Introduction
Liver regeneration refers to the liver repair
process that occurs following the loss of part of the liver due to
surgery, trauma, poisoning, infection and necrosis, or liver
transplantation. When liver regeneration occurs, hepatocytes in the
G0 phase are stimulated by various feedback signals to enter a
period of rapid growth. Liver regeneration has valuable
implications for hepatobiliary surgeries, such as the resection of
a liver mass, liver transplantation and split liver
transplantation, as the above-mentioned surgical procedures depend
largely on the ability of the liver to regenerate. Poor liver
regeneration may lead to the death of the patient, for example, via
small-for-size syndrome (SFSS) (1–4).
For that reason, understanding the mechanisms of liver regeneration
has great implications for clinical practice. A series of
interacting factors, including inflammatory cytokines (5), hepatocyte growth factor (HGF)
(6), cell cycle regulators,
cyclin-dependent kinases and metabolic regulators control the
hepatocyte cell cycle to ensure the rapid and orderly progression
of liver regeneration (7).
Studies have indicated that microRNAs (miRNAs or miRs) play a key
role in liver regeneration (8–10).
Despite the identification of many cytokines, miRNAs and growth
factors that regulate the expression of genes controlling
proliferation during liver regeneration (11), the exact molecular mechanisms
which are responsible for liver regeneration remain unclear, thus
prompting our interest to investigate the molecular mechanisms
underlying liver regeneration.
In recent years, the field of RNA biology has been
dominated by the study of miRNAs; however, several major biological
processes are also regulated by long non-coding RNAs (lncRNAs),
including carcinogenesis, development and differentiation (12,13). lncRNA molecules are >200
nucleotides in length and do not encode proteins. Studies have
reported that some lncRNAs, including lncRNA H19 and lncRNA LALR1,
play key roles during liver regeneration (14,15). lncRNA H19 promotes hepatocyte
proliferation during liver regeneration in both the rat and mouse
(15). Improving the
effectiveness of current regeneration techniques may help to
identify a potential therapeutic target to stimulate liver
regeneration.
Metastasis-associated lung adenocarcinoma transcript
1 (MALAT1) is one of the most widely studied lncRNAs and is
frequently overexpressed in malignant tumors; therefore, MALAT1 is
frequently regarded as a biomarker for a variety of tumors. A
previous study demonstrated that MALAT1 localizes in dynamic
structures known as nuclear speckles, which are key elements in the
collection and recruitment of splicing factors (16). Researchers have also found that
MALAT1 plays a variety of biological functions in hematopoietic
cells, including the regulation of the cell cycle (17). However, the biological role of the
lncRNA MALAT1 in liver regeneration and its mechanisms of action
regarding the regulation of hepatocyte proliferation remain
unclear.
In this study, we focused on the mechanisms
underlying the MALAT1-mediated modulation of hepatocyte
proliferation. We investigated the role of MALAT1 in the regulation
of liver regeneration with a murine model of 2/3 partial
hepatectomy (PH) and an in vitro hepatocyte proliferation
assay. The comprehensive dynamic changes in MALAT1 expression were
examined at multiple time points (including 0, 2, 6, 12, 15, 18,
24, 36, 48, 72, 120, 144 and 168 h) following 2/3 PH; we then aimed
to elucidate the mechanisms through which MALAT1 regulates liver
regeneration in the mouse. We found that MALAT1 promoted hepatocyte
proliferation and increased the expression of cyclin D1 by
activating the Wnt/β-catenin pathway. We also found that the
relative expression level of p53, which is a negative regulator of
MALAT1, was the opposite of MALAT1 expression following 2/3 PH. Our
study may have identified a potential therapeutic target for use
during liver regeneration.
Materials and methods
Animals and model of 2/3 PH
A total of 60 male BALB/c mice (8–12 weeks old,
weighing 23±2 g) were purchased from the Central Laboratory of
Animal Science, Wuhan University (Wuhan, China) and were maintained
in a specific pathogen-free facility with a 12-h light/dark cycle
and were fed standard laboratory chow with free access to water
ad arbitrium. All the animal protocols used in the study
were reviewed and approved by the Animal Experiment Center of Wuhan
University/Animal Biosafety Level-III Laboratory. 2/3 PH was
performed on 8-12-week-old mice using a previously described method
(18). Following a laparotomy,
the left and median liver lobes were resected in two steps. Five
mice were used for each 2/3 PH time point (0, 2, 6, 12, 15, 18, 24,
36, 48, 72, 120 and 168 h). The end of surgery was considered 0 h.
All the partial hepatectomies were performed in the early morning
hours. The mice were sacrificed (by cervical dislocation) at
defined time points following PH. The mice were sacrificed under
isoflurane anesthesia as previously described (18). The remaining parts of the livers
were surgically removed, cleaned with saline solution and were
snap-frozen in liquid nitrogen for RNA and protein analysis and
kept at −80°C until further use or fixed in 4% paraformaldehyde for
histological analysis and processed for immunohistochemistry. For
histological analysis, after being fixed in 4% paraformaldehyde for
48 h, the tissues were embedded in paraffin. The total body weight
was measured, and the remnant and regenerated liver tissues were
resected and weighed. The acquired data were expressed as a
percentage of the ratio between the remnant liver weight (A)
divided by the total body weight (B) ×100 (liver-to-body weight
ratio (%) = A/B ×100).
Histological analysis
At different time points following 2/3 PH, the
animals were sacrificed and the liver tissues were harvested. Liver
tissues were obtained from the remnant livers and then fixed
immediately in 10% buffered formalin phosphate solution, embedded
in paraffin and were then cut into 5-mm-thick sections. The
sections were stained with hematoxylin and eosin (H&E). The
H&E staining kit was obtained from Service Bio Co., Ltd.
(Wuhan, China Cat. no. G1005). All tissue sections were examined
with an Olympus BH-2 microscope (Olympus Optical Co. Ltd., Beijing,
China). Motic Images 2000 (Motic China Group Co. Ltd, Guangzhou,
China) was used for the characterization of histopathological
changes.
ELISA
The mice were sacrificed and blood samples were
collected via the post-orbital venous plexus at the mentioned
observation time points. Serum samples were separated and stored at
−80°C until further use. The serum levels of alanine
aminotransferase (ALT), aspartate aminotransferase (AST) and
albumin (ALB) were measured using respective ELISA kits (Nanjing
Jiancheng Biological Technology, Inc., Nanjing, China) as
previously described (19).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from the frozen tissues or
cells using TRIzol reagent (Invitrogen Life Technologies, Carlsbad,
CA, USA) according to the manufacturer's instructions. For RT-qPCR,
a reverse transcription kit (Takara Biotechnology Co., Ltd. Dalian,
China) was used to synthesize cDNA. A SYBR-Green PCR kit (Toyobo,
Osaka, Japan) was used to complete the RT-qPCR analyses. PCR
reactions were performed on a CFX96 system (Bio-Rad CFX96; Bio-Rad
Laboratories, Inc., Hercules, CA, USA). The results of RT-qPCR were
normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and
the 2−ΔΔCT values were normalized to the GAPDH levels.
All the RT-qPCR experiments were repeated at least 3 times. The
primer sequences used in this study are listed in Table I.
 | Table IPrimer and target sequences used in
this study. |
Table I
Primer and target sequences used in
this study.
| Genes | Sequences or target
sequences (5′→3′) |
|---|
| MALAT1-F |
TGCAGTGTGCCAATGTTTCG |
| MALAT1-R |
GGCCAGCTGCAAACATTCAA |
| GAPDH-F |
GGTGAAGGTCGGTGTGAACG |
| GAPDH-R |
CTCGCTCCTGGAAGATGGTG |
| HGF-F |
TCATTGGTAAAGGAGGCA |
| HGF-R |
GTCACAGACTTCGTAGCG |
| β-catenin-F |
ATGGCTTGGAATGAGACT |
| β-catenin-R |
TGAGGTCCTGGGCGTGT |
| Cyclin D1-F |
GAGGAGCAGAAGTGCGAAGA |
| Cyclin D1-R |
GCCGGATAGAGTTGTCAGTGTAG |
| Axin1-F |
GCTGCTATTGGAGACTGCT |
| Axin1-FR |
GTACCCGCCCATTGACT |
| GSK-3β-F |
TTATTTGACCGCATAGTTC |
| GSK-3β-R |
AAGCACCTGACTTTCCTC |
| APC-F |
CCTGTGGCAAGGAAACC |
| APC-R |
CTCGCTGAGCATCATCTGT |
| p53-F |
CAGCCCCCTCTCTGAGTAGT′ |
| p53-R |
ACCCTATGAGGGCCCAAGAT |
| MALAT1-siRNA-1 |
GCUCAGGACUUUGCAUAUATTUAUAUGCAAAGUCCUGAGCTT |
| MALAT1-siRNA-2 |
GCAGAAGAGUUGCUUCAUUTTAAUGAAGCAACUCUUCUGCTT |
| MALAT1-siRNA-3 |
GCGGAAUUGCUGGUAGUUUTTAAACUACCAGCAAUUCCGCTT |
|
MALAT1-siRNA-scramble |
UUCUCCGAACGUGUCACGUTTACGUGACACGUUCGGAGATTT |
Cell culture and transfection
NCTC 1469 and BNL CL.2 cells were obtained from the
China Center for Type Culture Collection (Wuhan, China). The NCTC
1469 cells were cultured with Dulbecco6s modified Eagle's medium
(DMEM H-21, 4.5 g/l glucose) supplemented with 15% horse serum. BNL
CL.2 cells were cultured with DMEM supplemented with 10% fetal
bovine serum (FBS). The growth media used in the present study
contained 5 U/ml penicillin and 5 μg/ml streptomycin
sulfate. Full-length MALAT1 (GenBank: NR_002847.2) was cloned into
the pcDNA3.1 vector (Invitrogen Life Technologies). An empty vector
(pcDNA3.1) was used as a control. siRNAs against MALAT1 were
designed and synthesized by Beijing View Solid Biotechnology
(Beijing, China). The siRNA target sequences used in this study are
listed in Table I. The MALAT1
expression levels were detected by RT-qPCR. Scramble siRNA was used
as a control. Transfection was conducted using a LipoJet™ in
vitro DNA and siRNA transfection kit (both from SignaGen
Laboratories, Gaithersburg, MD, USA) according to the
manufacturer's instructions. The hepatocytes were fixed with 4%
paraformaldehyde 48 h after transfection for further analysis.
NCTC 1469 cells were treated with HGF (Cat. no.
#315-23; PeproTech, Rocky Hill, NJ, USA) using a concentration
range of 5–20 ng/ml. It is recommended to reconstitute the
lophilized HGF in sterile 18MΩ-cm H2O not less than 100
μg/ml, which can then be further diluted to other aqueous
solutions.
Flow cytometric analysis
The cells transfected with siRNA or the pcDNA3.1
vector were collected at 48 h post-transfection. Following
double-labeling with Annexin V-FITC and propidium iodide (PI),
apoptosis was analyzed using a flow cytometer (FACScan®;
BD Biosciences, Franklin Lakes, NJ, USA) equipped with CellQuest
3.3 software. For the cell cycle analysis, 1×106 cells
were plated into a 60-mm dish and incubated for 24 h. The cells
were subsequently stained with 1 ml of staining solution [PI (50
μg/ml) and RNase A (20 μg/ml)] for 15 min using the
Cycletest™ PLUS DNA reagent kit (BD Biosciences) according to the
manufacturer's instructions.
Western blot analysis
The liver tissue samples and hepatocytes were
homogenized in lysis buffer (Beyotime Institute of Biotechnology,
Haimen, China) and incubated for 30 min on ice. For the detection
of β-catenin, cytoplasmic and nuclear protein extracts were
prepared from the cells using the NE-PER Nuclear and Cytoplasmic
Extraction Reagent kit (Cat. no. 78835; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) according to the manufacturer's
instructions. Total protein was electrophoresed by sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then
transferred onto polyvinylidene fluoride (PVDF) membranes
(Millipore, Billerica, MA, USA). The membranes were incubated with
the corresponding primary antibodies overnight at 4°C. The
following day, the membranes were incubated with secondary
antibodies [goat anti-rabbit IgG (H+L)-HRP (LK2001; Sungene Biotech
Co., Ltd., Tianjin, China)] for 1–2 h. Proteins were detected using
the ECL immunoblotting kit (Beyotime Institute of Biotechnology).
The fluorescent bands were observed in a dark room with infrared
light. The above procedure was performed according to the
manufacturer's instructions. The information for the antibodies is
listed in Table II.
 | Table IIInformation regarding the antibodies
used in this study. |
Table II
Information regarding the antibodies
used in this study.
| Antibody | WB | IHC | IF | Specificity | Company |
|---|
| GAPDH
(10494-1-AP) | 1:5,000 | | | Rabbit
polyclonal | Proteintech |
| β-catenin (no.
8480) | 1:1,000 | | 1:100 | Rabbit
monoclonal | Cell Signaling
Technology |
| Cyclin D1 (no.
2978) | 1:1,000 | | | Rabbit
monoclonal | Cell Signaling
Technology |
| Axin1 (no.
2087) | 1:1,000 | | | Rabbit
monoclonal | Cell Signaling
Technology |
| c-Myc (sc-788) | 1:200 | | | Rabbit
polyclonal | Santa Cruz
Biotechnology |
| Cyclin B1
(55004-1-AP) | 1:2,000 | | | Rabbit
polyclonal | Proteintech |
| Cyclin E1
(11554-1-AP) | 1:1,000 | | | Rabbit
polyclonal | Proteintech |
| GSK-3β (no.
12456) | 1:1,000 | | | Rabbit
polyclonal | Cell Signaling
Technology |
| APC (sc-896) | 1:200 | | | Rabbit
polyclonal | Santa Cruz
Biotechnology |
| p53 (sc-6243) | 1:200 | 1:200 | | Rabbit
polyclonal | Santa Cruz
Biotechnology |
| PCNA
(10205-2-AP) | 1:2,000 | 1:200 | | Rabbit
polyclonal | Proteintech |
| Histone H3 (no.
4499) | 1:2,000 | | | Rabbit
monoclonal | Cell Signaling
Technology |
Cell proliferation assay
A Cell Counting Kit-8 kit (CCK-8; Dojindo, Kumamoto,
Japan) was used for the cell proliferation assays. Approximately
3–5×104 cells were plated in triplicate in 96-well
plates and cultured in growth medium. The plate was incubated at
37°C and 5% CO2 for an appropriate length of time (4–6
h) until cell adherence. Ten microliters of CCK-8 solution were
then added to each well of the plate. The absorbance was measured
at 450 nm using a microplate reader (168-1000XC; Bio-Rad
Laboratories, Inc.) at the indicated time points.
5-Ethynyl-2′-deoxyuridine (EdU)
incorporation assay
The EdU incorporation assay was performed using an
EdU Apollo DNA in vitro kit (C10310-1; Guangzhou RiboBio
Co.,Ltd., Guangzhou, China) according to the manufacturer's
instructions. The hepatocytes were stained with EdU 48 h following
transfection. First, the cells were incubated with 100 ml of 50 mM
EdU/well for 2 h. The cells were then fixed with 4%
paraformaldehyde for 15 min at room temperature. Following fixation
and washing with phosphate-buffered saline (PBS), the cells were
incubated with 50 μl of 2 mg/ml glycine for 5 min. One
hundred microliters of permeabilization buffer (PBS containing 0.5%
Triton X-100) was added to each well, and the cells were incubated
with 100 μl 1X Apollo solution for 30 min at room
temperature in the dark. Then, 100 μl 1X Hoechst 33342
solution was added dropwise into each cell climbing slice for 15
min. The cell climbing slices were washed, dried, mounted and then
observed under a fluorescence microscope (Olympus BX53; Olympus,
Tokyo, Japan).
Immunohistochemistry (IHC) and
immunofluorescence (IF)
Paraffin-embedded tissue was cut into
4-μm-thick sections and processed for IHC in accordance with
a previously described protocol (20). For IF, the cells were fixed with
4% paraformaldehyde, permeabilized using 0.2% Triton X-100 and
incubated with primary and secondary antibodies according to the
manufacturer's instructions. The cells were counterstained with
4′,6-diamidino-2-phenylindole (DAPI) (Calbiochem, San Diego,
CA, USA) and imaged using a confocal laser scanning microscope
(Olympus FV1000; Olympus).
Statistical analysis
Quantitative data are presented as the means ±
standard deviation (SD) from at least 3 independent experiments.
Statistical significance was assessed using the Student's t-test,
χ2 test or Fisher's exact test, which were performed
using SPSS 22.0 statistical software (IBM SPSS, Inc., Chicago, IL,
USA). Differences were considered statistically significant when
P-values were <0.05.
Results
MALAT1 is upregulated during liver
regeneration
To determine the impact of MALAT1 on liver
regeneration, we first generated mouse model of 2/3 PH. To assess
rehabilitation from acute liver injury, the serum levels of ALT,
AST and ALB were determined 0, 24, 36, 48, 72 and 168 h following
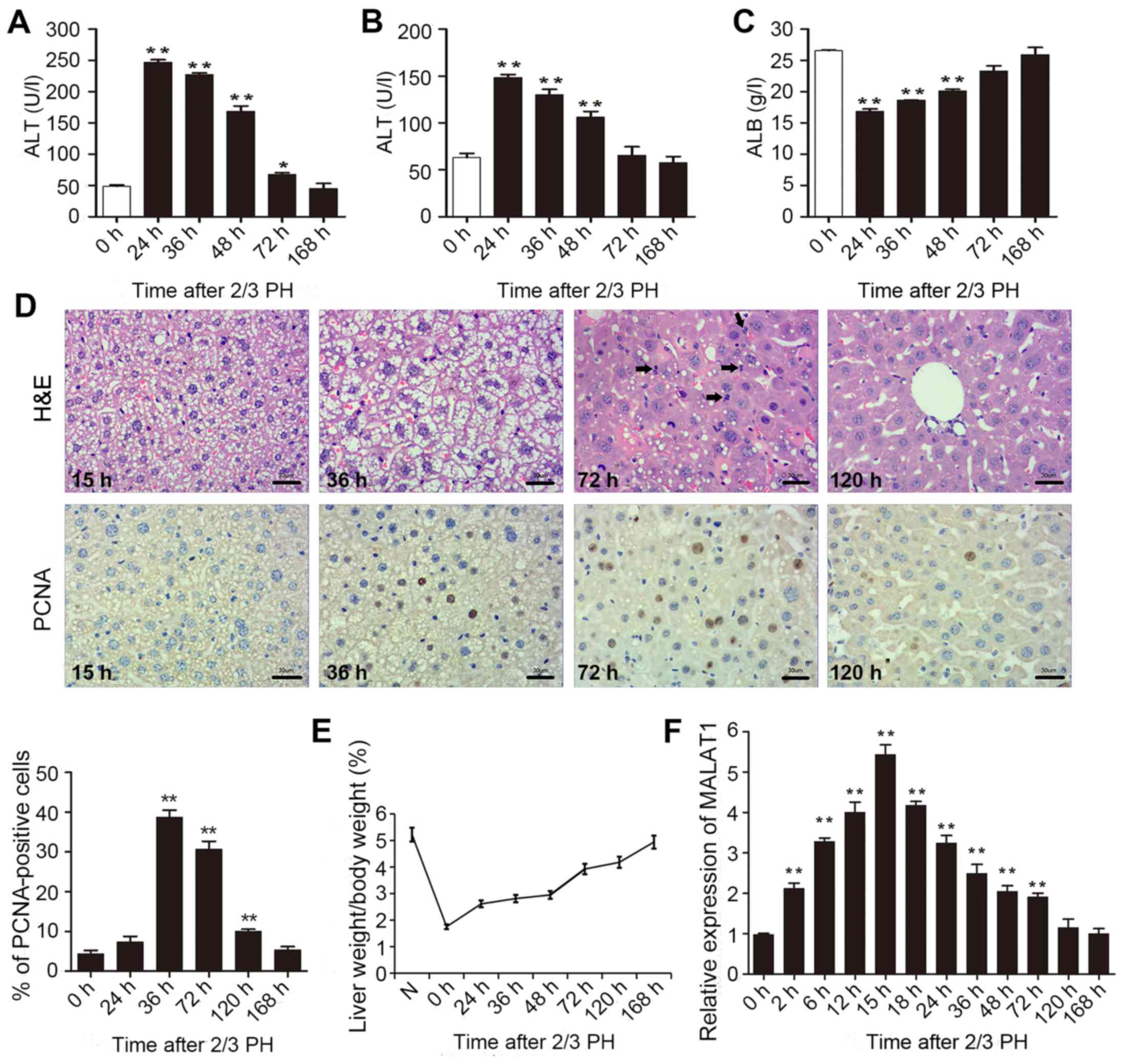
2/3 PH. As shown in Fig. 1A–C,
all the indicators returned to normal at 168 h following 2/3 PH.
H&E staining of the liver was carried out at different time
points. Following hepatectomy, we observed obvious cell edema at 15
and 36 h. Mitotic figures were clearly observed at 72 h, and the
hepatic lobe structure was close to normal at 120 h (Fig. 1D). Mouse hepatocyte proliferation
was determined in vivo by staining for proliferating cell
nuclear antigen (PCNA) (Fig. 1D)
using IHC. We found that the number of PCNA-positive nuclei reached
a peak at 36 h following 2/3 PH. The cell proliferative ability was
reduced at 72 and 120 h, and the proliferation at 24 and 168 h was
not significant (Fig. 1D). The
remnant liver/body weight ratio is shown in Fig. 1E.
2/3 PH in male BALB/c mice caused a >5-fold
increase in MALAT1 expression. MALAT1 expression increased from 2
h, peaked at 15 h following PH, and then returned to almost normal
levels 120 h following 2/3 PH (Fig.
1F). These results suggested that MALAT1 was specifically
overexpressed in hepatic tissue following 2/3 PH. Taken together,
this sharp increase indicates that MALAT1 may play a role in liver
regeneration.
lncRNA MALAT1 accelerates cell cycle
progression in hepatocytes and promotes proliferation in vitro
As it has been previously demonstrated that
hepatocytes from adult male mice enter the S phase at 36 h
following 2/3 PH (18) and that
the expression of MALAT1 reaches a maximum after 12–18 h of liver
regeneration, we hypothesized that MALAT1 may play an important
role in regulating the cell cycle, as the expression of MALAT1
peaked before 36 h.
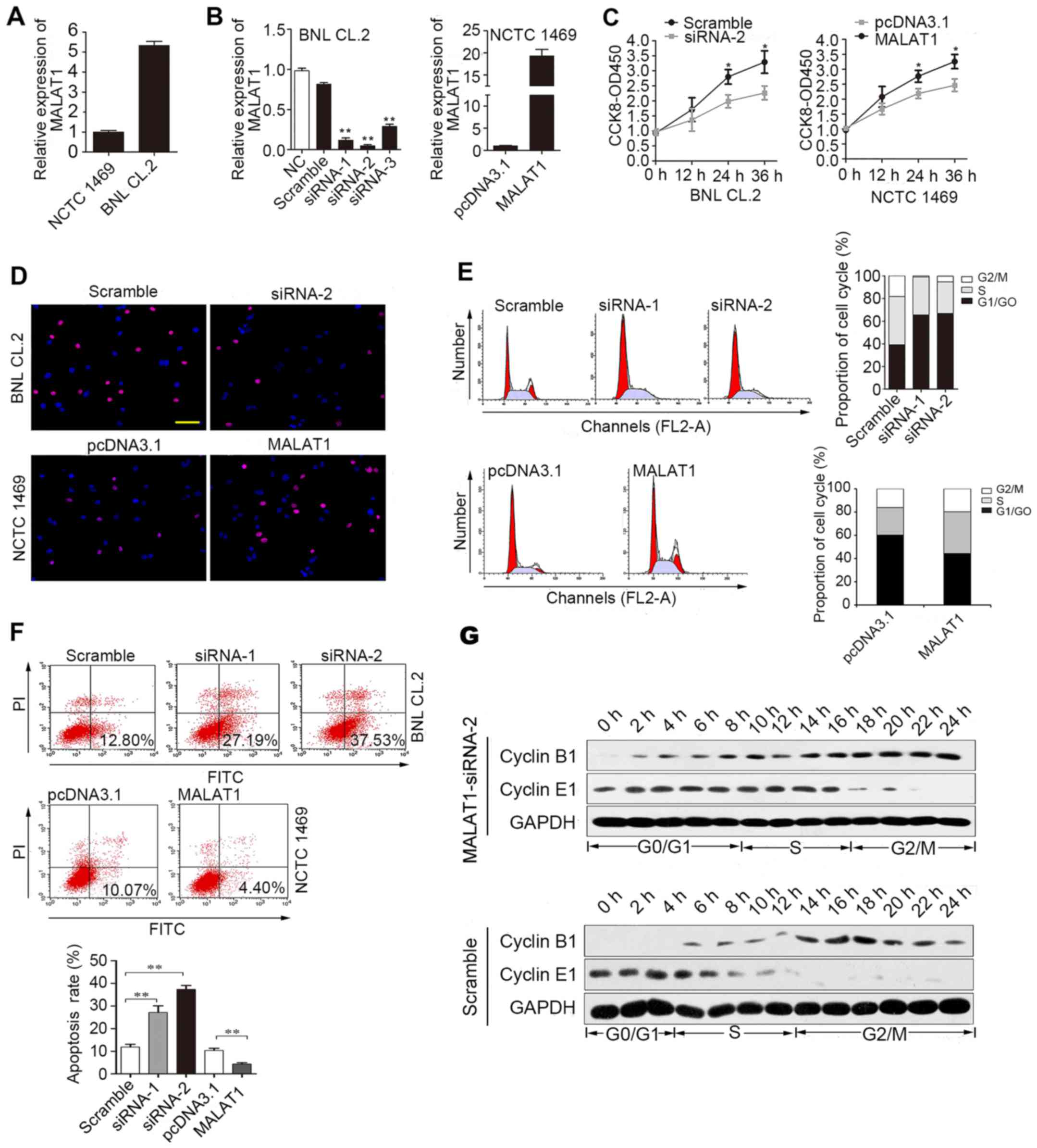
First, we analyzed MALAT1 expression in BNL CL.2 and
NCTC 1469 cells. MALAT1 expression was significantly higher in the
BNL CL.2 cells than in the NCTC 1469 cells (Fig. 2A). As shown in Fig. 2B, siRNA-2 was the most effective
at silencing MALAT1. MALAT1 expression was upregulated almost
18-fold following transfection of the NCTC 1469 cells with the
pcDNA3.1 vector carrying MALAT1. CCK-8 (Fig. 2C) and EdU immunofluorescence
assays (Fig. 2D) were used to
detect changes in the proliferative ability of the MALAT1-silenced
BNL CL.2 cells and MALAT1-overexpressing NCTC 1469 cells. The
experimental results revealed that cell proliferation was reduced
when MALAT1 was silenced in the BNL CL.2 cells and was increased
when MALAT1 was overexpressed in the NCTC 1469 cells. As shown in
Fig. 2D, the
MALAT1-overexpressing NCTC 1469 cells had more EdU-positive cells
than the control group, while the MALAT1-silenced BNL CL.2 cells
had fewer EdU-positive cells than the control cells. Moreover, FACS
analysis indicated that the MALAT1-silenced BNL CL.2 cells were
arrested in the G0/G1 phase of the cell cycle. Furthermore, the
number of cells in the G0/G1 phase decreased significantly and the
number of cells in the S phase increased significantly when MALAT1
was overexpressed in the NCTC 1469 cells (Fig. 2E). At the same time, the rate of
apoptosis increased when the BNL CL.2 cells were transfected with
MALAT1-siRNA-2, but decreased when the NCTC 1469 cells were
transfected with pcDNA3.1-MALAT1 (Fig. 2F). To further examine the role of
MALAT1 in cell cycle progression, the expression of cyclin E1 and
B1 was examined in the MALAT1-silenced BNL CL.2 cells. The results
revealed that the G0/G1 phase was extended when MALAT1 was silenced
in the BNL CL.2 cells (Fig. 2G).
Thus, the above-mentioned data indicate that MALAT1 facilitates
liver cell proliferation by accelerating cell cycle progression and
blocking liver cell apoptosis.
HGF increases MALAT1 expression
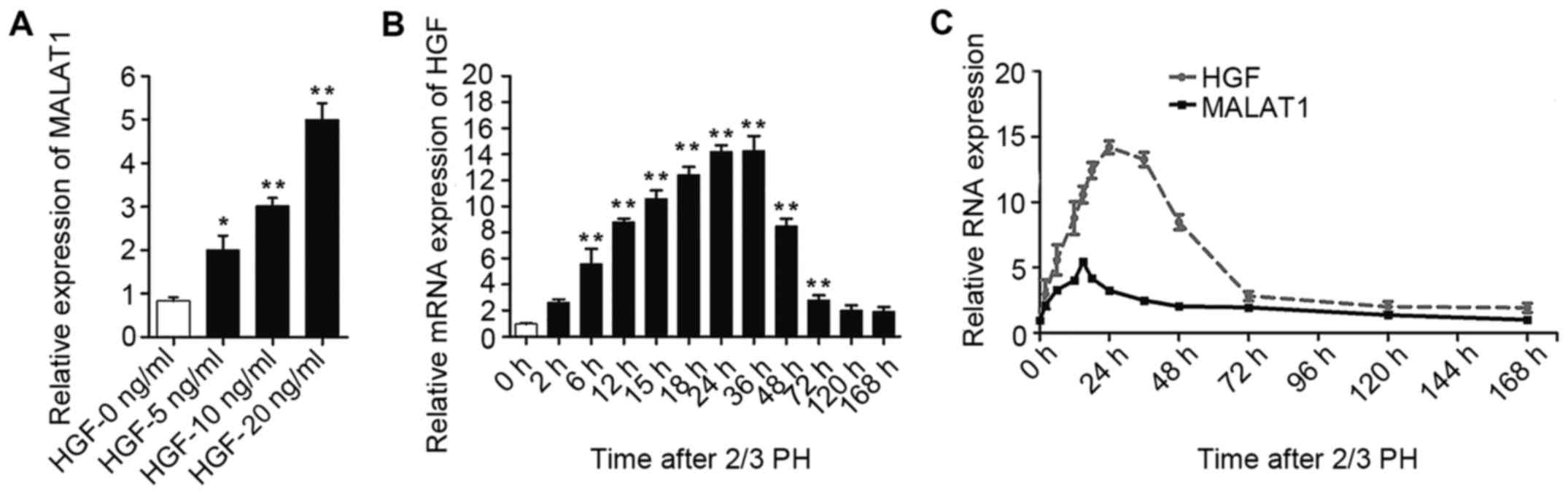
As MALAT1 expression increased during the early
stages after 2/3 PH and HGF rapidly increases following PH and
plays a substantial role during liver regeneration (6), we hypothesized that the increase in
MALAT1 expression may be mediated by HGF. To confirm our
hypothesis, the NCTC 1469 cells were treated with 5, 10 or 20 ng/ml
HGF to induce hepatocyte proliferation. As shown in Fig. 3A, MALAT1 expression gradually
increased following treatment with increasing concentrations of
HGF. The increased expression of HGF was detected in the BALB/c
mice subjected to 2/3 PH (Fig.
3B). Moreover, as shown in Fig.
3C, we identified a correlation between the HGF and MALAT1
levels in the liver tissue at different time points following 2/3
PH.
MALAT1 activates Wnt/β-catenin signaling
by inhibiting Axin1 and adenomatous polyposis coli (APC) in
vitro
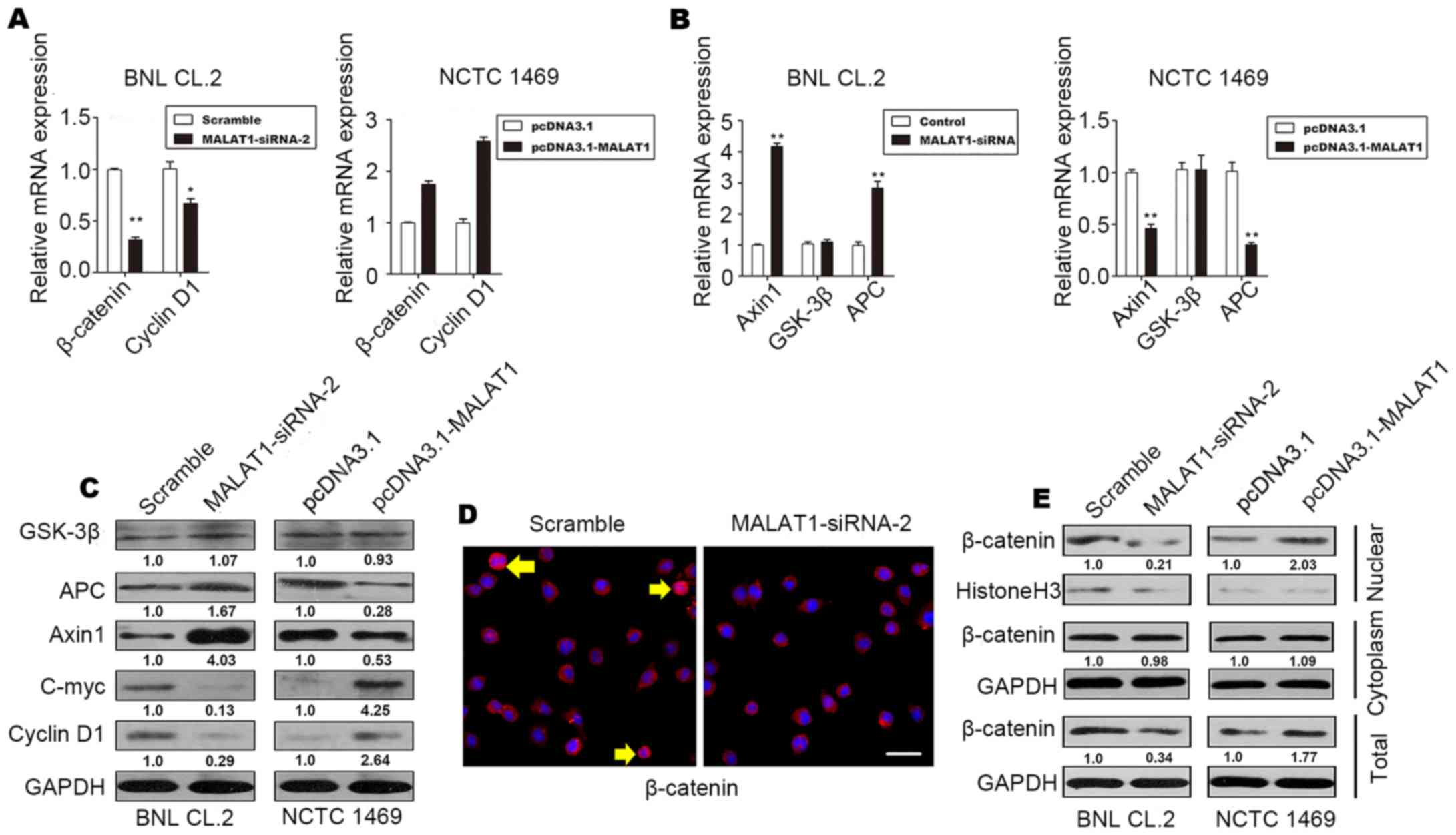
It has been demonstrated that the Wnt/β-catenin
pathway is associated with proliferation and differentiation
(21). Thus, to determine whether
MALAT1 accelerates hepatocyte proliferation by activating the
Wnt/β-catenin pathway, we evaluated the levels of cyclin D1,
β-catenin, the β-catenin-degradation component, Axin1, glycogen
synthase kinase (GSK)-3β and APC in MALAT1-silenced BNL CL.2 cells
and MALAT1-overexpressing NCTC 1469 cells. The results revealed an
increase in the protein and mRNA levels of β-catenin and cyclin D1
in the MALAT1-overexpressing NCTC 1469 cells (Fig. 4A, C and E). The protein and mRNA
levels of Axin1 and APC decreased when MALAT1 was overexpressed in
the NCTC 1469 cells (Fig. 4B and
C); however, there was no significant change in the level of
GSK-3β (Fig. 4B and C). The level
of c-Myc increased when MALAT1 was overexpressed, but declined
following the knockdown of MALAT1 (Fig. 4C).
As shown in Fig. 4D
and E, inactive β-catenin was located in the cytoplasm, and
total β-catenin staining decreased when MALAT1 was silenced in the
BNL CL.2 cells. In addition, western blot analysis of β-catenin
demonstrated that the level of β-catenin in the nucleus also
decreased significantly when MALAT1 was silenced. These effects
were reversed when MALAT1 was overexpressed. There was no
significant difference in the level of cytoplasmic β-catenin when
MALAT1 was silenced or overexpressed (Fig. 4E). β-catenin phosphorylation
occurs in a multiprotein complex that includes the GSK-3β,
β-catenin, Axin1 and APC proteins; this phosphorylation leads to
the degradation of cytoplasmic β-catenin (22,23).
On the whole, our data demonstrate that MALAT1
promotes cell cycle progression and accelerates hepatocyte
proliferation during liver regeneration by activating Wnt/β-catenin
signaling. By suppressing Axin1 and APC, MALAT1 indirectly reduces
the stability of the β-catenin degradation complex, decreasing the
level of inactive β-catenin, while increasing the level of active
β-catenin. Active β-catenin is released and begins functioning.
Subsequently, active β-catenin translocates to the nucleus to
controls the transcription of target genes, such as c-Myc and
cyclin D1. Finally, MALAT1 enhances hepatocyte proliferation by
promoting cell cycle progression in vitro.
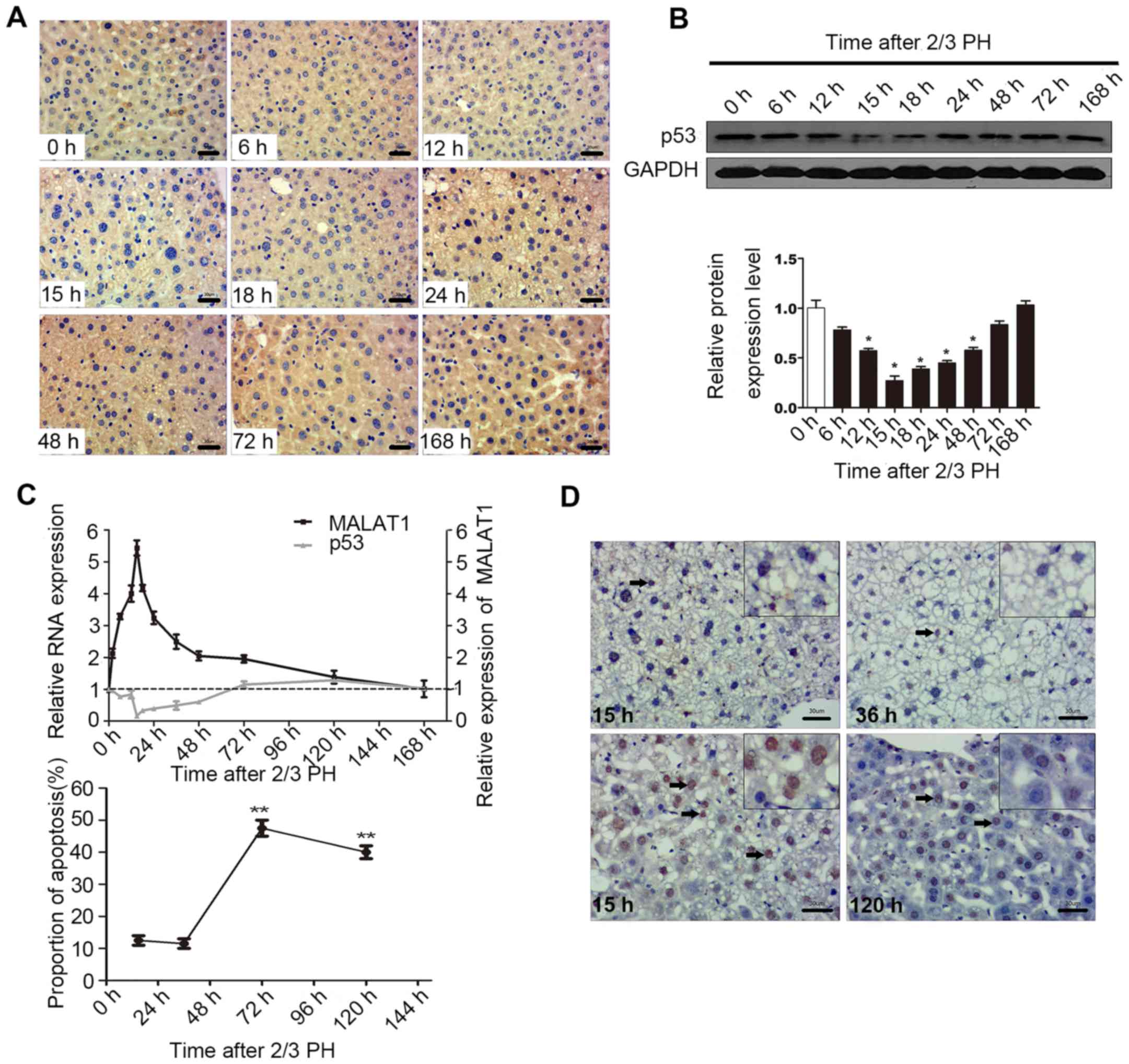
p53 regulates MALAT1 expression during
liver regeneration
p53 activation is involved in hepatocyte
proliferation following PH (24–26). A previous demonstrated shown that
the down-regulation of MALAT1 or the upregulation of p53 suppresses
the proliferation of murine hematopoietic cells, as p53
specifically binds to the core promoter region of MALAT1. The core
binding sequence is CGGCATGGCCGCCAAGGTCGCCGT GCCCT (27).
In this study, in order to determine the biological
relevance of the p53-mediated regulation of MALAT1 in vivo,
we examined the mRNA and protein levels of p53 during liver
regeneration. The results of IHC and western blot analysis
indicated that 2/3 PH decreased p53 expression in BALB/c mice, with
p53 expression being detectable at 6 h, with the lowest levels
being observed at 15 h, and the highest at 120 h. The levels of p53
at 72 and 168 h were close to the levels observed 0 h following PH
(Fig. 5A and B). A correlation
between MALAT1 and p53 may exist during liver regeneration
(Fig. 5C). Cellular functional
analysis indicated that MALAT1 had an effect on hepatocyte
apoptosis, and thus we specifically detected apoptosis at 15 h
following 2/3 PH. The proportion of cells undergoing apoptosis was
11% at 15 h, and the level of apoptosis may be related to p53 and
MALAT1 (Fig. 5D). Thus, we
hypothesized that MALAT1 may be negatively regulated by p53 during
liver regeneration.
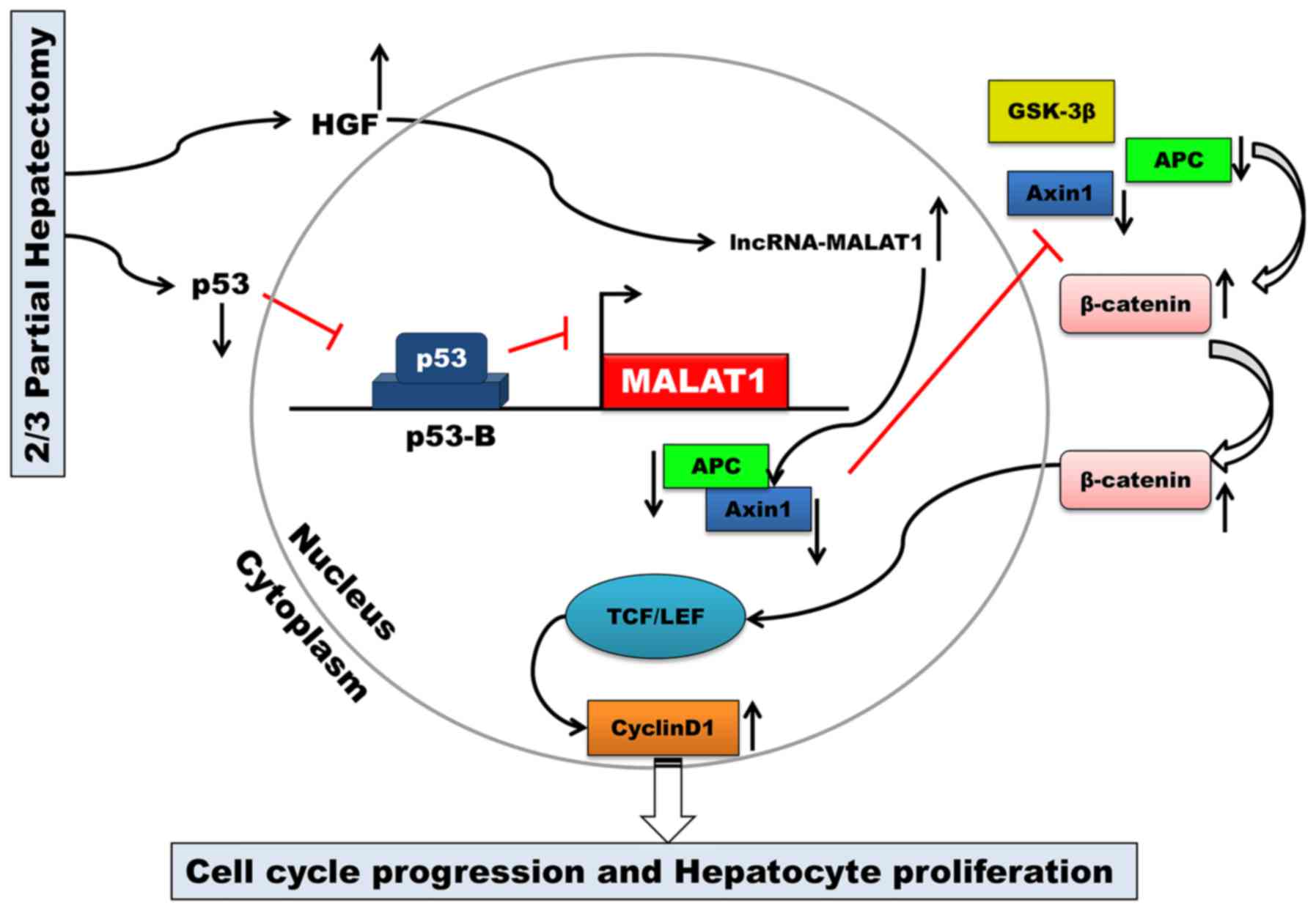
Discussion
The key finding of the present study was that MALAT1
is upregulated during mouse liver regeneration. MALAT1 may acts as
a regulatory factor in the cell cycle. Our results also
demonstrated that both HGF and p53 are closely associated with
MALAT1 during liver regeneration. MALAT1 can activate Wnt/β-catenin
signaling in vitro. Axin1 and APC were suppressed by MALAT1,
and the stability of the β-catenin degradation complex became weak.
Active β-catenin translocated from the cytoplasm to the nucleus,
where it then enhanced the transcription of c-Myc and cyclin D1.
Ultimately, MALAT1 facilitated hepatocyte proliferation by
promoting cell cycle progression from the G1 to the S phase
(Fig. 6).
By binding to the promoter of MALAT1, p53 can
negatively regulate the expression of MALAT1 (17,27). MALAT1 is an essential factor in
maintaining the proliferative potential of early-stage hepatocytes.
Combined with the results of our study, these results indicate that
MALAT1 may be transcriptionally regulated by p53 during liver
regeneration following 2/3 PH.
Although a number of cytokines, growth factors (such
as HGF) and miRNAs regulate proliferation-related genes during
liver regeneration (28,29), the role of lncRNAs in liver
regeneration remains largely unknown. A comprehensive analysis of
lncRNA expression during mouse liver regeneration has revealed
changes in the expression of multiple lncRNAs (14). In this study, we found that HGF
increased MALAT1 expression in vitro; however, the peak in
HGF expression was delayed compared to the peak in MALAT1
expression. The above-mentioned phenomenon indicates that HGF may
function as a regulatory factor for MALAT1. Between 15 and 24 h
following PH, the anti-MALAT1 effects override the HGF-mediated
increase in MALAT1, indicating that additional signaling molecules
and transcription factors may regulate the expression of MALAT1; we
aim to continue to investigate the mechanisms underlying the
association between HGF and MALAT1 in future studies. The HGF and
Wnt/β-catenin pathways both play key roles in accelerating
hepatocyte proliferation and can cooperate to stimulate hepatocyte
proliferation, which may be crucial in liver regeneration following
PH (30).
The complete liver regeneration process takes
approximately 7 days in mice; however, the mechanisms through which
the liver initiates and terminates regenerative growth are not yet
fully understood (31). Exploring
the biological functions of lncRNAs has recently come to the
foreground (12,32). Previous studies investigating the
non-coding RNAs in the liver have mainly focused on the association
between the presence of these RNAs and the development of
hepatocellular carcinoma (HCC) or the differential expression of
these RNAs in carcinomas and the adjacent tissue (33). Some genes associated with
hepatocyte proliferation and liver regeneration are also expressed
abnormally in HCC (34,35). Liver regeneration is a specific
cellular process that is both similar to and different from HCC
(36). To date, it remains
unclear as to whether the lncRNAs regulating cell proliferation in
HCC participate in liver regeneration. The overall mechanisms
warrant further investigation. Although partial liver
transplantation can minimize organ scarcity, the risk of death of a
healthy donor, the development of SFSS in both the donor and
recipient and technical complexity limit its applicability.
Compounds that interfere with the signaling pathways that regulate
proliferation are receiving increased attention and may be used to
prevent or reverse SFSS (37). In
conclusion, to the best of our knowledge, our research group is the
first to report a biological role of MALAT1 in hepatocyte
proliferation. MALAT1 is expected to become an indicator of
prognosis for a variety of liver diseases and liver resection.
References
|
1
|
Dutkowski P, Linecker M, DeOliveira ML,
Müllhaupt B and Clavien PA: Challenges to liver transplantation and
strategies to improve outcomes. Gastroenterology. 148:307–323.
2015. View Article : Google Scholar
|
|
2
|
Lehmann K, Tschuor C, Rickenbacher A, Jang
JH, Oberkofler CE, Tschopp O, Schultze SM, Raptis DA, Weber A, Graf
R, et al: Liver failure after extended hepatectomy in mice is
mediated by a 21-dependent barrier to liver regeneration.
Gastroenterology. 143:1609–1619. 2012. View Article : Google Scholar
|
|
3
|
Petrowsky H, Breitenstein S, Slankamenac
K, Vetter D, Lehmann K, Heinrich S, DeOliveira ML, Jochum W,
Weishaupt D, Frauenfelder T, et al: Effects of pentoxifylline on
liver regeneration: a double-blinded, randomized, controlled trial
in 101 patients undergoing major liver resection. Ann Surg.
252:813–822. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Michalopoulos GK: Liver regeneration after
partial hepatectomy: critical analysis of mechanistic dilemmas. Am
J Pathol. 176:2–13. 2010. View Article : Google Scholar :
|
|
5
|
Selzner N, Selzner M, Tian Y, Kadry Z and
Clavien PA: Cold ischemia decreases liver regeneration after
partial liver transplantation in the rat: a
TNF-alpha/IL-6-dependent mechanism. Hepatology. 36:812–818. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pediaditakis P, Lopez-Talavera JC,
Petersen B, Monga SP and Michalopoulos GK: The processing and
utilization of hepatocyte growth factor/scatter factor following
partial hepatectomy in the rat. Hepatology. 34:688–693. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fausto N, Campbell JS and Riehle KJ: Liver
regeneration. Hepatology. 43(Suppl 1): S45–S53. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shu J, Kren BT, Xia Z, Wong PY, Li L,
Hanse EA, Min MX, Li B, Albrecht JH, Zeng Y, et al: Genomewide
microRNA down-regulation as a negative feedback mechanism in the
early phases of liver regeneration. Hepatology. 54:609–619. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Song G, Sharma AD, Roll GR, Ng R, Lee AY,
Blelloch RH, Frandsen NM and Willenbring H: MicroRNAs control
hepatocyte proliferation during liver regeneration. Hepatology.
51:1735–1743. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Castro RE, Ferreira DM, Zhang X, Borralho
PM, Sarver AL, Zeng Y, Steer CJ, Kren BT and Rodrigues CM:
Identification of microRNAs during rat liver regeneration after
partial hepatectomy and modulation by ursodeoxycholic acid. Am J
Physiol Gastrointest Liver Physiol. 299:G887–G897. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Forbes SJ and Newsome PN: Liver
regeneration - mechanisms and models to clinical application. Nat
Rev Gastroenterol Hepatol. 13:473–485. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dey BK, Mueller AC and Dutta A: Long
non-coding RNAs as emerging regulators of differentiation,
development, and disease. Transcription. 5:e9440142014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ørom UA, Derrien T, Beringer M, Gumireddy
K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q,
et al: Long noncoding RNAs with enhancer-like function in human
cells. Cell. 143:46–58. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xu D, Yang F, Yuan JH, Zhang L, Bi HS,
Zhou CC, Liu F, Wang F and Sun SH: Long noncoding RNAs associated
with liver regeneration 1 accelerates hepatocyte proliferation
during liver regeneration by activating Wnt/β-catenin signaling.
Hepatology. 58:739–751. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yamamoto Y, Nishikawa Y, Tokairin T, Omori
Y and Enomoto K: Increased expression of H19 non-coding mRNA
follows hepatocyte proliferation in the rat and mouse. J Hepatol.
40:808–814. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Spector DL and Lamond AI: Nuclear
speckles. Cold Spring Harb Perspect Biol. 3:pii: a000646. 2011.
View Article : Google Scholar
|
|
17
|
Tripathi V, Shen Z, Chakraborty A, Giri S,
Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A and
Prasanth KV: Long noncoding RNA MALAT1 controls cell cycle
progression by regulating the expression of oncogenic transcription
factor B-MYB. PLoS Genet. 9:e10033682013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mitchell C and Willenbring H: A
reproducible and well-tolerated method for 2/3 partial hepatectomy
in mice. Nat Protoc. 3:1167–1170. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bockhorn M, Goralski M, Prokofiev D,
Dammann P, Grünewald P, Trippler M, Biglarnia A, Kamler M, Niehues
EM, Frilling A, et al: VEGF is important for early liver
regeneration after partial hepatectomy. J Surg Res. 138:291–299.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chang L, Li C, Guo T, Wang H, Ma W, Yuan
Y, Liu Q, Ye Q and Liu Z: The human RNA surveillance factor UPF1
regulates tumorigenesis by targeting Smad7 in hepatocellular
carcinoma. J Exp Clin Cancer Res. 35:82016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tan X, Behari J, Cieply B, Michalopoulos
GK and Monga SP: Conditional deletion of beta-catenin reveals its
role in liver growth and regeneration. Gastroenterology.
131:1561–1572. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li VS, Ng SS, Boersema PJ, Low TY,
Karthaus WR, Gerlach JP, Mohammed S, Heck AJ, Maurice MM, Mahmoudi
T and Clevers H: Wnt signaling through inhibition of β-catenin
degradation in an intact Axin1 complex. Cell. 149:1245–1256. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ha NC, Tonozuka T, Stamos JL, Choi HJ and
Weis WI: Mechanism of phosphorylation-dependent binding of APC to
beta-catenin and its role in beta-catenin degradation. Mol Cell.
15:511–521. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang L, Liu L, He Z, Li G, Liu J, Song Z,
Jin H, Rudolph KL, Yang H, Mao Y, et al: Inhibition of wild-type
p53-induced phosphatase 1 promotes liver regeneration in mice by
direct activation of mammalian target of rapamycin. Hepatology.
61:2030–2041. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen S, Zheng J, Hao Q, Yang S, Wang J,
Chen H, Chen L, Zhou Y, Yu C, Jiao B and Cai Z: p53-insensitive
PUMA down-regulation is essential in the early phase of liver
regeneration after partial hepatectomy in mice. J Hepatol.
52:864–871. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Stepniak E, Ricci R, Eferl R, Sumara G,
Sumara I, Rath M, Hui L and Wagner EF: c-Jun/AP-1 controls liver
regeneration by repressing p53/p21 and p38 MAPK activity. Genes
Dev. 20:2306–2314. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ma XY, Wang JH, Wang JL, Ma CX, Wang XC
and Liu FS: Malat1 as an evolutionarily conserved lncRNA, plays a
positive role in regulating proliferation and maintaining
undifferentiated status of early-stage hematopoietic cells. BMC
Genomics. 16:6762015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ng R, Song G, Roll GR, Frandsen NM and
Willenbring H: A microRNA-21 surge facilitates rapid cyclin D1
translation and cell cycle progression in mouse liver regeneration.
J Clin Invest. 122:1097–1108. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yuan Q, Loya K, Rani B, Möbus S,
Balakrishnan A, Lamle J, Cathomen T, Vogel A, Manns MP, Ott M, et
al: MicroRNA-221 overexpression accelerates hepatocyte
proliferation during liver regeneration. Hepatology. 57:299–310.
2013. View Article : Google Scholar
|
|
30
|
Apte U, Zeng G, Muller P, Tan X, Micsenyi
A, Cieply B, Dai C, Liu Y, Kaestner KH and Monga SP: Activation of
Wnt/beta-catenin pathway during hepatocyte growth factor-induced
hepatomegaly in mice. Hepatology. 44:992–1002. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cox AG and Goessling W: Regenerative
biology: take the brakes off for liver repair. Nature. 506:299–300.
2014. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Meola N, Pizzo M, Alfano G, Surace EM and
Banfi S: The long noncoding RNA Vax2os1 controls the cell cycle
progression of photoreceptor progenitors in the mouse retina. RNA.
18:111–123. 2012. View Article : Google Scholar :
|
|
33
|
He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv
XW and Li J: Long noncoding RNAs: novel insights into hepatocelluar
carcinoma. Cancer Lett. 344:20–27. 2014. View Article : Google Scholar
|
|
34
|
Bandiera S, Pfeffer S, Baumert TF and
Zeisel MB: miR-122 - a key factor and therapeutic target in liver
disease. J Hepatol. 62:448–457. 2015. View Article : Google Scholar
|
|
35
|
John K, Hadem J, Krech T, Wahl K, Manns
MP, Dooley S, Batkai S, Thum T, Schulze-Osthoff K and Bantel H:
MicroRNAs play a role in spontaneous recovery from acute liver
failure. Hepatology. 60:1346–1355. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gandhi CR, Chaillet JR, Nalesnik MA, Kumar
S, Dangi A, Demetris AJ, Ferrell R, Wu T, Divanovic S, Stankeiwicz
T, et al: Liver-specific deletion of augmenter of liver
regeneration accelerates development of steatohepatitis and
hepatocellular carcinoma in mice. Gastroenterology. 148:379–391.
2015. View Article : Google Scholar
|
|
37
|
Huang W, Ma K, Zhang J, Qatanani M,
Cuvillier J, Liu J, Dong B, Huang X and Moore DD: Nuclear
receptor-dependent bile acid signaling is required for normal liver
regeneration. Science. 312:233–236. 2006. View Article : Google Scholar : PubMed/NCBI
|