfate gel (SDS; Beyotime)
and then transferred onto 0.45 nm polyvinylidene fluoride membranes
(PVDF; Millipore, Billerica, MA, USA). After being blocked with 5%
non-fat powdered milk in Tris-buffered saline containing 0.1%
Tween-20 (TBST) for 1.5 h, the membranes were incubated at 4°C
overnight with moloclonal rabbit anti-IRF1 (D5E4) or anti-IRF5
(E1N9G) antibodies (1:1,000; Cell Signaling Technology, Danvers,
MA, USA) or rabbit anti-β-actin antibody (1:1,000, YT0099;
ImmunoWay Biotechnology, Co., Newark, DE, USA) as the primary
antibodies. This was followed by incubation with horseradish
peroxidase-conjugated AffiniPure goat anti-rabbit secondary
antibody (1:2,000, ZB-2301; ZSGB-BIO, Beijing, China) at room
temperature for 2 h. The immunoreactive complexes were visualized
by enhanced chemiluminescence (ECL) (Millipore). The intensities of
the protein bands were quantified using Bio-Rad Quantity One
software (Bio-Rad Laboratories, Inc., Hercules, CA, USA). β-actin
antibody was used to normalize the results.
Total RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
The U937 cells were stimulated with PMA or IFN-γ +
LPS for 2, 4, 6, 8, 12 and 24 h. Total RNA was isolated from the
macrophages using TRIzol reagent (Takara Bio, Inc., Otsu, Japan)
and a total of 1 µg of RNA was subjected to reverse
transcription reactions using the PrimeScript™ RT reagent kit (no.
RR047A; Takara Bio, Inc.) in accordance with the manufacturer's
instructions. qPCR was conducted with SYBR Premix Ex Taq™ II (no.
RR820A; Takara Bio, Inc.) on the Bio-Rad CFX-Connext Real-Time PCR
Detection system (Bio-Rad, Philadelphia, PA, USA) with the
following steps: 95°C for 10 sec, 59°C for 30 sec, and 72°C for 30
sec for 39 cycles. The primers specific for our target genes are
listed in Table I. β-actin was
used as an internal control for normalization. The data were
analyzed using the 2−ΔΔCt method. Each experiment was
repeated 3 times. All RT-qPCR reactions were performed in
triplicate.
 | Table ISequences of oligonucleotide primers
used for RT-qPCR. |
Table I
Sequences of oligonucleotide primers
used for RT-qPCR.
| Gene | Sequence | Orientation | Amplification size
(bp) |
|---|
| β-actin
(NM_001101.3) |
CTGGGACGACATGGAGAAAA | Sense | 564 |
|
AAGGAAGGCTGGAAGAGTGC | Antisense | |
| p40
(NM_002187.2) |
CTCTGGCAAAACCCTGACC | Sense | 85 |
|
GCTTAGAACCTCGCCTCCTT | Antisense | |
| p35
(NM_000882.3) |
ACCAGGTGGAGTTCAAGACC | Sense | 134 |
|
TGGCACAGTCTCACTGTTGA | Antisense | |
| IL-10
(NM_000572.2) |
GATGCCTTCAGCAGAGTGAA | Sense | 93 |
|
ACCCTTAAAGTCCTCCAGCA | Antisense | |
| IFNB1
(NM_002176.2) |
AGGACAGGATGAACTTTGAC | Sense | 183 |
|
TGATAGACATTAGCCAGGAGGTT | Antisense | |
| IRF5
(NM_001098627.2) |
AGGGCTTCAATGGGTCAAC | Sense | 141 |
|
ACGCCTTCGGTGTATTTCC | Antisense | |
| IRF1
(NM_002198.2) |
GCTGGGACATCAACAAGGAT | Sense | 164 |
|
CCTGCTCTGGTCTTTCACCT | Antisense | |
| IL-6
(NM_000600.3) |
ATGTGTGAAAGCAGCAAAGAG | Sense | 111 |
|
CACCAGGCAAGTCTCCTCA | Antisense | |
| IL-23p19
(NM_016584) |
AATCCTTCGCAGCCTCCA | Sense | 105 |
|
TGAGTGCCATCCTTGAGC | Antisense | |
| TNF-α
(NM_000594) |
CGAGTGACAAGCCTGTAGCC | Sense | 172 |
|
TTGAAGAGGACCTGGGAGTAG | Antisense | |
Neutralization of IFN-β
To determine the effects of IFN-β on the
polarization of macrophages, anti-IFN-β antibody (no Ab6979; Abcam
Inc., Cambridge, MA, USA) was utilized to neutralize IFN-β secreted
in the supernatant according to the manufacturer's instructions.
Briefly, the anti-IFN-β antibody was added to the medium after the
U937 cells were treated with PMA. Three hours later, the cells were
stimulated with IFN-γ and LPS for different periods of time for the
next experiment.
Small interfering RNA (siRNA)-mediated
gene knockdown
The unpolarized macrophages (M0) were treated with
siRNA specific to IRF1 or IFNB1 (RioboBio Co., Guangzhou, China).
Non-targeting siRNA served as the control (siC). Three siRNA
sequences were designed for the siRNAs specific to IRF1 or IFNB1.
The one that had the highest silencing efficiency was used in the
following experiments. siRNA transfection was performed using the
RNAiMAX reagent (no. 13778100; Invitrogen Trading Co., Ltd,
Shanghai, China) according to the manufacturer's instructions.
Briefly, RNAiMAX reagent and siRNA were diluted with Optimedium,
respectively, and then mixed gently in an equal volume at room
temperature for 20 min. Subsequently, 500 µl mixture and 1.5
ml complete RPMI-1640 medium without penicillin and streptomycin
were added to each well of a 6-well plate. The cells were counted
under a microscope before they were added to the 6-well plate and
approximately 1×106 cells were added to each well. After
6 h, the medium was changed and the cells were stimulated with
IFN-γ (20 ng/ml) and LPS (100 ng/ml) as mentioned above for the
following experiment. The silencing efficiency was evaluated by
RT-qPCR and western blot analysis or ELISA.
Preparation of conditioned medium
(CM)
To evaluate the antitumor effects of M1 macrophages
with different treatments on HCC, we collected the CM as follows:
the U937 cells were treated with PMA as mentioned above. Following
differentiation, the macrophages were washed slightly with PBS and
the following additives were added to the medium: PBS served as the
control; 20 ng/ml IFN-γ and 100 ng/ml LPS were used to generate M1
macrophages; siRNA against IRF1 or IFNB1 and negative control siRNA
were used prior to IFN-γ/LPS stimulation; 2.7 ng/ml neutralized
monoclonal anti-IFN-β monoclonal antibody was used prior to
IFN-γ/LPS stimulation. The cells were transfected with the siRNA
for 6 h or subjected to anti-IFN-β antibody neutralization for 3 h.
The medium was removed and the cells were washed with PBS
carefully. The cells were then stimulated with IFN-γ/LPS for 24 h.
The supernatant was collected from 6 groups of cells, labeled as
follows: i) CM-control; ii) CM-M1; iii) CM-siRNA control (siC); iv)
CM-siRNA against IRF1 (siIRF1); v) CM-siRNA against IFNB1
(siIFNB1); and vi) CM-anti-IFN-β antibody (Ab). For culture with
HCC cells, all the CM were mixed with an equal volume of complete
DMEM (80% DMEM + 20% FBS + 1% penicillin and streptomycin).
Cell proliferation assay
To examine the effects of CM on the the
proliferation of HCC cells, the viable cells were monitored using
the Cell Counting Kit-8 (CCK-8) (Dojindo, Tokyo, Japan). Briefly,
the HCC cells were seeded in 96-well plates at a density of
3×103 cells/well and were cultured in the different CMs
for 24, 48, 72, 96 and 120 h. The viable cells were examined by
CCK-8 assay according to the manufacturer's instructions. Five
reduplicative wells were used for each group. Each experiment was
repeated 3 times.
Analysis of apoptosis
To evaluate the effects of CM on the apoptosis of
HCC cells, flow cytometry was applied. Briefly, the SMMC-7721 and
HepG2 cells (5×105) were cultured in 6-well plates and
treated with different CM for 72 h. The cells were washed twice
with cold PBS after being harvested, and then re-suspended with 400
µl cold PBS. Annexin V-FITC and PI (BD Biosciences, San
Jose, CA, USA) were added to each well. The cells were incubated in
the dark at room temperature for 15 min and were examined using an
influx cell sorter flow cytometer (BD Biosciences) immediately.
Each experiment was repeated 3 times.
Transwell invasion assay
To evaluate the effects of CM on the invasion of HCC
cells, the Transwell invasion assay was performed using 24-well 8
µm pore size Transwell plates (Millipore) coated with
Matrigel (1:3 dilution) (BD Biosciences). Briefly, a total of
1×105 cells suspended in 200 µl serum-free medium
containing 0.1% BSA was added to the upper chamber. The lower
chamber was filled with 600 µl CM. Following incubation for
24 h, the non-invaded cells in the upper side of the chamber were
carefully removed by scraping. The cells that had invaded into the
lower side of the chamber were fixed with 0.4% paraformaldehyde for
20 min and stained with 0.1% crystal violet for 10 min. Images were
then captured using a Nikon Eclipse 80i microscope (Nikon, Tokyo,
Japan) at ×100 magnification. Five fields for each group were
randomly selected to be subjected to statistical analysis.
Statistical analysis
All experiments were repeated at least 3 times
independently. The data are presented as the means ± standard error
of the mean (SEM) in this study. Statistical analyses were
performed using SPSS software version 17.0 software (SPSS, Inc.,
Chicago, IL, USA). An independent sample t-test or one-way ANOVA
were used to determine the differences between 2 groups. A value of
P<0.05 was considered to indicate a statistically significant
difference.
Results
U937 cells stimulated with IFN-γ and LPS
readily acquire an M1 status
U937 is a monocytic tumor cell line, which
extensively serves as the precursor of macrophages with some
specific treatments (10,35). The genetic profile typical of the
M1 phenotype, which includes the IL-12p35, IL-12p40, IL-23p19, IL-6
and TNF-α genes, and the M2 cytokine, IL-10, were detected using
RT-qPCR. The protein levels of IL-12p70 and IL-10 were detected by
ELISA. As expected, the U937 cells stimulated with IFN-γ and LPS
exhibited a higher mRNA expression of IL-12p35, IL-12p40, IL-23p19,
IL-6 and TNF-α than the unstimulated (PMA alone-treated) cells
(P<0.01; Fig. 1A). The IFN-γ-
and LPS-stimulated U937 cells exhibited a higher secretion of
IL-12p70 than the PMA-treated cells [not detected (n.d.)] (Fig. 1B). Of note, the M2 associated
cytokine, IL-10, was also expressed at higher levels in the IFN-γ-
and LPS-stimulated cells than in the PMA-treated ones (Fig. 1B).
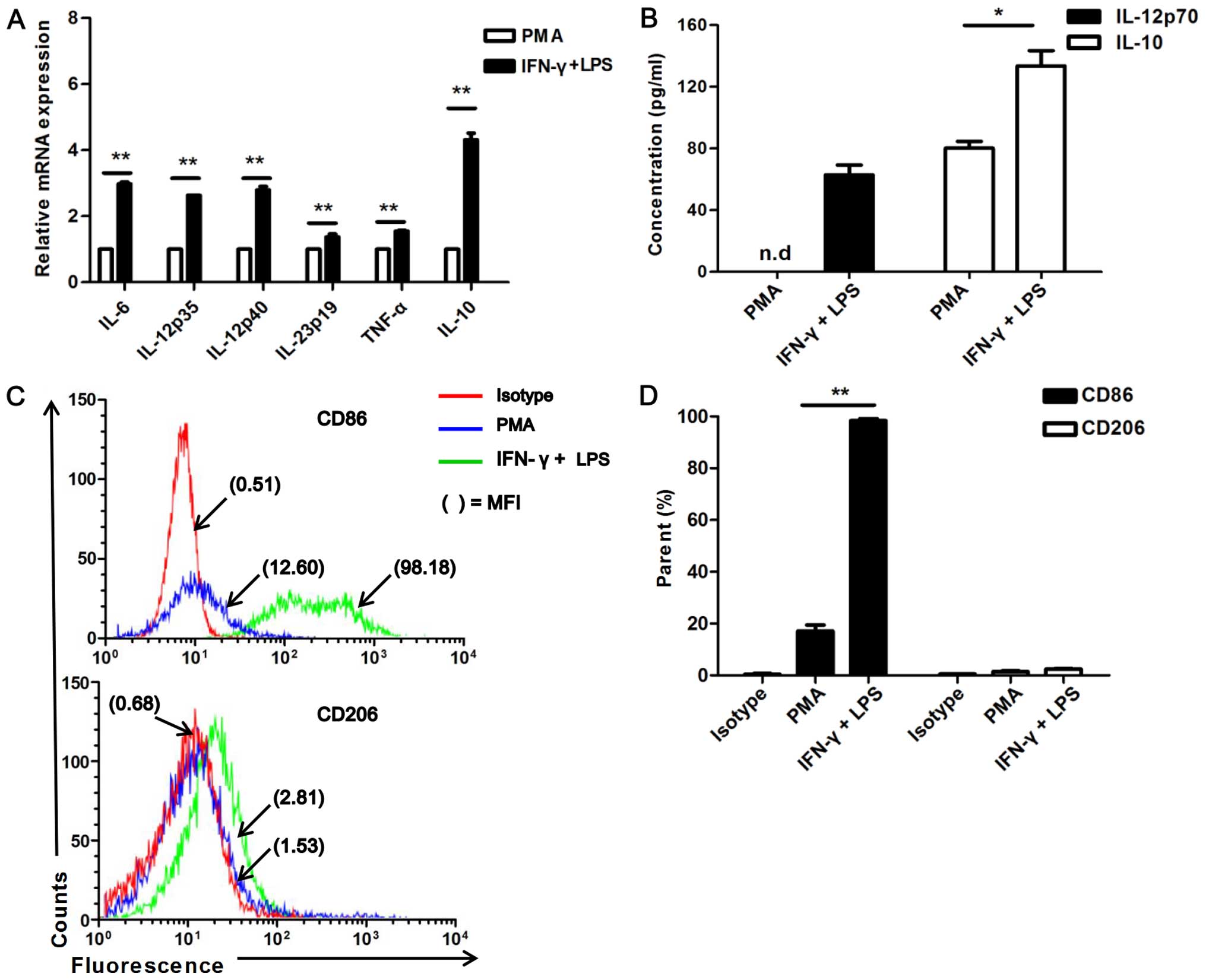 | Figure 1Expression of cytokines and markers
of U937-macrophages stimulated with interferon-γ (IFN-γ) and
lipopolysaccharide (LPS). (A) RT-qPCR analysis of M1-associated
genes, including IL-12p40, IL-12p35, IL-23p19, IL-6 and TNF-α, and
the M2-associated gene, IL-10, in U937 unstimulated (only treated
with 5 ng/ml PMA; labeled as PMA) or stimulated with IFN-γ (20
ng/ml) and LPS (100 ng/ml) (labeled as IFN-γ + LPS) for 8 h,
**P<0.01. (B) ELISA of the IL-12p70 and IL-10
secretion levels in PMA or IFN-γ + LPS group,
*P<0.05. (C) The raw flow cytometry fluorescence data
are representative of 3 independent experiments for CD86 and CD206
expression; staining profiles of PMA or IFN-γ + LPS-stimulated U937
cells. The percentage of positive cells and mean fluorescence
intensities (MFIs) are denoted. (D) Histograms shows surface
staining for CD86 and CD206 expression on PMA or IFN-γ +
LPS-stimulated U937 cells, **P<0.001. Data were
calculated from 3 independent experiments. |
Furthermore, the surface markers, CD86 (M1-specific)
and CD206 (M2-specific), were analyzed by flow cytometry. A higher
expression of CD86 was detected in response to IFN-γ and LPS
stimulation compared to the unstimulated (PMA alone-treated) cells
(P<0.001; Fig. 1C and D).
However, no significant difference in CD206 expression was observed
between the IFN-γ/LPS stimulated and unstimulated (PMA
alone-treated) U937 cells, and it was poorly expressed in all cell
groups (Fig. 1C and D). These
data collectively demonstrate that U937 cells can be polarized to
an M1 status by stimulation with IFN-γ/LPS.
Upregulation of IRF1, IFN-β and IRF5 in
U937-M1 macrophages
Studies have shown that IRF1 and IFN-β are crucial
molecules involved in the IFN-γ- and LPS-initiated activation of
signaling pathways (the IFN-γ/STAT1/IRF1 and LPS/TLR4/TRAM/TRIF
pathways) (17,18,23). Recent studies have also revealed
that IRF5 plays a crucial role in the regulation of M1 macrophages
(28,29). Therefore, we wished to determine
whether these 3 molecules are upregulated in the U937-M1 model. As
shown in Fig. 2A, it was observed
that the mRNA expression of IRF1, IFNB1 and IRF5 was significantly
upregulated by stimulation with IFN-γ and LPS (P<0.05 and
P<0.01). IRF1 and IFNB1 exhibited a similar tendency in
expression; both reached peak levels at 4 h of stimulation with
IFN-γ and LPS. However, IRF5 expression reached peak levels at 6 h.
In addition, the protein levels of IFN-β, IRF1 and IRF5 were
examined by ELISA or western blot analysis. Consistent with
above-mentioned increase in the mRNA levels, increased protein
expression levels of IRF1, IFN-β and IRF5 were detected in the
IFN-γ/LPS-stimulated U937 cells compared with the unstimulated (PMA
alone-treated) cells (Fig. 2B and
C). Collectively, these results indicate that IRF1, IFN-β and
IRF5 are upregulated in U937-M1 macrophages. These results prompted
us to further investigate the roles they play in the M1
polarization of macrophages and in M1-mediated antitumor
effects.
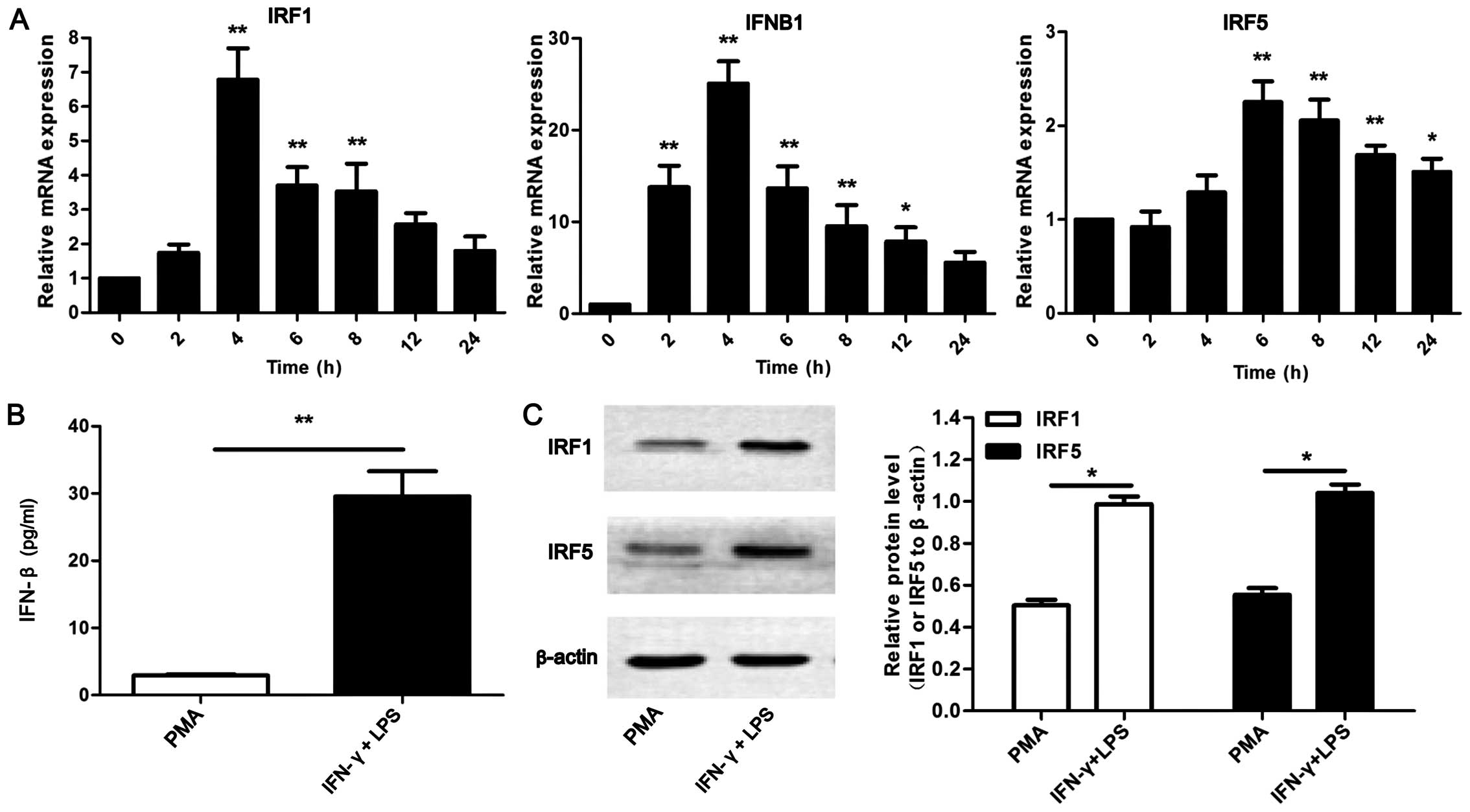 | Figure 2High expression of interferon
regulatory factor (IRF)1, interferon-β (IFN-β) and IRF5 in M1
macrophages stimulated with interferon-γ (IFN-γ) and
lipopolysaccharide (LPS). (A) RT-qPCR analysis of IRF1, IFN-β and
IRF5 mRNA in PMA or IFN-γ + LPS stimulated U937 cells for 2, 4, 6,
8, 12 and 24 h. Results are presented relative to those of
unstimulated macrophages (0 h), *P<0.05,
**P<0.01. (B) ELISA of IFN-β secretion in supernatant
in PMA or IFN-γ + LPS treated U937 cells for 24 h,
**P<0.01. (C) Western blot analysis of IRF1 and IRF5
in cell lysate from PMA or IFN-γ + LPS treated U937 cells for 24 h,
**P<0.01. Actin was used as an internal control for
both RT-qPCR and western blot analysis. Western blot analysis data
were calculated from 3 individual experiments. |
IRF1 affects U937-M1 macrophage
polarization status
Given that IRF1 can be upregulated several fold in
IFN-γ-stimulated macrophages or in dendritic cells (DCs) as opposed
to resting DCs or macrophages (20) and given its role in IL-12
regulation (20), and taking our
above-mentioned findings into consideration, the role of IRF1 in M1
polarization was investigated following transfection of the cells
with siRNA targeteing IRF1 (siIRF1). The silencing efficiency of 3
siIRF1s was determined at the mRNA and protein level by RT-qPCR and
western blot analysis (Fig. 3A).
siIRF1-2 had the highest silencing efficiency (P<0.01); thus,
siIRF1-2 (termed siIRF1) was used in the following experiments.
Marked differences in the levels of phenotypic markers were
observed between the siIRF1-transfected U937-M1 macrophages and the
siC (control)-transfected cells (Fig.
3B). The siIRF1-transfected U937-M1 cells exhibited a generally
downregulated expression of M1 genes, including IL-12p35, IL-12p40,
IL-23p19, IL-6 and TNF-α, but an enhanced expression of the M2
gene, IL-10, compared with the siC-transfected U937-M1 cells
(P<0.01; Fig. 3B).
Furthermore, we detected the amount of IL-12p70 and IL-10 secreted
in supernatant by ELISA, and observed that upon IRF1 knockdown, no
production of IL-12p70 was detected. However, an enhanced
production of IL-10 was observed in the siIRF1-transfected U937-M1
macrophages compared to the siC-transfected U937-M1 cells
(P<0.05; Fig. 3C).
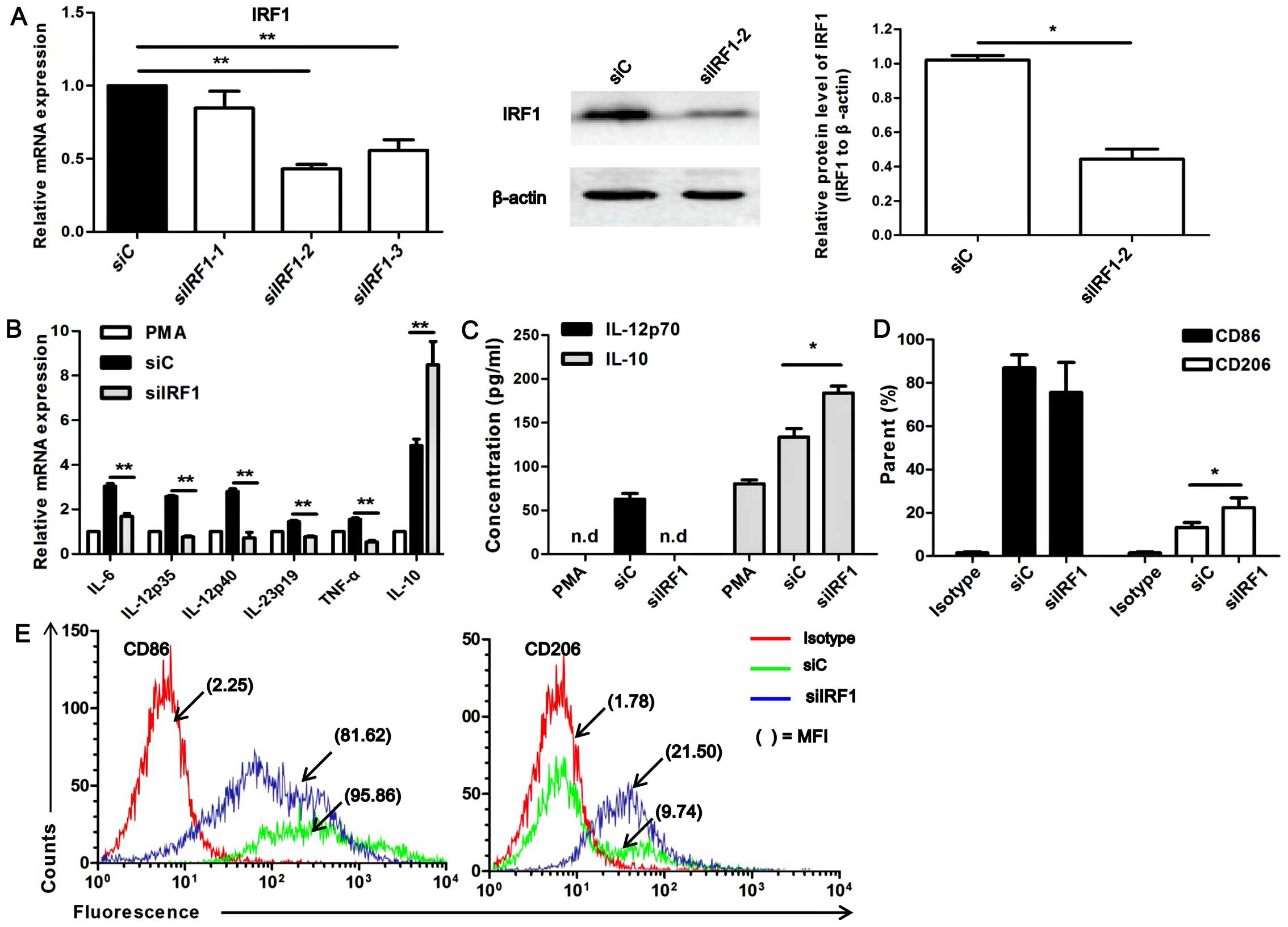 | Figure 3Interferon regulatory factor 1 (IRF1)
regulates the expression of macrophage polarization–specific
cytokines. (A) RT-qPCR analysis of the silencing efficiency of 3
siRNAs targeting IRF1, **P<0.01. Western blot
analysis of the silencing efficiency of siIRF1-2,
*P<0.05. The western blot analysis chart is a
representative of 3 independent experiments. (B) RT-qPCR analysis
of the mRNA expression of IL-12p40, IL-12p35, IL-23p19, IL-6, TNF-α
and IL-10 in M1 macrophages transfected with siIRF1-2 (siIRF1) or
the control siRNA (siC), **P<0.01. (C) ELISA of
IL-12p70 and IL-10 secretion in M1 macrophages transfected with
siIRF1-2 (siIRF1) or siC, *P<0.05,
**P<0.01; n.d, not detected. (D) Histograms showing
surface staining for CD86 and CD206 expression on siIRF1- or
siC-transfected U937-M1 cells, *P<0.05. Data were
calculated from 3 independent experiments. (E) The raw flow
cytometry fluorescence data are representative of 3 independent
experiments for CD86 and CD206 expression; staining profiles of
U937-M1 cells transfected with siIRF1 or siC. The percentage of
positive cells and mean fluorescence intensities (MFIs) are
denoted. |
Moreover, the M1-specific marker, CD86, and the
M2-specific marker, CD206, were analyzed by flow cytometry. As
shown in Fig. 3D and E, an
increased expression of CD206 was detected in the
siIRF1-transfected U937-M1 cells compared with siC-transfected
U937-M1 cells (P<0.05; Fig. 3D and
E). Although the reduction in CD86 expression did not reach a
level of significance between the siIRF1-transfected M1 macrophages
and the siC-transfected M1 macrophages, an obvious decreasing trend
in its expression was observed in the siIRF1-transfected M1
macrophages. These data suggest that IRF1 is involved in the M1
polarization of macrophages, as indicated by the expression of
M1/M2-associated markers and cytokines.
Inhibition of IFN-β affects the M1 status
induced by stimulation with IFN-γ and LPS
A recent study reported that IFN-β is expressed in
high levels in classically polarized macrophages [(THP-CAM) (M1)],
whereas it is expressed in low levels in alternatively activated
macrophages [(THP-AAM) (M2)] (37). It has also been demonstrated that
the expression of the M1-specific marker, IL-12p70, is impaired in
IFNAR1−/− GM-BMM (M1), and is enhanced by exogenous
IFN-β in GM-BMM (24). In this
study, we inhibited IFN-β with anti-IFN-β neutralizing antibody or
siRNA targeting the IFNB1 gene (siIFNB1). The neutralizing and
silencing efficiency were examined by RT-qPCR and ELISA,
respectively (Fig. 4A and B). We
found that siIFNB1-3 had the highest silencing efficiency
(P<0.001; Fig. 4A); thus,
siIFNB1-3 (termed siIFNB1) was used in the following experiments.
The secretion level of IFN-β was decreased in the
siIFNB1-transfected U937-M1 cells and in the cells in which IFN-β
had been neutralized (Ab) compared with the control siRNA
(siC)-transfected U937-M1 cells (P<0.05; Fig. 4B). It was observed that with the
use of either siIFNB1 or IFN-β neutralizing antibody (Ab), the
U937-M1 cells exhibited a significantly reduced expression of
M1-associated genes, including IL-12p35, IL-12p40, IL-23p19, IL-6
and TNF-α, but an enhanced expression of the M2-associated gene,
IL-10, compared with the siC-transfected U937 cells (P<0.05 and
P<0.01; Fig. 4C and D). As
regards IL-12p70, its secretion was significantly decreased in the
IFN-β-neutralized U937-M1 cells, and its secretion levels were even
undetectable in the siIFNB1-transfected U937-M1 cells. However, as
regards IL-10, its secretion was increased by 2-fold in the
IFN-β-neutralized U937-M1 cells and in the siIFNB1-transfected
U937-M1 cells compared with the siC-transfected and
IFN-γ/LPS-stimulated U937-M1 cells (P<0.05 and P<0.01;
Fig. 4G and H).
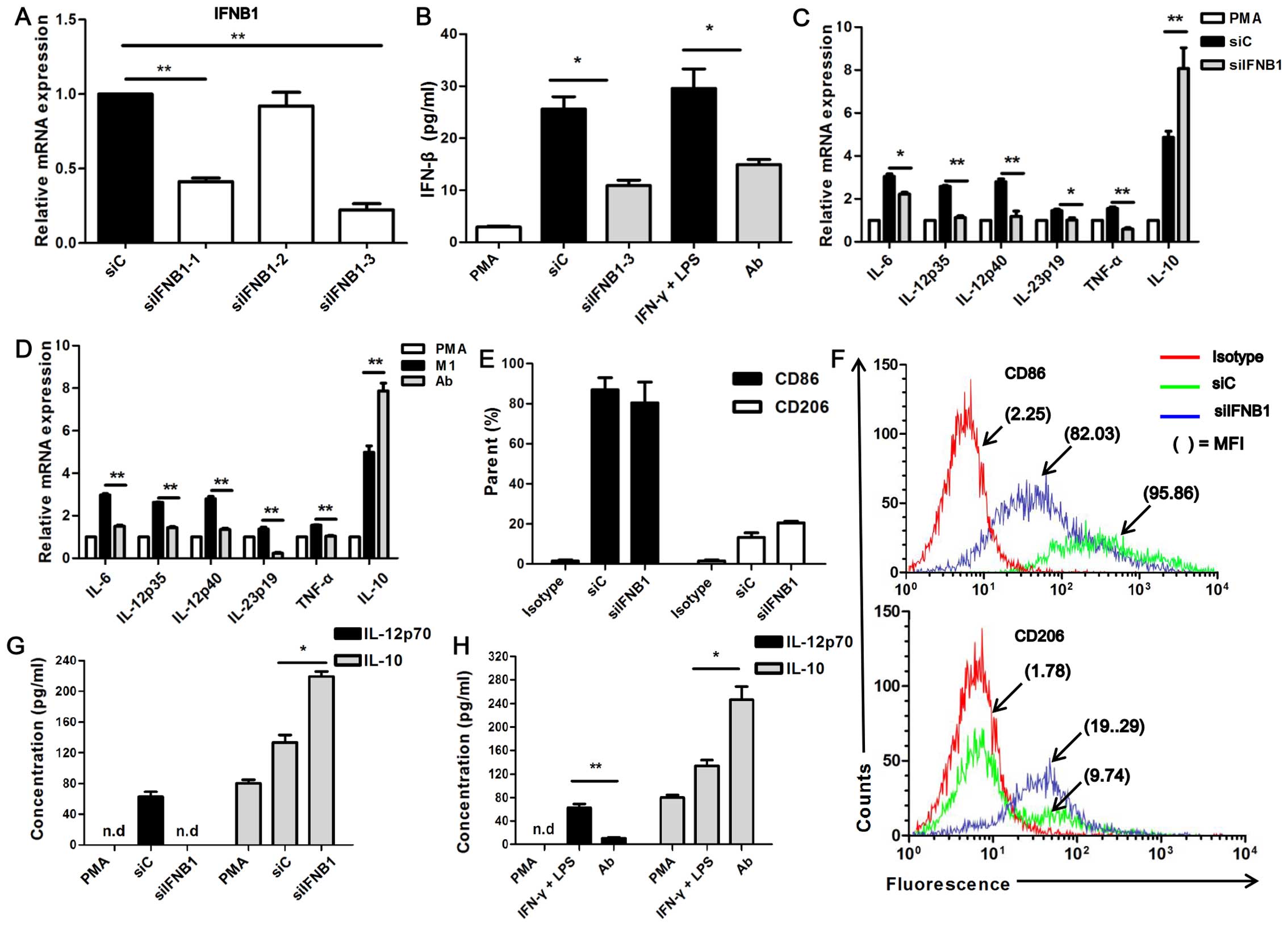 | Figure 4Interferon-β (IFN-β) affects the
expression of macrophage polarization-specific cytokines. (A)
RT-qPCR analysis of the silencing efficiency of siIFNB1,
**P<0.01. (B) ELISA of the silencing efficiency of
siIFNB1-3 or neutralizing efficiency of anti-IFN-β,
*P<0.05. (C and D) RT-qPCR analysis of mRNA
expression of IL-12p40, IL-12p35, IL-23p19, IL-6, TNF-α, and IL-10
in M1 macrophages transfected with siIFNB1-3 (siIFNB1) or siC, or
neutralized with anti-IFN-β antibody (Ab), *P< 0.05,
**P<0.01. (E) Histograms showing surface staining for
CD86 and CD206 expression on siIFNB1- or siC-transfected U937-M1
cells. Data were calculated from 3 independent experiments. (F) The
raw flow cytometry fluorescence data are representative of 3
independent experiments for CD86 and CD206 expression on U937-M1
transfected with siIFNB1 or siC. The percentage of positive cells
and mean fluorescence intensities (MFIs) are denoted. (G and H)
ELISA of the IL-12p70 and IL-10 secretion by M1 macrophages
transfected with siIFNB1 or siC or neutralized with anti-IFN-β
antibody. Data were calculated from 3 independent experiments.
*P<0.05, **P<0.01; n.d, not
detected. |
The expression of CD86 and CD206 did not reach a
level of significance between the siIFNB1-transfected M1 and the
siC-transfected M1 macrophages; however, an obvious increasing
trend in CD206 expression and a decreasing trend in CD86 expression
was observed in the siIFNB1-transfected M1 macrophages (Fig. 4E and F). The impaired expression
of M1-associated markers and the enhanced expression of
M2-associated markers observed following the inhibition of IFN-β in
the U937-M1 macrophages suggests that endogenous IFN-β is required
to maintain the M1 polarization status.
Association between IRF1 and IFN-β in the
regulation of M1 polarization
To further investigate the underlying mechanism of
M1 polarization associated with IRF1 and IFN-β, the expression of
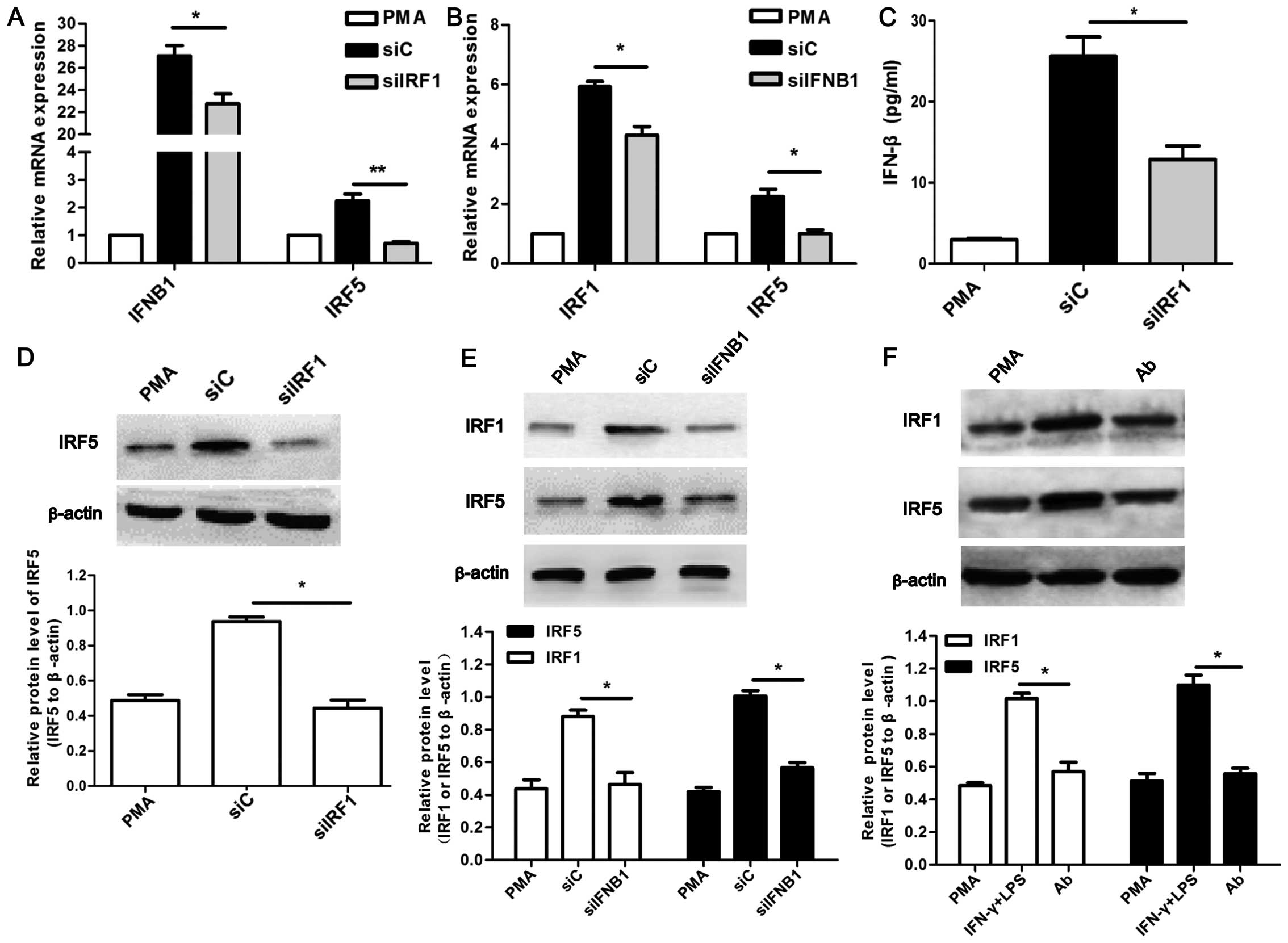
IFN-β in the siIRF1-transfected U937-M1 cells was detected. The
expression of IRF1 in the IFN-β-neutralized U937-M1 was also
examined. As shown in Fig. 5A and
C, the siIRF1-transfected U937-M1 cells exhibited a decreased
expression of IFN-β at both the mRNA and protein level (P<0.05).
Concordantly, the decreased expression of IRF1 at both the mRNA and
protein level was observed in the siIFNB1-transfected U937-M1 cells
(P<0.05; Fig. 5B and E) and in
the IFN-β-neutralized (Ab) U937-M1 cells (Fig. 5F).
Furthermore, the decreased mRNA and protein
expression of IRF5 was observed in the U937-M1 cells in which IRF1
was inhibited (P<0.05; Fig. 5A and
D). Surprisingly, we observed similar results with the
expression of IRF5 in the cells in which IFN-β was inhibited
(P<0.05; Fig. 5B, E and
F).
Therefore, these results suggest that IRF1 and IFN-β
interact with each other, which bridges the signaling pathways
activated by IFN-γ and LPS in M1 polarized macrophages.
Furthermore, both IRF1 and IFN-β may regulate the expression of
IRF5, which in turn contributes to the M1 polarization of
macrophages. However, the detailed mechanisms involved require
further investigation.
IRF1 and IFN-β play significant roles in
M1-mediated antitumor effects
Several studies have demonstrated that, M1
macrophages can combat tumors. They can suppress proliferation,
prevent invasion and promote the apoptosis of cancer cells through
the secretion of certain cytokines (45–49). Our observation of the impaired M1
status (as shown above) promoted us to investigate whether
M1-mediated antitumor effects would be affected under such
treatment conditions. HepG2 and SMMC-7721 cells were cultured with
various CM. The M1-mediated antitumor effects were then examined
with respect to proliferation, apoptosis and invasion ability.
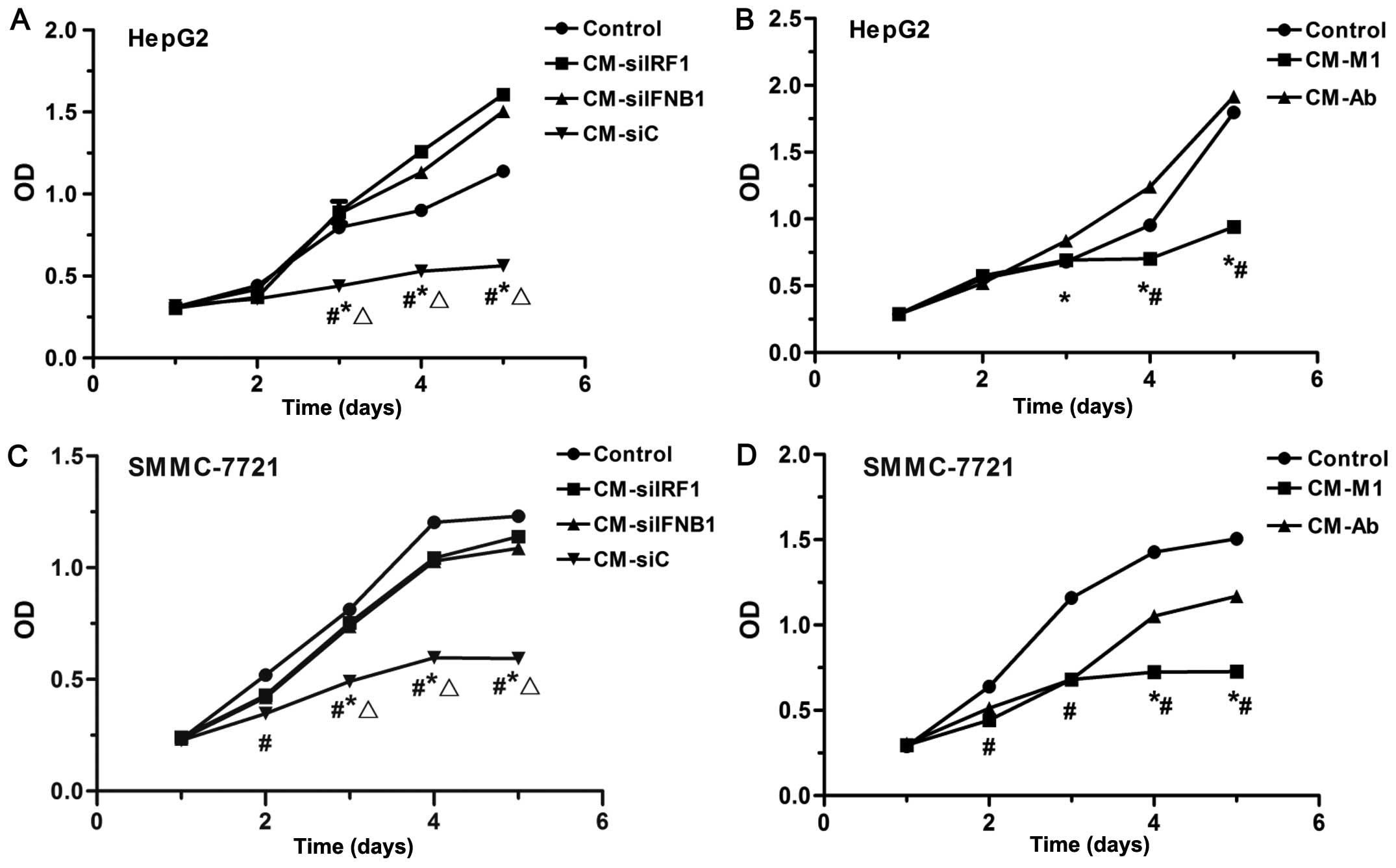
To examine the effects of various CM on the
proliferation of HCC cells, CCK-8 assay was performed. As shown in
Fig. 6, CM collected from the M1
macrophages (CM-M1) and siC-transfected M1 macrophages (CM-siC)
significantly inhibited the proliferation of HepG2 cells
(P<0.05; Fig. 6A and B) and
SMMC-7721 (P<0.05; Fig. 6C and
D). By contrast, the CM collected from M1 macrophages in which
IRF1 or IFN-β was inhibited (CM-siIRF1, CM-siIFNB1 or CM-Ab) partly
restored the proliferation of SMMC-7721 cells (P<0.05; Fig. 6C and D), and even promoted the
proliferation of HepG2 cells (P<0.05; Fig. 6A and B). These data suggest that
IRF1 and IFN-β play important roles in the M1-mediated
anti-proliferative effects on HCC cells.
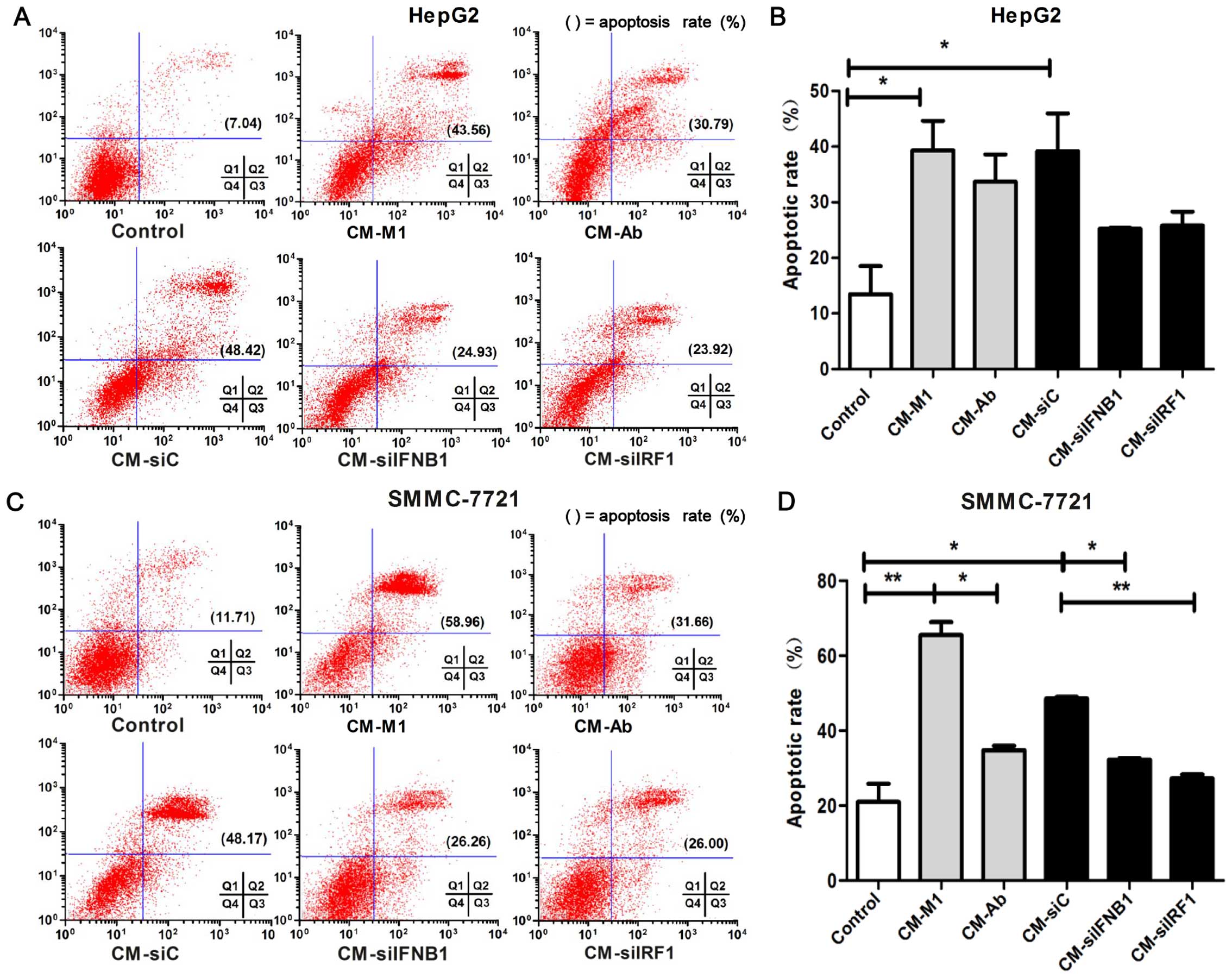
Flow cytometry was performed to determine the
effects of IRF1 and IFN-β on the apoptosis of HCC cells. As shown
in Fig. 7, an increased apoptotic
rate was observed in the cells cultured with CM-M1 and CM-siC
compared the control cells cultured with DMEM (HepG2 cells,
P<0.05; Fig. 7B; and SMMC-7721
cells, P<0.01; Fig. 7D).
However, the pro-apoptotic effect was partly inhibited in the
SMMC-7721 cells cultured with CM-siIRF1 or CM-siIFNB1 or CM-Ab
(P<0.01; Fig. 7D). Although
the apoptosis of the HepG2 cells did not reach a level of
significance between the cells cultured with CM-siC vs. CM-siIFNB1
(P=0.056) and CM-siC vs. CM-siIRF1 (P=0.065), an obvious decreasing
trend in apoptosis was observed in the HepG2 cells cultured with M1
cells in which IFNB1 and IRF1 was inhibited compared with the
controls (Fig. 7B). These data
indicate that IRF1 and IFN-β may play a role in the M1-mediated
pro-apoptotic effects on HCC cells.
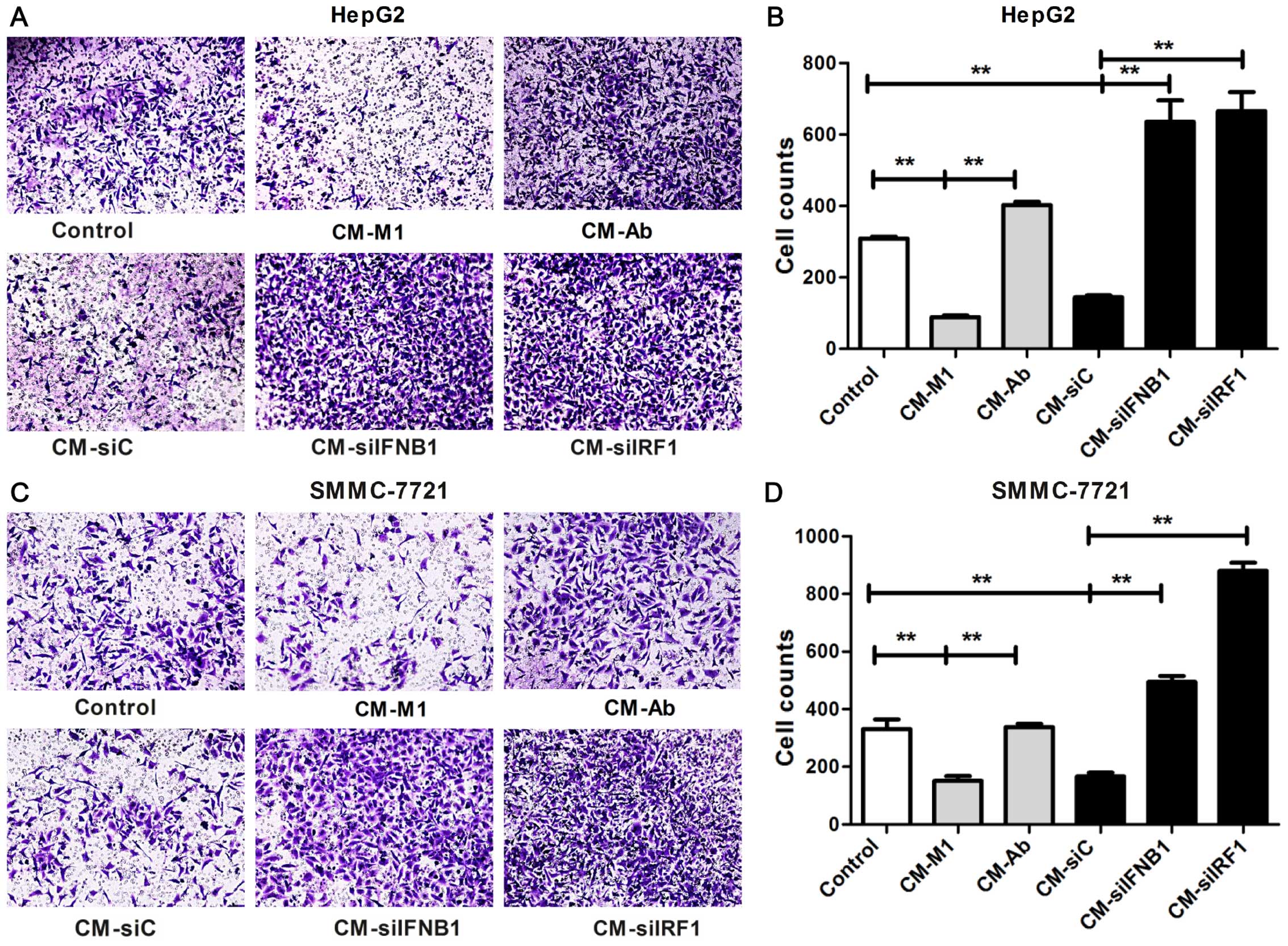
The invasion ability was examined by Transwell
invasion assay. As shown in Fig.
8, in the HCC cells cultured with CM-M1 and CM-siC, the number
of invading cells was significantly reduced compared with the
control cells cultured in DMEM (P<0.01; HepG2, Fig. 8B; SMMC-7721, Fig. 8D). By contrast, these inhibitory
effects on the invasion of HCC cells were completely reversed in
the HCC cells cultured with CM-siIRF1, CM-siIFNB1 or CM-Ab
(P<0.01; HepG2, Fig. 8B;
SMMC-7721, Fig. 8D). These data
suggest that IRF1 and IFN-β contribute to the M1-mediated
inhibitory effects on the invasion ability of HCC cells.
Collectively, the results of our above-mentioned
experiments suggest that U937-M1 macrophages exert tumoricidal
effects against HepG2 and SMMC-7721 cells, and we further
identified the crucial roles of IRF1 and IFN-β in the M1-mediated
antitumor effects.
Discussion
Macrophages display remarkable plasticity and
undergo alterations in their phenotypes in response to distinct
environmental cues (1). Two major
subsets, including classically activated (M1) and alternatively
activated (M2) macrophages, have long been recognized (3,4,38).
They can play contrasting roles in tumorigenesis depending on the
different phenotypes. It has been well established that M1
macrophages possess antitumor properties, whereas M2 macrophages
are characterized by pro-tumor effects (14,15,39). Therefore, a comprehensive
understanding of the underlying mechanisms of macrophage
polarization is necessary for developing antitumor strategies.
Several signaling pathways related to the
polarization of macrophages have been identified. A major locus for
macrophage polarization is at the transcriptional level of
regulation. The key regulators, including STAT1, activator protein
1 (AP1), IRF1/3/5/9 and hypoxia-inducible factor (HIF)-1α, play
crucial roles in the M1 polarization of macrophages. Other
modulators, such as STAT6, peroxisome proliferator-activated
receptor (PPAR)-γ, IRF4, HIF-2α, Kruppel-like factor 4 (KLF4) and
CCAAT-enhancer-binding protein (C/EBP)β play a significant role in
the M2 polarization of macrophages (16,40).
Interestingly, it has been revealed that synergistic
stimulation with IFN-γ and LPS is necessary for the polarization of
human M1 macrophages, as oppposed to stimulation with either factor
alone (27). Thus, it is
necessary to explore the underlying mechanisms of the M1
polarization of macrophages under the synergistic effects of IFN-γ
and LPS.
IRFs are transcriptional mediators, which can be
induced by bacteria or viruses. In mammals, IRFs consist of 9
members, including IRF1, IRF2, IRF3, IRF4, IRF5, IRF6, IRF7, IRF8
(ICSBP) and IRF9 (ISGF3γ/p48). They play prominent roles in
antiviral defense, immune response, tumor suppression, SLE
susceptibility, cell differentiation and apoptosis (40–42). In particular, IRF1/5/8 has been
shown too regulate the M1 polarization of macrophages, whereas IRF4
is involves in M2 polarization (43). The mechanisms of macrophage
polarization as regards IRFs are not yet fully understood.
It has been reported that IRF1 can be induced by
IFN-γ in M1 macrophages (19),
and that it is responsible for the expression of M1-associated
cytokine IL-12 subunits and iNOS (20,21). On the other hand, the production
of IFN-β is dependent on the LPS-induced activation of the
TRIF-dependent pathway (23).
Furthermore, it has been demonstrated that endogenous IFN-β is also
necessary for the production of IL-12p70 in GM-BMM (20,24). It has also been revealed that
IFN-β is expressed in high levels in THP-M1 macrophages, whereas it
is expressed in low levels in THP-M2 macrophages (37). We noted that IRF1 was originally
discovered to regulate the transcription of IFN-β in fibroblasts
infected with virus (25).
However, the connection between IRF1 and IFN-β in M1 macrophages is
not yet well clarified.
To address this issue, we referred to several
studies in the literature (10,16,35), and established the M1 macrophage
model with the monocytic tumor cell line, U937 (U937-M1) in the
present study. The results demonstrated that U937 cells could be
readily polarized into the M1 status, as indicated by the high
expression of CD86 and several pro-inflammatory cytokines, but the
low expression of CD206. Surprisingly, we noted that the M2
associated cytokine, IL-10, was also upregulated in U937-M1
macrophages. Although there has been some controversy, IL-10 is
generally considered an M2-associated cytokine marker (4,5,8,9).
It has been previously reported that IFN-γ and LPS stimulate
macrophages to produce IL-10 (8,44).
We speculate that IL-10, as an anti-inflammatory cytokine, may play
a role in the resolution of inflammation to avoid the M1-mediated
excessive pro-inflammatory response.
The present study indicated that IRF1 and IFN-β play
crucial roles in the regulation of the M1 polarization of
macrophages. In the M1 macrophages in which the IRF1 gene was
silenced, not only IL-12 production was impaired, but also the
expression of other pro-inflammatory cytokines, such as IL-6,
IL-23p19, and TNF-α. Simultaneously, IFN-β played a similar role as
IRF1 in M1-associated gene regulation, which was investigated by
the use of siRNA and neutralizing monoclonal IFN-β antibody.
Interestingly, the M2-associated markers, CD206 and IL-10, were
further significantly increased in the M1 macrophages in which IRF1
or IFN-β was inhibited. These results collectively indicate that
both IRF1 and IFN-β affect the expression of M1/M2-associated
markers, which in turn may affect the M1 polarization of
macrophages.
It has been reported that IRF1 regulates certain
genes by binding to the IRF-E and ISRE sites (32), such as IRF5 (31) and IFN-β (25,32). As a cytokine, IFN-β plays a
functional role mainly by binding to its receptor on the cell
membrane and initiating downstream signaling. It has also been
found that IFN-β induces IRF1 expression in RAW264.7 and peritoneal
macrophages through receptor recognized pathways (26). Based on this evidence, we
investigated the association between IRF1 and IFN-β in the context
of the M1 polarization of macrophages. We found that the knockdown
of IRF1 in U937-M1 cells exhibited reduced the production of IFN-β.
Similarly, the neutralization of IFN-β or IFNB1 knockdown in
U937-M1 cells led to a decreased expression of IRF1. These data
suggest that IRF1 and IFN-β may interact with each other, which
bridges the two pathways initiated by IFN-γ and LPS in M1
macrophages.
What should be noted is that our detected
M1/M2-associated cytokines (IL-12p35, IL-12p40, IL-23p19, IL-6,
TNF-α and IL-10) are also regulated by IRF5. It has been
demonstrated that IRF5 is directly recruited to the promoters and
promotes the expression of M1-associated genes, but suppresses
M2-associated gene expression (28–30). In the current study, IRF5 was
upregulated by the stimulation of U937-M1 cells with IFN-γ and LPS.
To determine whether IRF5 plays a role in IRF1- and
IFN-β-associated activities, we detected IRF5 expression in U937-M1
cells in which IRF1 or IFN-β was inhibited. Interestingly, IRF5 was
impaired in the U937-M1 in which IRF1 or IFN-β was inhibited. These
observations suggest that IRF5 is involved in IRF1- and
IFN-β-mediated activities. The association between IRF1, IFN-β and
IRF5 may involve more complex mechanisms of the M1 polarization of
macrophages. Thus, further studies are warranted to investigate the
detailed mechanisms.
In this study, the role of IRF1 and IFN-β in
M1-mediated antitumor effects on HCC cells was also explored. HepG2
and SMMC-7721 cells were incubated with CM collected from the
supernatant of M1 macrophages in which IRF1 or IFN-β was inhibited.
The results demonstrated that U937-M1 macrophages exerted
anti-tumor effects on HepG2 and SMMC-7721 cells, including
anti-proliferative, pro-apoptotic and anti-invasive effects.
However, the inhibition of IRF1 or IFN-β in the U937-M1 macrophages
attenuated these antitumor effects. Our functional experiments
further proved that IRF1 and IFN-β play significant roles in the
antitumor effects mediated by M1 macrophages. We speculate that the
IRF1- and IFN-β-mediated antitumor effects may due to the
regulation of M1/M2-associated cytokines, which have been reported
to responsible for antitumor/pro-tumor effects (45–49).
In conclusion, in the present study, we provide
evidence that IRF1 and IFN-β may cooperate with each other to take
part in the M1 polarization of macrophages, as well as in the
regulation of IRF5, consequently affecting the M1-mediated
antitumor effects. Our data may provide a novel target for targeted
cancer therapy.
Acknowledgments
This study was supported by the National Natural
Science Foundation of China (grant no. 81172016). We would like to
thank Dr Jian Zhang, Dr Shengkai Yan, Dr Shengxiang Ge and Dr
Keyang Chen for their valuable suggestions.
References
|
1
|
Das A, Sinha M, Datta S, Abas M, Chaffee
S, Sen CK and Roy S: Monocyte and macrophage plasticity in tissue
repair and regeneration. Am J Pathol. 185:2596–606. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Murray PJ and Wynn TA: Protective and
pathogenic functions of macrophage subsets. Nat Rev Immunol.
11:723–737. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lawrence T and Natoli G: Transcriptional
regulation of macrophage polarization: enabling diversity with
identity. Nat Rev Immunol. 11:750–761. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mosser DM and Edwards JP: Exploring the
full spectrum of macrophage activation. Nat Rev Immunol. 8:958–969.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Duluc D, Corvaisier M, Blanchard S, Catala
L, Descamps P, Gamelin E, Ponsoda S, Delneste Y, Hebbar M and
Jeannin P: Interferon-gamma reverses the immunosuppressive and
protumoral properties and prevents the generation of human
tumor-associated macrophages. Int J Cancer. 125:367–373. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Labonte AC, Tosello-Trampont AC and Hahn
YS: The role of macrophage polarization in infectious and
inflammatory diseases. Mol Cells. 37:275–285. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chistiakov DA, Bobryshev YV, Nikiforov NG,
Elizova NV, Sobenin IA and Orekhov AN: Macrophage phenotypic
plasticity in atherosclerosis: the associated features and the
peculiarities of the expression of inflammatory genes. Int J
Cardiol. 184:436–445. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Melton DW, McManus LM, Gelfond JA and
Shireman PK: Temporal phenotypic features distinguish polarized
macrophages in vitro. Autoimmunity. 48:161–176. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Biswas SK and Mantovani A: Macrophage
plasticity and interaction with lymphocyte subsets: cancer as a
paradigm. Nat Immunol. 11:889–896. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chabot S, Charlet D, Wilson TL and Yong VW
and Yong VW: Cytokine production consequent to T cell - microglia
interaction: the PMA/IFN gamma-treated U937 cells display
similarities to human microglia. J Neurosci Methods. 105:111–120.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dall'Asta M, Derlindati E, Ardigò D,
Zavaroni I, Brighenti F and Del Rio D: Macrophage polarization: The
answer to the diet/inflammation conundrum? Nutr Metab Cardiovasc
Dis. 22:387–392. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ohashi W, Hattori K and Hattori Y: Control
of macrophage dynamics as a potential therapeutic approach for
clinical disorders involving chronic inflammation. J Pharmacol Exp
Ther. 354:240–250. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Heusinkveld M and van der Burg SH:
Identification and manipulation of tumor associated macrophages in
human cancers. J Transl Med. 9:2162011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bögels M, Braster R, Nijland PG, Gül N,
van de Luijtgaarden W, Fijneman RJ, Meijer GA, Jimenez CR, Beelen
RH and van Egmond M: Carcinoma origin dictates differential skewing
of monocyte function. OncoImmunology. 1:798–809. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pollard JW: Macrophages define the
invasive microenvironment in breast cancer. J Leukoc Biol.
84:623–630. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tugal D, Liao X and Jain MK:
Transcriptional control of macrophage polarization. Arterioscler
Thromb Vasc Biol. 33:1135–1144. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gough DJ, Levy DE, Johnstone RW and Clarke
CJ: IFNgamma signaling - does it mean JAK-STAT? Cytokine Growth
Factor Rev. 19:383–394. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jaruga B, Hong F, Kim WH and Gao B:
IFN-gamma/STAT1 acts as a proinflammatory signal in T cell-mediated
hepatitis via induction of multiple chemokines and adhesion
molecules: a critical role of IRF-1. Am J Physiol Gastrointest
Liver Physiol. 287:G1044–G1052. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Martinez FO, Gordon S, Locati M and
Mantovani A: Transcriptional profiling of the human
monocyte-to-macrophage differentiation and polarization: new
molecules and patterns of gene expression. J Immunol.
177:7303–7311. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Negishi H, Fujita Y, Yanai H, Sakaguchi S,
Ouyang X, Shinohara M, Takayanagi H, Ohba Y, Taniguchi T and Honda
K: Evidence for licensing of IFN-gamma-induced IFN regulatory
factor 1 transcription factor by MyD88 in Toll-like
receptor-dependent gene induction program. Proc Natl Acad Sci USA.
103:15136–15141. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu J, Guan X, Tamura T, Ozato K and Ma X:
Synergistic activation of interleukin-12 p35 gene transcription by
interferon regulatory factor-1 and interferon consensus
sequence-binding protein. J Biol Chem. 279:55609–55617. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ouyang X, Negishi H, Takeda R, Fujita Y,
Taniguchi T and Honda K: Cooperation between MyD88 and TRIF
pathways in TLR synergy via IRF5 activation. Biochem Biophys Res
Commun. 354:1045–1051. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Toshchakov V, Jones BW, Perera PY, Thomas
K, Cody MJ, Zhang S, Williams BR, Major J, Hamilton TA, Fenton MJ
and Vogel SN: TLR4, but not TLR2, mediates IFN-beta-induced
STAT1alpha/beta-dependent gene expression in macrophages. Nat
Immunol. 3:392–398. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fleetwood AJ, Dinh H, Cook AD, Hertzog PJ
and Hamilton JA: GM-CSF- and M-CSF-dependent macrophage phenotypes
display differential dependence on type I interferon signaling. J
Leukoc Biol. 86:411–421. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Reis LF, Harada H, Wolchok JD, Taniguchi T
and Vilcek J: Critical role of a common transcription factor,
IRF-1, in the regulation of IFN-beta and IFN-inducible genes. EMBO
J. 11:185–193. 1992.PubMed/NCBI
|
|
26
|
Guo Z, Garg S, Hill KM, Jayashankar L,
Mooney MR, Hoelscher M, Katz JM, Boss JM and Sambhara S: A distal
regulatory region is required for constitutive and IFN-beta-induced
expression of murine TLR9 gene. J Immunol. 175:7407–7418. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu J, Cao S, Herman LM and Ma X:
Differential regulation of interleukin (IL)-12 p35 and p40 gene
expression and interferon (IFN)-gamma-primed IL-12 production by
IFN regulatory factor 1. J Exp Med. 198:1265–1276. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Krausgruber T, Blazek K, Smallie T,
Alzabin S, Lockstone H, Sahgal N, Hussell T, Feldmann M and Udalova
IA: IRF5 promotes inflammatory macrophage polarization and TH1-TH17
responses. Nat Immunol. 12:231–238. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dalmas E, Toubal A, Alzaid F, Blazek K,
Eames HL, Lebozec K, Pini M, Hainault I, Montastier E, Denis RG, et
al: Irf5 deficiency in macrophages promotes beneficial adipose
tissue expansion and insulin sensitivity during obesity. Nat Med.
21:610–618. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Feng D, Sangster-Guity N, Stone R,
Korczeniewska J, Mancl ME, Fitzgerald-Bocarsly P and Barnes BJ:
Differential requirement of histone acetylase and deacetylase
activities for IRF5-mediated proinflammatory cytokine expression. J
Immunol. 185:6003–6012. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mancl ME, Hu G, Sangster-Guity N,
Olshalsky SL, Hoops K, Fitzgerald-Bocarsly P, Pitha PM, Pinder K
and Barnes BJ: Two discrete promoters regulate the alternatively
spliced human interferon regulatory factor-5 isoforms. Multiple
isoforms with distinct cell type-specific expression, localization,
regulation, and function. J Biol Chem. 280:21078–21090. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tan RS, Taniguchi T and Harada H:
Identification of the lysyl oxidase gene as target of the
antioncogenic transcription factor, IRF-1, and its possible role in
tumor suppression. Cancer Res. 56:2417–2421. 1996.PubMed/NCBI
|
|
33
|
Zimmermann A, Trilling M, Wagner M,
Wilborn M, Bubic I, Jonjic S, Koszinowski U and Hengel H: A
cytomegaloviral protein reveals a dual role for STAT2 in
IFN-(gamma) signaling and antiviral responses. J Exp Med.
201:1543–1553. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Steen HC and Gamero AM: The role of signal
transducer and activator of transcription-2 in the interferon
response. J Interferon Cytokine Res. 32:103–110. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Taniguchi K, Hikiji H, Okinaga T,
Hashidate-Yoshida T, Shindou H, Ariyoshi W, Shimizu T, Tominaga K
and Nishihara T: Essential role of lysophosphatidylcholine
acyltransferase 3 in the induction of macrophage polarization in
PMA-treated U937 cells. J Cell Biochem. 116:2840–2848. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Maeß MB, Wittig B, Cignarella A and
Lorkowski S: Reduced PMA enhances the responsiveness of transfected
THP-1 macrophages to polarizing stimuli. J Immunol Methods.
402:76–81. 2014. View Article : Google Scholar
|
|
37
|
El Fiky A, Perreault R, McGinnis GJ and
Rabin RL: Attenuated expression of interferon-β and interferon-λ1
by human alternatively activated macrophages. Hum Immunol.
74:1524–1530. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Stout RD, Watkins SK and Suttles J:
Functional plasticity of macrophages: in situ reprogramming of
tumor-associated macrophages. J Leukoc Biol. 86:1105–1109. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Siveen KS and Kuttan G: Role of
macrophages in tumour progression. Immunol Lett. 123:97–102. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jensen MA and Niewold TB: Interferon
regulatory factors: critical mediators of human lupus. Transl Res.
165:283–295. 2015. View Article : Google Scholar :
|
|
41
|
Paun A and Pitha PM: The IRF family,
revisited. Biochimie. 89:744–753. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Salloum R and Niewold TB: Interferon
regulatory factors in human lupus pathogenesis. Transl Res.
157:326–331. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Günthner R and Anders HJ:
Interferon-regulatory factors determine macrophage phenotype
polarization. Mediators Inflamm. 2013:7310232013. View Article : Google Scholar :
|
|
44
|
Jaguin M, Houlbert N, Fardel O and
Lecureur V: Polarization profiles of human M-CSF-generated
macrophages and comparison of M1-markers in classically activated
macrophages from GM-CSF and M-CSF origin. Cell Immunol. 281:51–61.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Duechler M, Peczek L, Zuk K, Zalesna I,
Jeziorski A and Czyz M: The heterogeneous immune microenvironment
in breast cancer is affected by hypoxia-related genes.
Immunobiology. 219:158–165. 2014. View Article : Google Scholar
|
|
46
|
Nicolini A, Carpi A and Rossi G: Cytokines
in breast cancer. Cytokine Growth Factor Rev. 17:325–337. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang YC, He F, Feng F, Liu XW, Dong GY,
Qin HY, Hu XB, Zheng MH, Liang L, Feng L, et al: Notch signaling
determines the M1 versus M2 polarization of macrophages in
antitumor immune responses. Cancer Res. 70:4840–4849. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Buijs JT, Stayrook KR and Guise TA: TGF-β
in the bone microenvironment: role in breast cancer metastases.
Cancer Microenviron. 4:261–281. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Drabsch Y and ten Dijke P: TGF-β signaling
in breast cancer cell invasion and bone metastasis. J Mammary Gland
Biol Neoplasia. 16:97–108. 2011. View Article : Google Scholar : PubMed/NCBI
|