Introduction
A number of individuals worlwide are known to suffer
from cockroach (CR) allergies and the CR allergies are considered
to play an important role in IgE-mediated type I hypersensitivity
since 1964 (1). German CR
[Blattella germanica (Bla g)], American CR [Periplaneta
Americana (Per a)] and smoky brown CR (Periplaneta
fuliginosa) are the main species which are known to cause
allergies worldwide (2). In
China, a number of patients with allergies are found to positive
for CR allergens in the skin prick test (SPT); 25.7% are shown to
be positive for the American CR and 18.7% are shown to be positive
for the German CR (3).
The adult American CR has 22 IgE binding components
with pooled sera from patients, including the proteins of 23, 28,
35, 38, 40, 49, 72, 78 and 97 kDa as major allergens (4). Some of these allergens namely, Per a
1 (5), Per a 2 (previously known
as Cr PI) (6), Per a 3 (7), Per a 4 (8), Per a 5 (9), Per a 6 (10), Per a 7 (11), Per a 9 (previously known as
Periplaneta americana arginine kinase) (12,13), Per a 10 (14), Per a 11 (15) and Per a 12 (15) have been characterized. Among these
allergens, Per a 10 was isolated from American CR extract using a
benzamidine sepharose column and characterized by immunobiochemical
methods. It is a serine proteinase with a molecular weight of
approximately 28 kDa (14). Per a
10 was recognized as a major allergen, showing IgE reactivity with
>80% of cockroach sensitized patients by skin tests and
immunoblot (14). Further studies
found that Per a 10 can modulate CD40 expression on the dendritic
cell surface through the nuclear factor-κB (NF-κB) pathway
(16) and can promote the
dendritic cell type 2 phenotype via the upregulation of CD86,
increased high interleukin-6 (IL-6) secretion and reduced IL-12
secretions (17).
Recent studies have suggested that CR immunotherapy
may be a promising treatment strategy with immunomodulatory and
clinical effects (18,19). However, patients with CR allergies
often exhibit complex sensitization patterns to multiple
CR-associated proteins and an immunodominant allergen has not yet
been identified (20). To realize
the full potential of this treatment modality, further
investigations of CR allergens are warranted. The identification of
B cell epitopes (IgE-binding epitopes) of an allergen is helpful in
designing sequences for more accurate and safer peptide-based
allergen diagnosis and immunotherapeutic agents. The IgE-binding
epitopes of Per a 1 (21), Per a
2 (22), Per a 3 (23), Per a 4 (8) and Per a 6 (10) have previously been identified by
experimental or bioinformatics methods. T cell epitopes have been
successfully identified based on computer simulation over the past
decade. Extracellular peptides have to bind to major
histocompatibility complex (MHC) class II to stimulate T lymphocyte
responses. Thus, T cell epitopes have been predicted indirectly by
the identification of MHC-binding molecules (20). In a previous study (13), using in silico analysis, we
performed epitope prediction and functional analysis of the Per a 9
allergen from the American CR. In the present study, in
continuation of our investigation on Amiercan CR allerens, we
firstly cloned, expressed the American CR major allergen Per a 10
and identified the B and T cell epitopes of the Per a 10 allergen
using an in silico approach. Our findings suggest the
potential utility of these allergens and epitopes in the
development of peptide-based vaccines for the prevention and/or
treatment of CR allergies.
Materials and methods
Patients and samples
As in our previous study (13), in this study, 16 patients with
allergic rhinitis who were found to have positive SPT results and
positive results for the serum IgE test for the American CR extract
(by using ImmunoCAP assay; Pharmacia Diagnostics AB, Uppsala,
Sweden) and 6 healthy controls (HC) who showed negative results for
the serum IgE test were recruited. Blood samples were obtained from
all participants after obtaining written informed consent and this
study was approved by the Ethics Committee of the First Affiliated
Hospital of Nanjing Medical University, Nanjing, China.
Molecular cloning of the American CR Per
a 10 gene
Total RNA was isolated from female CRs using TRIzol
reagent (Invitrogen, Carlsbad, CA, USA). Total RNA was quantified
by measuring the absorbance ratios at 260/280 nm. Total RNA was
reverse transcribed into cDNA using oligo(dT) by reverse
transcriptase using a commercial cDNA synthesis kit according to
the manufacturer's instructions (Takara Biotech Co., Dalian,
China). The gene encoding Per a 10 was amplified by PCR using
primers based on the nucleotides sequence of the Per a 10 gene
(AY792954.1; forward, 5′-ATGC TTCGCTACCTGGTACTT-3′ and reverse,
5′-TTAGTTGACTCCAGTCTGTTC-3′). The PCR conditions were as follows:
98°C/5 min (1 cycle), 98°C/10 sec, 50°C/15 sec and 72°C/1 min (35
cycles), and 72°C/5 min (1 cycle). The purified PCR product was
cloned into the pMD18-T vector (Takara Biotech Co.) and transformed
into the Escherichia coli (E. coli) strain DH5α. The
inserts were confirmed by DNA sequencing.
Expression and purification of Per a 10
in E. coli
As previously described with some modifications
(13), the Per a 10 gene was
subcloned into the pET-22b(+) vector (Novagen, Madison, WI, USA)
using the NdeI and XhoI sites and confirmed by DNA
sequencing. The recombinant pET22b(+)-Per a 10 plasmid was
transformed into the Rosette-gami E. coli host strain. The
selected pET22b(+)-Per a 10-transformed Rosette-gami E. coli
was inoculated into 5 ml of LB-ampicillin broth, and incubated at
37°C overnight. Five milliliters of the culture were inoculated
into 500 ml of fresh LB-ampicillin broth and incubated at 28°C
while shaking at 200 rpm until the optical density (OD) at
A600nm reached 0.5.
Isopropyl-β-D-thiogalactopyranoside (IPTG) was added to a final
concentration of 1 mM and the culture was incubated for a further 7
h. The bacterial cells were harvested by centrifugation at 4,000 ×
g at 4°C for 20 min, and were lysed in lysis buffer by sonication
at 20 kHz, 2 min pulse-on, 3 min pulse-off. The recombinant Per a
10 was mainly contained in inclusion bodies. The inclusion bodies
were collected by centrifugation at 12,000 × g at 4°C for 20 min.
Following solubilization of the inclusion bodies by 6 M urea, the
supernatant was loaded onto a Nickel column (Genscript, Nanjing,
China), washed with running buffer containing 50 mM Tris-HCl, 300
mM NaCl (pH 8.0), and eluted with elution buffer containing 50 mM
Tris-HCl, 300 mM NaCl, 250 mM imidazole. The eluted fractions were
collected and dialyzed with 6-4-2-1-0.5-0 M urea, each for 2 h.
Immunoreactivity of recombinant Per a 10
with serum from patients with CR allergies
A 96-well plate was coated with recombinant 100
μl Per a 10 (10 μg/ml) in carbonate-bicarbonate
buffer (0.05 M, pH 9.6) overnight at 4°C. Human serum samples (1:20
dilution in PBS-Tween-20 with 2% BSA) were then added to the plates
and incubated for 2 h at room temperature. Following IgE binding,
the plates were incubated with horseradish peroxidase-labeled goat
anti-human IgE (1:2,500 dilution) (KPL, Inc., Gaithersburg, MD,
USA), and the color was developed with tetramethylbenzidine (TMB)
peroxidase substrate. The plates were read on a microplate reader
(Eon; BioTek, Winooski, Vermont, USA) at an absorbance of 405 nm.
The cut-off value of the enzyme-linked immunosorbent assay (ELISA)
was calculated as the mean of the negative controls plus 2 SDs, as
previously described (9).
Immunoblot analysis of the IgE reactivity
of Per a 10
Immunoblots for the detection of IgE reactivity of
Per a 10 were performed as described previously (24,25). Per a 10 (5 μg) was
conducted to a sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE; gel concentration of 15%) under reduced
conditions and then transferred onto nitrocellulose membranes. The
nitrocellulose membranes were firstly incubated with sera from
patients with American CR allergies (1:5 to 1:20 in PBS-Tween-20
with 1% BSA, 10% normal goat serum) for 90 min, and then incubated
with peroxidase-labeled anti-human IgE monoclonal antibody
(074-1004, KPL, Inc., Gaithersburg, MD, USA). The positive protein
bands were visualized by incubating the membranes with TMB
peroxidase substrate. Sera from 2 non-allergic healthy subjects
were used as the negative controls.
Basophil activation test
The expression of CD63 and CCR3 on the basophil
surface is considered as an indicator of basophil activation
(26,27). Briefly, and as previously
described (13), peripheral blood
mononucleated cells (PBMCs) from 4 healthy volunteers were
separated by Ficoll-Paque density gradient, and treated with 10 ml
LS solution (1.3 M NaCl, 0.005 M KCl and 0.01 lactic acid, pH 3.9)
for 2 min at 8°C. Following neutralization with 12% Tris (pH 10.9),
non-specific IgE on basophils was stripped off. Subsequently, the
basophils were passively sensitized with the sera of patients with
American CR allergies or from the healthy non-allergic healthy
controls (n=4, 1 in 10 dilution, 2 h at 37°C) (same patients and
controls as described above) as previously described (23). The sensitized basophils were
challenged with Per a 10 (1.0 μg/ml) for 15 min at 37°C.
CCR3-PE-labelled antibody (85-12-1939-42; eBioscience Inc., San
Diego, CA, USA) and anti-human CD63-FITC antibody (HH-MHCD63014;
Invitrogen, Camarillo, CA, USA) were added to cells for 15 min at
37°C. Flow cytometric analysis of CD63 and CCR3 was performed at
488 nm on a FACSAria flow cytometer (Becton-Dickinson, Franklin
Lakes, NJ, USA) and analyzed by FACSDiva software.
Sequence retrieval Per a 10 homolog and
phylogenetic analysis
The complete amino acid sequence of Per a 10 was
used as query to search for homologous sequences by tBLASTn in NCBI
(blast.ncbi.nlm.nih.gov/Blast.cgi) (10,28–30). The phylogenetic tree of Per a 10
and its homolog was obtained by the maximum-likelihood (ML) method
on the basis of the JTT amino acid sequence distance implemented in
MEGA 5.1, the reliability was evaluated by the bootstrap method
with 1,000 replications (28–32).
Physiochemical analysis and
post-translational patterns and motifs of Per a 10
As previously described (13), physiochemical analysis, including
molecular weight, theoretical pI, amino acid composition,
instability index, aliphatic index and the grand average of
hydropathicity (GRAVY) of Per a 10 was performed using the
ProtParam tool (http://web.expasy.org/protparam/) (33).
Secondary structure prediction of Per a
10
As previously described (13), the secondary structure of Per a 10
was assessed by PSIPRED (bioinf.cs.ucl.ac.uk/psipred) (34) and NetSurfP ver. 1.1 (www.cbs.dtu.dk) (35).
Homology modeling and validation
As previously described (13), the best homologous templates for
Per a 10 were selected using the PSI-BLAST server (http://blast.ncbi.nlm.nih.gov/Blast.cgi)
in NCBI and Swiss-model server (http://swissmodel.expasy.org/) and used for homology
modeling. The modeled protein structure of Per a 10 was built by
SWISS-MODEL (http://swissmodel.expasy.org/). An initial structural
model was generated and checked for recognition of errors in 3D
structure by PROCHECK (36),
ERRAT (verification of protein structures: patterns of nonbonded
atomic interactions) (33,37)
and VERIFY_3D (a method to identify protein sequences that fold
into a known three-dimensional structure. Assessment of protein
models with 3-dimensional profiles) programs in Structural Analysis
and Verification Server (http://nihserver.mbi.ucla.edu/SAVES/) (33,37).
In silico prediction of B cell epitopes
of Per a 10
As previously described (13), 3 immunoinformatics tools, namely
the DNAStar Protean system (38),
bioinformatics predicted antigenic peptides (BPAP) system
(http://imed.med.ucm.es/Tools/antigenic.pl) and the
BepiPred 1.0 server (http://www.cbs.dtu.dk/services/BepiPred/) were used to
predict the B cell epitopes of Per a 10 (39,40). The ultimate consensus epitope
results were obtained by combining the results of the 3 tools
(41). If the results of all 3
methods were non-epitope, the consensus result was then considered
0% epitope. Similarly, if the predicted results had only one or no
non-epitope, the consensus result was considered 67 or 100%
epitope, respectively. Finally, the regions whose consensus epitope
result was 67 or 100% were chosen as the final potential epitope
regions.
In silico prediction of T cell
epitopes
As previously described (13), for HLA-DR-based T cell epitope
prediction, the artificial neural network-based alignment
(NN-align) method NetMHCIIpan-3.0 (http://www.cbs.dtu.dk/services/NetMHCIIpan/) was
applied (42). We used HLA-DR
101, HLA-DR 301, HLA-DR 401 and HLA-DR 501. The ultimate
HLA-DR-based T cell epitope results were obtained by combining
those 4 results together that if 3 of them showed epitope, and then
the consensus result was epitope. For HLA-DQ alleles, NetMHCII-2.2
(http://www.cbs.dtu.dk/services/NetMHCII/) was used
(43). We only used
HLA-DQA10501-DQB10201, HLA-DQA10301-DQB10302,
HLA-DQA10401-DQB10402, and HLA-DQA10102-DQB10602. As a result, the
ultimate consensus epitope results were obtained by combining the
results of the HLA-DR-based T cell epitope and HLA-DQ-based T cell
epitope. B cell and T cell epitopes identified by computational
tools were mapped onto linear sequence and on the three dimensional
model of Per a 10 to determine their position and secondary
structure elements involved.
Statistical analysis
Data are expressed as the means ± SE for the
indicated number of independently performed duplicated experiments.
Statistical significance between means was analyzed by one-way
ANOVA or the Student's t-test utilizing the SPSS 13.0 version. A
valoue of P<0.05 was considered to indicate a statistically
significant difference.
Results
Molecular cloning of the Per a 10
allergen of the American CR
The cDNA encoding the Per a 10 gene was amplified by
PCR. It is 771 bp gene and encodes a 256 amino acids protein. The
sequence homology with the published one (Accession no. AY792954.1)
was 100% (256/256) at the protein level. Among the 256 amino acid
protein, MLRYLVLASLIACSLS is a signal peptide, and AVPKAKRPRLDGR is
a pro-peptide, which would be removed in the mature Per a 10. So
the mature Per a 10 contained 227 amino acids.
Expression and purification of Per a 10
in E. coli
The American CR allergen Per a 10 was subcloned into
the pET-22b(+) vector and transformed into the Rosette-gami E.
coli host strain. Per a 10 was firstly expressed at
temperatures of 16°C, 28°C or 37°C (Fig. 1A). It was found that Per a 10 was
expressed mainly in the inclusion body and 28°C was the optimal
temperature condition for its expression. Thus, the condition of 1
mM IPTG and a temperature of 28°C was selected for the large
expression and purification of Per a 10. The dissolved Per a 10
inclusion body was purified by a Ni column. After the successful
renaturation of purified Per a 10, approximately 1.4 mg recombinant
Per a 10 was obtained from 500 ml of cell culture. The purity of
the purified Per a 10 was identified by SDS-PAGE. It showed a
single band with an apparent molecular weight of 30 kDa (Fig. 1B).
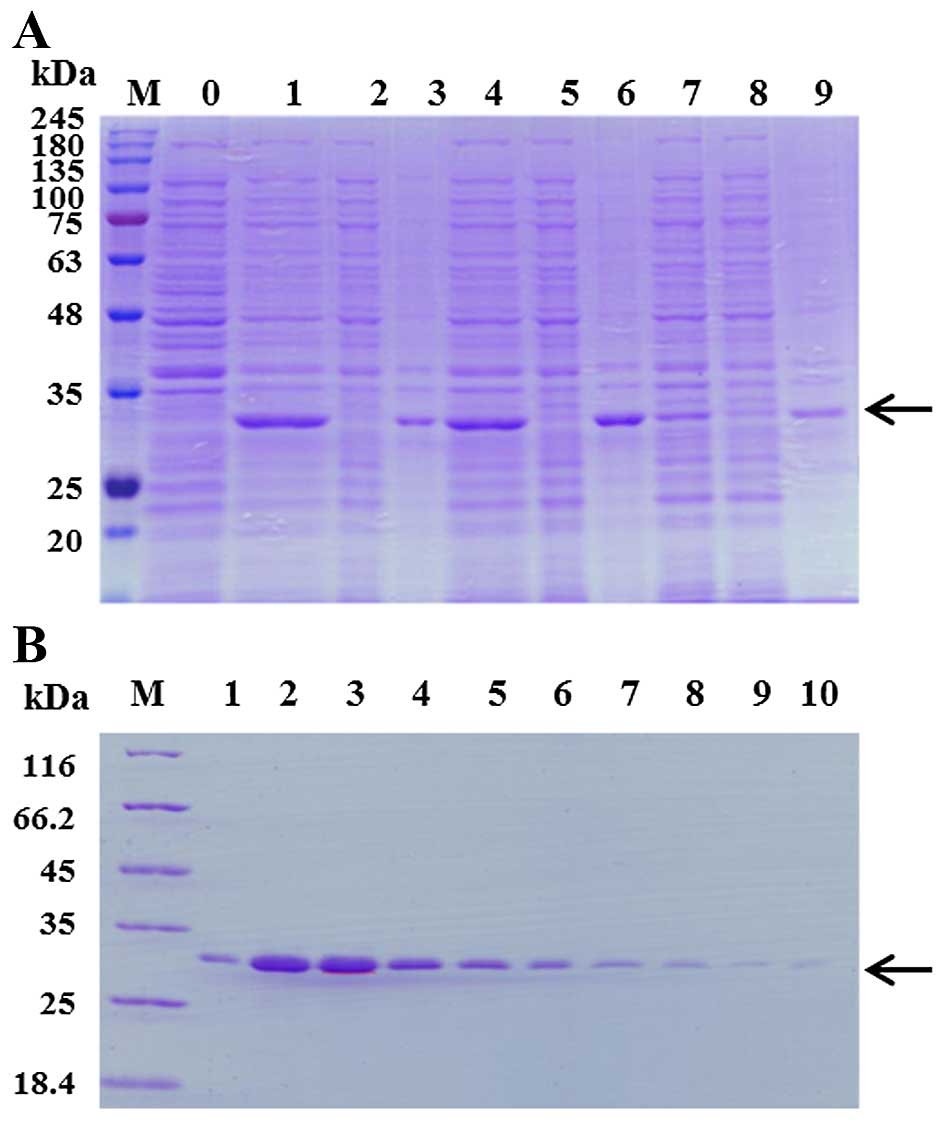 | Figure 1Expression and purification of Per a
10 in E. coli. (A) Per a 10 expressed at 16°C, 28°C or 37°C
was analyzed by sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE). Lane M, protein makers; lane 1, the
total protein of un-induced bacteria; lane 2, the total protein of
bacteria induced by 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG)
at 37°C; lane 3, the supernatant of the bacteria induced by 1 mM
IPTG at 37°C; lane 4, the precipitant of the bacteria induced by 1
mM IPTG at 37°C; lane 5, the total protein of bacteria induced by 1
mM IPTG at 28°C; lane 6, the supernatant of the bacterial induced
by 1 mM IPTG at 28°C; lane 7, the precipitant of the bacterial
induced by 1 mM IPTG at 28°C; lane 8, the total protein of bacteria
induced by 1 mM IPTG at 16°C; lane 9, the supernatant of the
bacteria induced by 1 mM IPTG at 16°C; lane 10, the precipitant of
the bacteria induced by 1 mM IPTG at 16°C. The arrow represents Per
a 10 protein. (B) The purification of Per a 10 expressed in E.
coli. Lane M, protein makers; lanes 1–10, washing with 250 mM
imidazole. The arrow represents Per a 10 protein. |
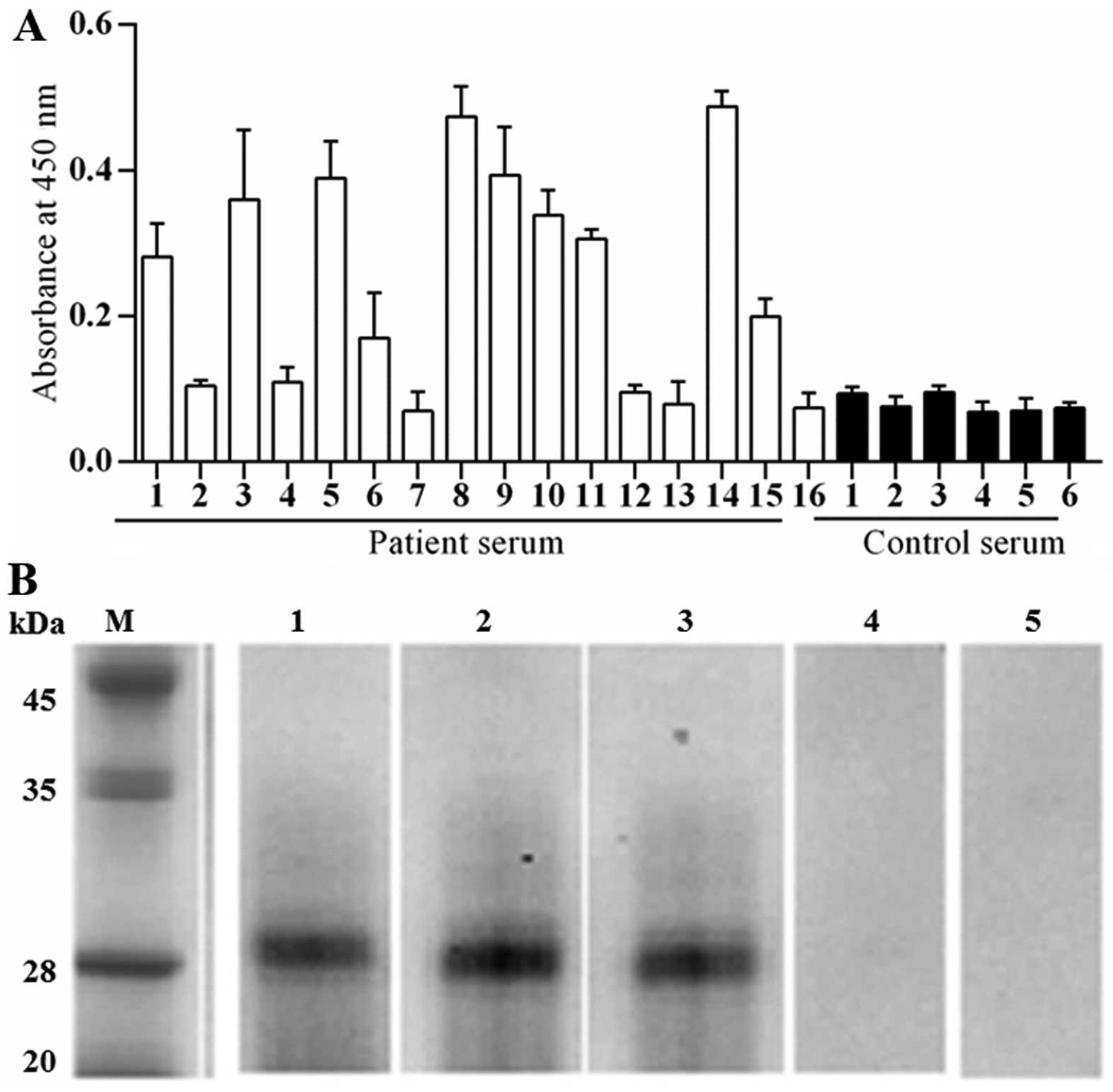
Immuno-reactivity of Per a 10 to IgE
In order to determine the allergenicity of Per a 10,
the ability of Per a 10 to bind IgE in sera from patients with
American CR allergies was determined by a direct ELISA technique.
The sera from patients 1, 3, 5, 6, 8, 9, 11, 14 and 15 showed
positive IgE reactivity to Per a 10. The results revealed that 9
out of 16 (56.3%) of the sera from these patients reacted to Per a
10 (Fig. 2A). The IgE binding
activity of Per a 10 in a representative group of 3 patients and 2
healthy controls was assessed by western blot analysis and the
results are illustrated in Fig.
2B.
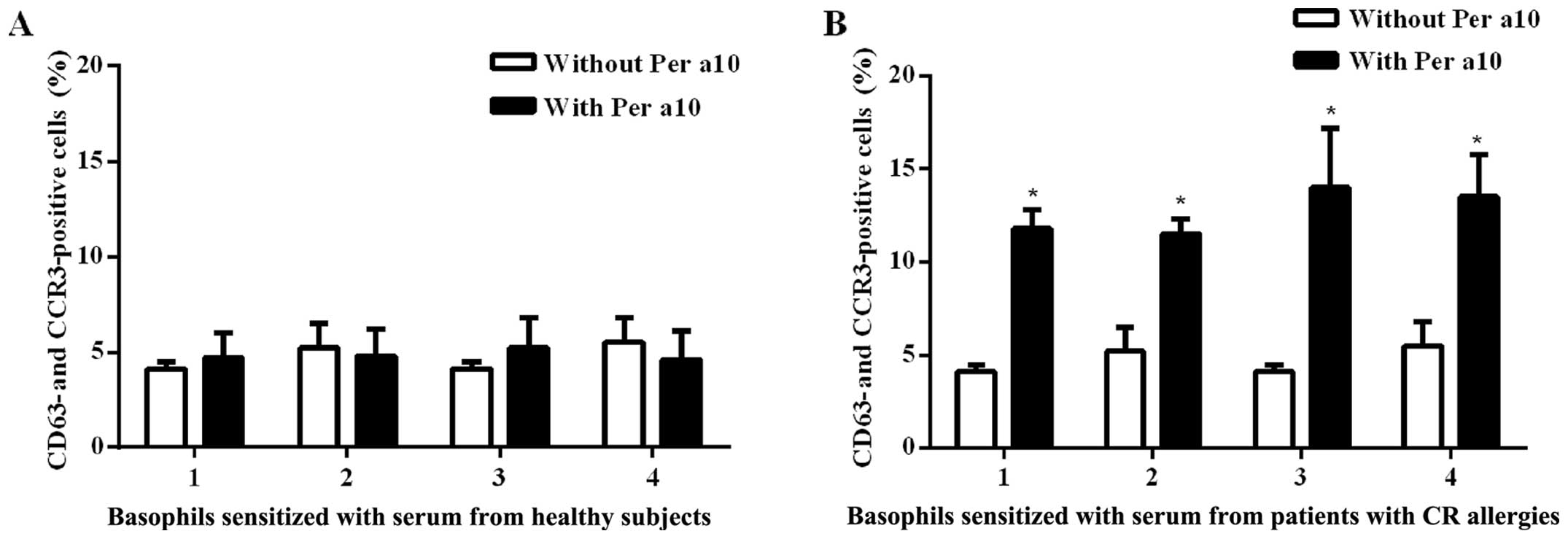
Per a 10 induces human basophil
activation
Per a 10 at 1.0 μg/ml induced an
approximately 2.3-fold increase in the number of CD63 and CCR3
double-positive cells following incubation with passively
sensitized basophils from the sera of patients with American CR
allergies. We found that Per a 10 had no effect on the basophils
sensitized by the sera from the healthy controls (Fig. 3). These results are similar to
those of our previous study on the Per a 9 American CR allergen,
where we had also found that thye basophils sensitized by the sera
from the healthy controls were not affected, whereas the basophils
sensitized by sera from the allergic patients exhibited a 4.2-fold
increase in the number of CD63 and CCR3 double-positive cells
(13).
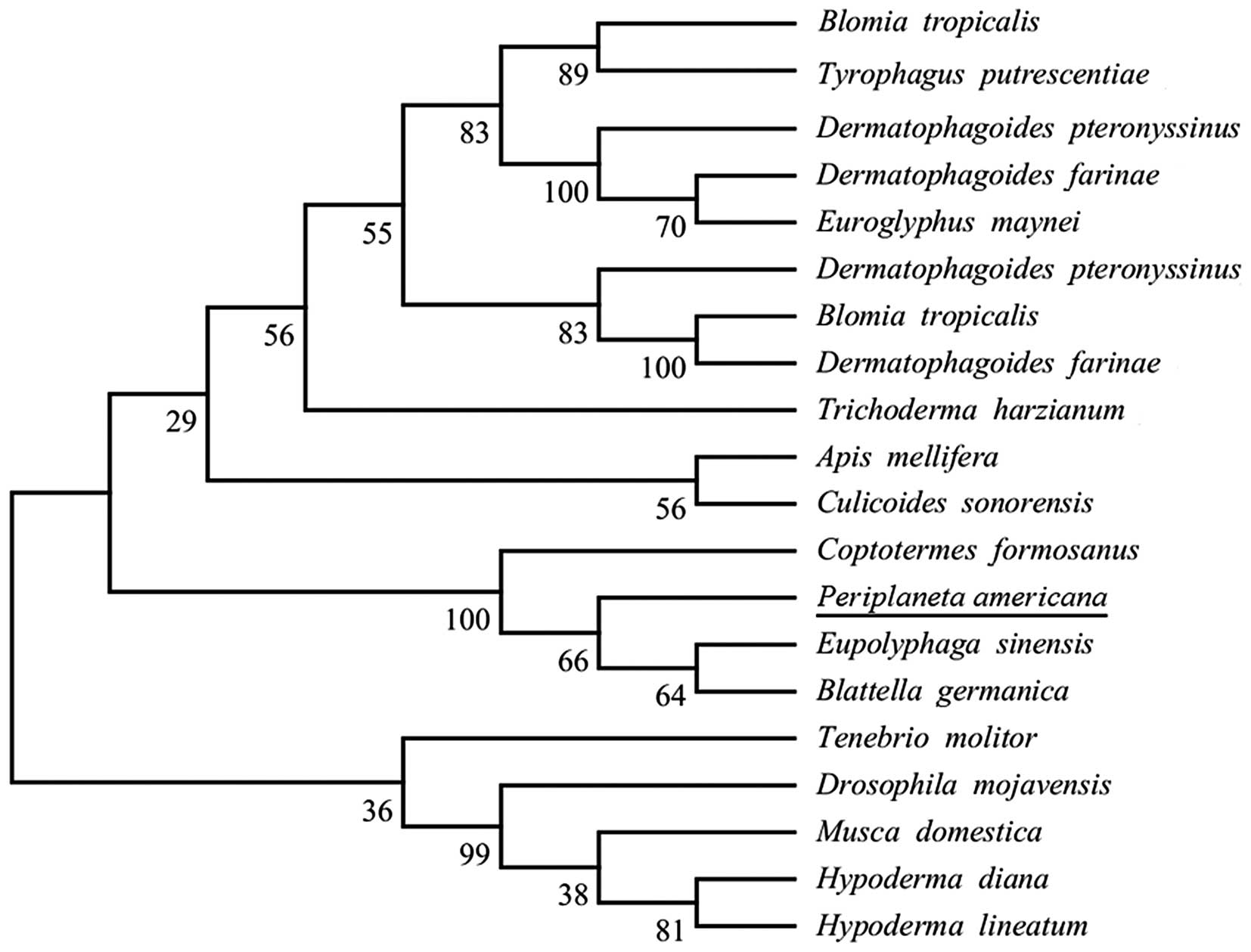
Sequence retrieval and phylogenetic
analysis
Uniprot and tBLASTn were used to search the
homologous sequences of Per a 10. As a result, 20 sequences were
obtained. Moreover, the results of phylogenetic analysis of Per a
10 and its homologous sequences inferred by the ML method are shown
in Fig. 4.
Physiochemical analyses of Per a 10
The primary structure of Per a 10 contained 227
amino acids and the molecular weight was 23295.6. The theoretical
pI was 4.28 and the aliphatic index was 76.04. The GRAVY was 0.001,
indicting that Per a 10 exhibited a hydrophilic character. The
instability index was 34.61 (<40), indicating that the Per a 10
protein was stable.
Structural analysis of Per a 10
Secondary structure prediction with PSIPRED
identified 2 α-helices and 14 β-sheets in Per a 10. PredictProtein
predicted 2 α-helices and 12 β-sheets. Moreover, NetSurfP v1.1 also
predicted 2 α-helices and 12 β-sheets. In total, all secondary
structure prediction yielded the same results: the secondary
structure of Per a 10 contained 2 α-helices and 14 β-sheets and the
results are presented in Table
I.
 | Table IThe predicated secondary structure of
Per a 10. |
Table I
The predicated secondary structure of
Per a 10.
| Secondary
structural prediction methods | α-helices | β-sheets |
|---|
| PSIPRED | 147–153,
217–225 | 5–6, 15–19, 24–32,
35–39, 51–55, 58–60, 65–74, 86–92, 105–106, 117–122, 138–145,
184–187, 190–196, 208–212 |
| NetSurfP
ver1.1 | 147–153,
213–225 | 14–20, 24–30,
35–42, 50–59, 65–74, 87–92, 102–106, 117–122, 138–145, 165–167,
190–196, 209–212, |
| PredictProtein | 148–151,
213–224 | 14–20, 24–32,
35–43, 51–59, 65–79, 87–92, 101–106, 117–122, 137–146, 162–168,
185–197, 209–212 |
| Overall
results | 147–153,
217–225 | 14–20, 24–32,
35–39, 51–55, 58–60, 65–74, 87–92, 117–122, 137–145, 184–187,
185–196, 208–212. |
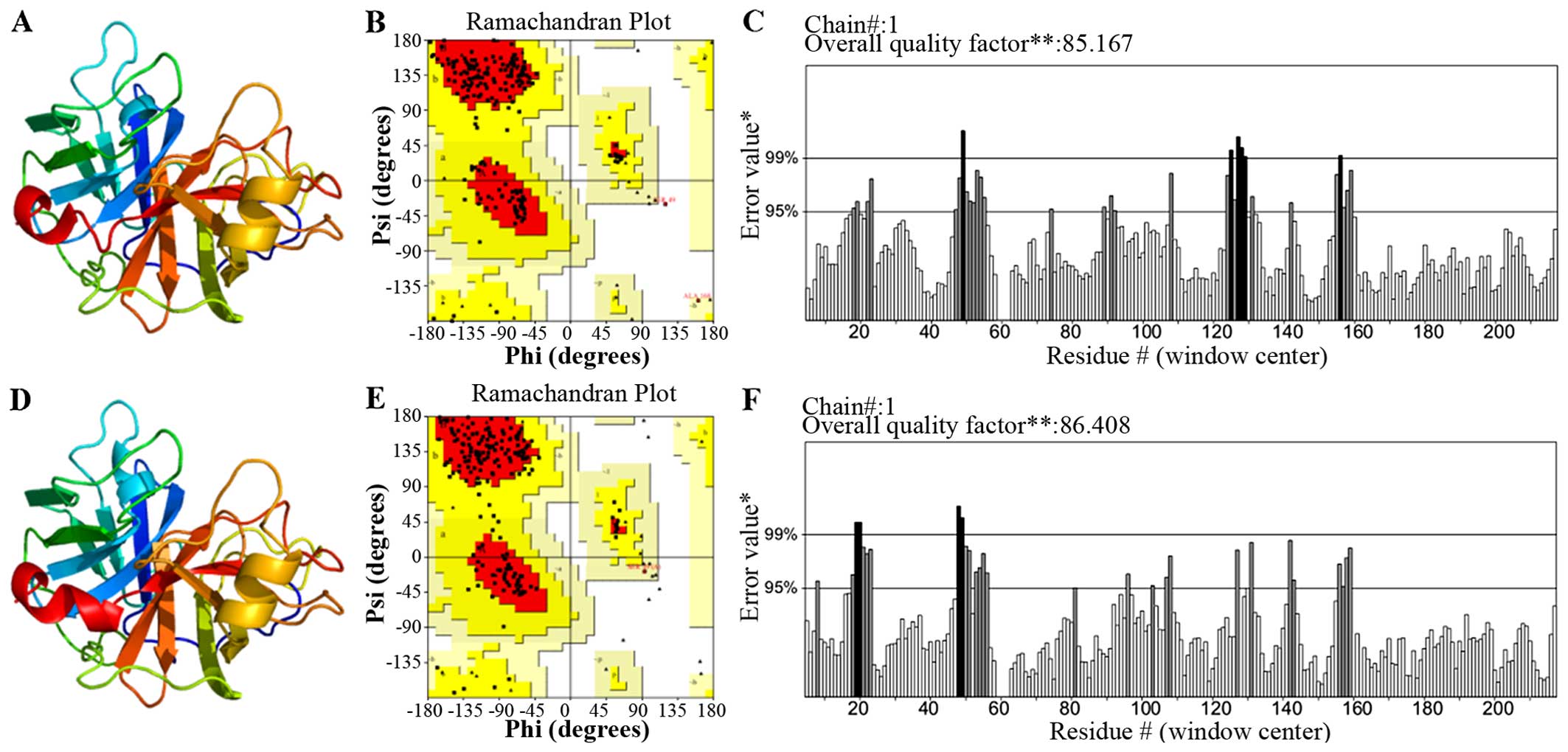
Homology modeling and validation of Per a
10
The search for the proteins Per a 10 with a known
tertiary structure in the PDB yielded pacific salmon anionic
Trypsin (PDB Accession no. 1MBQ; https://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid=1MBQ)
showing the highest sequence identity (44% with Per a 10). As a
result, the 1MBQ template was used for homology modeling. The
tertiary structure of Per a 10 constructed by the homology model is
shown in Fig. 5A. As indicated by
the Ramachandran plot (Fig. 5B),
81.9% of the residues in the model structure were within the most
favored regions, 17.5% of the residues were in the allowed region,
and 0.6% of the residues were in the disallowed region. As
indicated by the ERRAT program, the results (Fig. 5C) revealed that the overall
quality factor was 85.167, which indicated that the structure had a
relatively high resolution. As indicated by the VERIFY_3D program,
the results revealed that 90.54% of the residues had an average 3D
(atomic model)-1D (amino acid sequence) score of >0.2, which is
not sufficient. Thus, we optimized this modeled structure by Chiron
(http://troll.med.unc.edu/chiron/login.php). The
optimized Per a 10 structure is shown in Fig. 6D. The results of the Ramachandran
plot revealed that 79.7% of the residues in the model structure
were within the most favored regions, 20.3% of the residues were in
the allowed region, and 0% of the residues were in the disallowed
region (Fig. 5D). As indicated by
the ERRAT program, the result (Fig.
5E) showed that the overall quality factor is 86.408 which mean
the structure has a relatively high resolution. As indicated by the
VERIFY_3D program (Fig. 5F), the
results indicated that 94.14% of the residues had an average 3D
(atomic model)-1D (amino acid sequence) score of >0.2, and this
results was favourable. Based on these validations, the optimized
Per a 10 structure was adopted for further analysis.
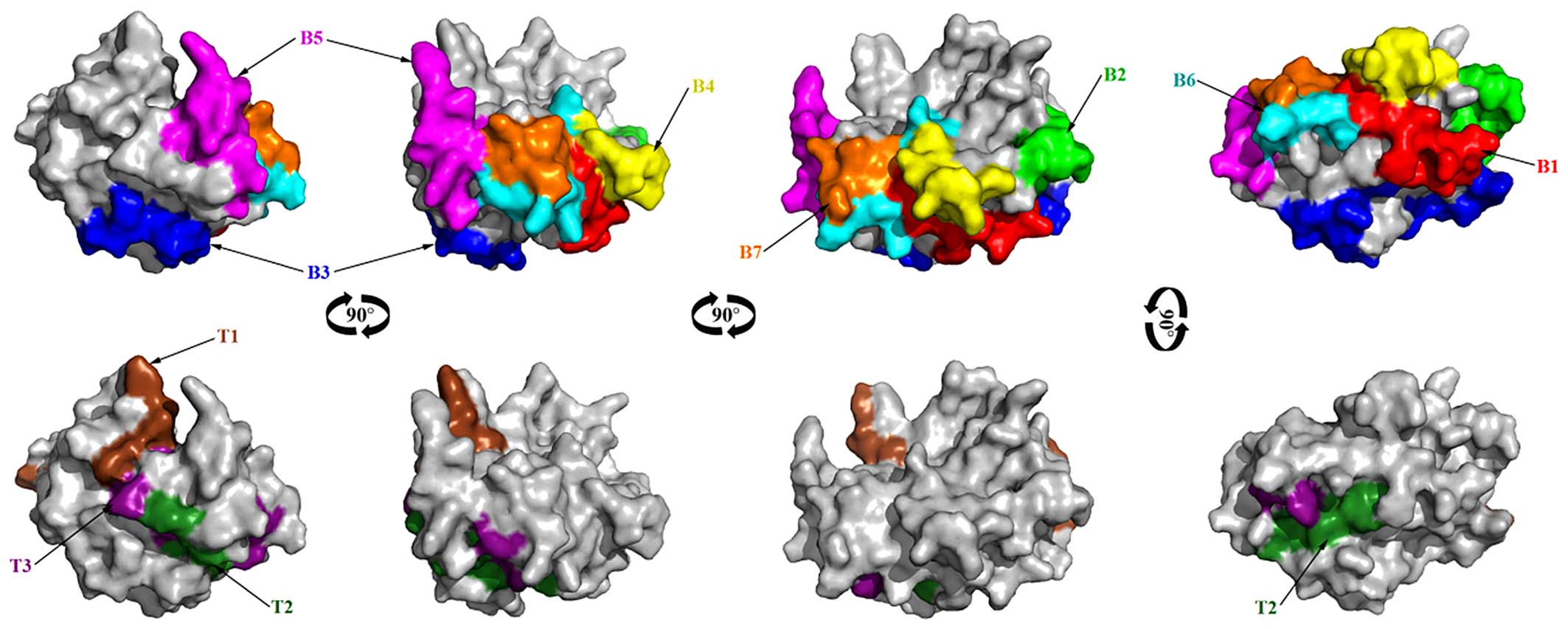
B cell epitope prediction of Per a
10
Bases on surface accessibility, fragment flexibility
and hydrophobicity analysis of Per a 10, the final predicting
regions of Per a 10 by DNAstar were obtained as: 2–10, 55–65,
109–115, 125–133, 147–160, 170–182, 201–208 and 223–227. The
predicted results of the BPAP system were 8–20, 22–51, 63–74,
78–96, 98–110, 113–120, 132–147, 149–156, 162–168, 182–203 and
205–218. The predicted results of the BepiPred 1.0 server were
1–12, 46–67, 74–80, 95–134, 149–160, 169–186, 201–211 and 223–227.
The ultimate results of the 3 immunoinformatics tools finally
predicted 8 peptides (2–12, 55–67, 98–120, 125–133, 149–160,
170–182, 201–208 and 223–227) and these peptides are shown in
Fig. 6.
T cell epitope prediction
For HLA-DR-based T cell epitope prediction of Per a
10, the final predicting regions of HLA-DR 101, HLA-DR 301, HLA-DR
401 and HLA-DR 501 are shown in Table II and the ultimate results of
HLA-DR-based T cell epitope prediction finally predicted 2 peptides
(83–91 and 139–147). For HLA-DQ alleles, the final results of
HLA-DQA10101-DQB10501, HLA-DQA10301-DQB10302,
HLA-DQA10401-DQB10402, and HLA-DQA10102-DQB10602 are also shown in
Table II and the ultimate
results of these 4 methods finally predicted 2 peptides, 84–92 and
162–170. As a result, Per a 10 was predicted to have 3 T cell
epitope sequences, namely 83–92 (a combination of peptide 83–91 and
84–92), 139–147 and 162–170 as shown in Fig. 6.
 | Table IIThe T cell epitope prediction of Per
a 10. |
Table II
The T cell epitope prediction of Per
a 10.
| HLA types | Location of the
prediction results |
|---|
| HLA-DR101 | 1–9, 14–22, 17–25,
18–26, 20–28, 22–30, 24–32, 25–33, 26–34, 28–36, 31–39, 35–43,
42–50, 43–51, 45–53, 53–61, 65–73, 69–77, 70–78, 72–80, 80–88,
81–89, 85–93, 87–95, 89–97, 96–104, 99–107, 102–110, 105–113,
107–115, 117–125, 136–144, 137–145, 184–192, 186–194, 187–195,
191–199, 206–214, 209–217, 210–218, 213–221 |
| HLA-DR301 | 8–16, 72–80,
81–89 |
| HLA-DR401 | 83–91, 87–105,
89–97, 136–144, 139–147, 184–192, 185–193 |
| HLA-DR501 | 18–26, 35–43,
46–54, 52–60, 83–91, 85–93, 89–97, 139–147, 209–217, 210–218 |
|
HLA-DQA10101-DQB10501 | 78–86, 81–89,
184–202 |
|
HLA-DQA10501-DQB10201 | 13–21, 38–46,
78–86, 80–88, 157–165 |
|
HLA-DQA10301-DQB10302 | 82–90, 84–92,
90–98, 162–170 |
|
HLA-DQA10401-DQB10402 | 11–19, 84–92,
162–170, 195–203 |
|
HLA-DQA10102-DQB10602 | 20–28, 25–33,
26–34, 31–39, 34–42, 37–45, 43–51, 47–55, 61–69, 66–74, 67–75,
84–92, 87–95, 90–98, 97–105, 102–110, 132–140, 162–170, 165–173,
190–198, 217–225 |
| The final predicted
T cell epitopes | 83–92, 139–147,
162–170 |
Discussion
To better understand the Per a 10-mediated CR
allergies and with an aim improve the diagnosis and treatment of CR
allergies, we cloned and purified the American CR allergen, Per a
10, in an E. coli expression system. Per a 10 was a serine
proteinase, isolated from American CR extract using a benzamidine
sepharose column and characterized by immunobiochemical methods and
represents a major American CR allergen, with IgE reactivity to 80%
of sera from patients with CR allergies, as shown by skin tests and
immunoblot analysis (14).
Our results showing that 9 out of 16 (56.3%) of sera
from patients with CR allergies react to Per a 10, confirmed that
Per a 10 is a major allergen of the American CR. It has <80% IgE
reactivity in the Thai population (14). In this study, we confirmed the IgE
reactivity of Per a 10 by a basophil activation test, which is a
more advanced technique for the determination of allerginicity of
allergens. It was found that Per a 10 is an active allergen of the
American CR and that it is able to activate basophils which are
sensitized by sera from patients with American CR allergies. The
availability of recombinant allergens has increased our
understanding of IgE-mediated allergies and may help to improve the
diagnosis and treatment of these diseases (44). In our case, recombinant Per a 10
may be used for the further functional and clinical analysis.
Allergen-specific IgE is the key molecule for the
development of allergic symptoms. The identification of B cell
epitopes (IgE-binding epitopes) of an allergen is valuable for the
accurate and safer peptide-based allergen diagnosis and
immunotherapeutic agents. The in silico prediction of B cell
epitopes has already become a useful tool for selecting B cell
epitopes from immunological relevant proteins (45) and correlates well with the
experimental approach (46). Many
algorithms have been developed to predict B cell epitopes on a
protein sequence based on the propensity values of amino acid
properties of hydrophilicity, antigenicity, segmental mobility,
flexibility and accessibility (47). In the present study, we predicted
the B cell linear epitopes of Per a 10 allergens by the 3
sequence-based tools (DNAStar protean system, BPAP and BepiPred 1.0
server) and predicted 8 peptides (2–12, 55–67, 98–120, 125–133,
149–160, 170–182, 201–208 and 223–227) as potential B cell linear
epitopes.
T cell epitopes are principally predicted indirectly
by identifying the binding of peptide fragments to the MHC
complexes. However, the binding grooves of MCH-II molecules are
open at both ends, allowing various lengths of peptides to bind. On
the other hand, the same MHC molecule can accommodate a variety of
binding sequences. These two properties make the development of
accurate predictive algorithms for MHC-class II binding complicated
(40). Some algorithms have
substantially improved their accuracy to predict T cell epitopes of
immunological relevant proteins. However, most algorithms target
HLA-DR molecules, but not HLA-DP and HLA-DQ molecules, even though
they are important for antigen presentation. In this study,
NetMHCIIpan-3.0 was applied for HLA-DR-based T cell epitope
prediction of Per a 10 and finally predicted 2 peptides (83–91 and
139–147).
NetMHCII-2.2 was used for HLA-DQ-based T cell
epitope prediction of Per a 10 and predicted 3 T cell epitope
sequences, including 84–92, 139–147 and 162–170. As a result, the
ultimate consensus epitope results were obtained by combining the
results of the HLA-DR-based T cell epitope and HLA-DQ-based T cell
epitope. As a result, Per a 10 was predicted to have 3 T cell
epitope sequences, namely 83–92, 139–147 and 162–170.
Acknowledgments
This study was sponsored by grants from the Special
Fund for Forestry-Scientific Research in the Public Interest (no.
201304103), grants from the National Natural Science Foundation of
China (nos. 81571568, 81400037, 31340073 and 81273274); Jiangsu
Province's Key Provincial Talents Program (no. RC201170); the
Priority Academic Program Development of Jiangsu Higher Education
Institutions (PAPD); the Six Talents Peak projects of Jiangsu
Province (to J.-F.W.).
References
|
1
|
Bernton HS and Brown H: Insect allergy
preliminary studies of the cockroach. J Allergy. 35:506–513. 1964.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Arruda LK, Vailes LD, Ferriani VPL, Santos
AB, Pomés A and Chapman MD: Cockroach allergens and asthma. J
Allergy Clin Immunol. 107:419–428. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sun BQ, Lai XX, Gjesing B, Spangfort MD
and Zhong NS: Prevalence of sensitivity to cockroach allergens and
IgE cross-reactivity between cockroach and house dust mite
allergens in Chinese patients with allergic rhinitis and asthma.
Chin Med J (Engl). 123:3540–3544. 2010.
|
|
4
|
Thangam Sudha V, Arora N, Sridhara S, Gaur
SN and Singh BP: Biopotency and identification of allergenic
proteins in Periplaneta americana extract for clinical
applications. Biologicals. 35:131–137. 2007. View Article : Google Scholar
|
|
5
|
He S, Zhang Z, Zhang H, Wei J, Yang L,
Yang H, Sun W, Zeng X and Yang P: Analysis of properties and
proinflammatory functions of cockroach allergens Per a 1.01s. Scand
J Immunol. 74:288–295. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu HQ, Liu ZG, Ran PX, Zhou ZW and Gao B:
Expression, purification, and immunological characterization of Cr
PI. Protein Pept Lett. 14:881–885. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mindykowski B, Jaenicke E, Tenzer S, Cirak
S, Schweikardt T, Schild H and Decker H: Cockroach allergens Per a
3 are oligomers. Dev Comp Immunol. 34:722–733. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tan YW, Chan SL, Ong TC, Yit Y, Tiong YS,
Chew FT, Sivaraman J and Mok YK: Structures of two major allergens,
Bla g 4 and Per a 4, from cockroaches and their IgE binding
epitopes. J Biol Chem. 284:3148–3157. 2009. View Article : Google Scholar
|
|
9
|
Wei JF, Yang H, Li D, Gao P and He S:
Preparation and identification of Per a 5 as a novel American
cockroach allergen. Mediators Inflamm. 2014:5914682014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen H, Yang HW, Wei JF and Tao AL: In
silico prediction of the T-cell and IgE-binding epitopes of Per a 6
and Bla g 6 allergens in cockroaches. Mol Med Rep. 10:2130–2136.
2014.PubMed/NCBI
|
|
11
|
Yang H, Kong X, Wei J, Liu C, Song W,
Zhang W, Wei W and He S: Cockroach allergen Per a 7 down-regulates
expression of Toll-like receptor 9 and IL-12 release from P815
cells through PI3K and MAPK signaling pathways. Cell Physiol
Biochem. 29:561–570. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sookrung N, Chaicumpa W, Tungtrongchitr A,
Vichyanond P, Bunnag C, Ramasoota P, Tongtawe P, Sakolvaree Y and
Tapchaisri P: Periplaneta americana arginine kinase as a major
cockroach allergen among Thai patients with major cockroach
allergies. Environ Health Perspect. 114:875–880. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang H, Chen H, Jin M, Xie H, He S and Wei
JF: Molecular cloning, expression, IgE binding activities and in
silico epitope prediction of Per a 9 allergens of the American
cockroach. Int J Mol Med. 38:1795–1805. 2016.
|
|
14
|
Sudha VT, Arora N, Gaur SN, Pasha S and
Singh BP: Identification of a serine protease as a major allergen
(Per a 10) of Periplaneta americana. Allergy. 63:768–776. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fang Y, Long C, Bai X, Liu W, Rong M, Lai
R and An S: Two new types of allergens from the cockroach,
Periplaneta americana. Allergy. 70:1674–1678. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Goel C, Kalra N, Dwarakanath BS, Gaur SN
and Arora N: Per a 10 protease activity modulates CD40 expression
on dendritic cell surface by nuclear factor-kappaB pathway. Clin
Exp Immunol. 180:341–351. 2015. View Article : Google Scholar :
|
|
17
|
Goel C, Govindaraj D, Singh BP, Farooque
A, Kalra N and Arora N: Serine protease Per a 10 from Periplaneta
americana bias dendritic cells towards type 2 by upregulating CD86
and low IL-12 secretions. Clin Exp Allergy. 42:412–422. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
He W, Jimenez F, Martinez H, Harper NL,
Manoharan MS, Carrillo A, Ingale P, Liu YG, Ahuja SS, Clark RA, et
al: Cockroach sensitization mitigates allergic rhinoconjunctivitis
symptom severity in patients allergic to house dust mites and
pollen. J Allergy Clin Immunol. 136:658–666. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bassirpour G and Zoratti E: Cockroach
allergy and allergen-specific immunotherapy in asthma: Potential
and pitfalls. Curr Opin Allergy Clin Immunol. 14:535–541. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nielsen M, Lund O, Buus S and Lundegaard
C: MHC class II epitope predictive algorithms. Immunology.
130:319–328. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sookrung N, Khetsuphan T, Chaisri U,
Indrawattana N, Reamtong O, Chaicumpa W and Tungtrongchitr A:
Specific B-cell epitope of Per a 1: A major allergen of american
cockroach (Periplaneta americana) and anatomical localization.
Allergy Asthma Immunol Res. 6:325–332. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lee MF, Chang CW, Song PP, Hwang GY, Lin
SJ and Chen YH: IgE-binding epitope mapping and tissue localization
of the major American cockroach allergen Per a 2. Allergy Asthma
Immunol Res. 7:376–383. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu CH, Lee MF and Tseng CY: IgE-binding
epitopes of the American cockroach Per a 3 allergen. Allergy.
58:986–992. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
An S, Chen L, Wei JF, Yang X, Ma D, Xu X,
Xu X, He S, Lu J and Lai R: Purification and characterization of
two new allergens from the venom of Vespa magnifica. PLoS One.
7:e319202012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
An S, Ma D, Wei JF, Yang X, Yang HW, Yang
H, Xu X, He S and Lai R: A novel allergen Tab y 1 with inhibitory
activity of platelet aggregation from salivary glands of
horseflies. Allergy. 66:1420–1427. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sanz ML, Gamboa PM, Antépara I, Uasuf C,
Vila L, Garcia-Avilés C, Chazot M and De Weck AL: Flow cytometric
basophil activation test by detection of CD63 expression in
patients with immediate-type reactions to betalactam antibiotics.
Clin Exp Allergy. 32:277–286. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sainte-Laudy J, Vallon C and Guérin JC:
Analysis of membrane expression of the CD63 human basophil
activation marker. Applications to allergologic diagnosis. Allerg
Immunol (Paris). 26:211–214. 1994.In French.
|
|
28
|
Yang L, Luo Y and Wei J: Integrative
genomic analyses on Ikaros and its expression related to solid
cancer prognosis. Oncol Rep. 24:571–577. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang L, Luo Y, Wei J and He S: Integrative
genomic analyses on IL28RA, the common receptor of interferon-λ1,
-λ2 and -λ3. Int J Mol Med. 25:807–812. 2010.PubMed/NCBI
|
|
30
|
Yang L, Wei J and He S: Integrative
genomic analyses on interferon-λs and their roles in cancer
prediction. Int J Mol Med. 25:299–304. 2010.PubMed/NCBI
|
|
31
|
Wang M, Wei X, Shi L, Chen B, Zhao G and
Yang H: Integrative genomic analyses of the histamine H1 receptor
and its role in cancer prediction. Int J Mol Med. 33:1019–1026.
2014.PubMed/NCBI
|
|
32
|
Ding Z, Yang HW, Xia TS, Wang B and Ding
Q: Integrative genomic analyses of the RNA-binding protein, RNPC1,
and its potential role in cancer prediction. Int J Mol Med.
36:473–484. 2015.PubMed/NCBI
|
|
33
|
Li X, Yang HW, Chen H, Wu J, Liu Y and Wei
JF: In silico prediction of T and B cell epitopes of Der f 25 in
Dermatophagoides farinae. Int J Genomics. 2014:4839052014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
McGuffin LJ, Bryson K and Jones DT: The
PSIPRED protein structure prediction server. Bioinformatics.
16:404–405. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Petersen B, Petersen TN, Andersen P,
Nielsen M and Lundegaard C: A generic method for assignment of
reliability scores applied to solvent accessibility predictions.
BMC Struct Biol. 9:512009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Laskowski RA, MacArthur MW and Thornton
JM: Validation of protein models derived from experiment. Curr Opin
Struct Biol. 8:631–639. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Maganti L, Manoharan P and Ghoshal N:
Probing the structure of Leishmania donovani chagasi DHFR-TS:
Comparative protein modeling and protein-ligand interaction
studies. J Mol Model. 16:1539–1547. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Burland TG: DNASTAR's Lasergene sequence
analysis software. Methods Mol Biol. 132:71–91. 2000.
|
|
39
|
Larsen JE, Lund O and Nielsen M: Improved
method for predicting linear B-cell epitopes. Immunome Res.
2(2)2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang X and Yu X: An introduction to
epitope prediction methods and software. Rev Med Virol. 19:77–96.
2009. View Article : Google Scholar
|
|
41
|
Zheng LN, Lin H, Pawar R, Li ZX and Li MH:
Mapping IgE binding epitopes of major shrimp (Penaeus monodon)
allergen with immunoinformatics tools. Food Chem Toxicol.
49:2954–2960. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Karosiene E, Rasmussen M, Blicher T, Lund
O, Buus S and Nielsen M: NetMHCIIpan-3.0, a common pan-specific MHC
class II prediction method including all three human MHC class II
isotypes, HLA-DR, HLA-DP and HLA-DQ. Immunogenetics. 65:711–724.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Nielsen M and Lund O: NN-align. An
artificial neural network-based alignment algorithm for MHC class
II peptide binding prediction. BMC Bioinformatics. 10:2962009.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Schmidt M and Hoffman DR: Expression
systems for production of recombinant allergens. Int Arch Allergy
Immunol. 128:264–270. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li GF, Wang Y, Zhang ZS, Wang XJ, Ji MJ,
Zhu X, Liu F, Cai XP, Wu HW and Wu GL: Identification of
immunodominant Th1-type T cell epitopes from Schistosoma japonicum
28 kDa glutathione-S-transferase, a vaccine candidate. Acta Biochim
Biophys Sin (Shanghai). 37:751–758. 2005. View Article : Google Scholar
|
|
46
|
Nair S, Kukreja N, Singh BP and Arora N:
Identification of B cell epitopes of alcohol dehydrogenase allergen
of Curvularia lunata. PLoS One. 6:e200202011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Pomés A: Relevant B cell epitopes in
allergic disease. Int Arch Allergy Immunol. 152:1–11. 2010.
View Article : Google Scholar :
|