Introduction
Leucine-rich glioma inactivated (LGI)3 is a secreted
protein of the LGI family that is predominantly expressed in the
brain in a developmentally regulated manner (1). LGI3 has been shown to be regulated
by neuronal restrictive silencer element and AP-2 at the
transcription level (1). We
previously reported that LGI3 regulates neuronal exocytosis and
differentiation (2,3). Apart from the brain, LGI3 has been
shown to be expressed in diverse tissues, including the skin and
adipose tissues (4,5). In a previous study, we demonstrated
that the ultraviolet B (UVB) irradiation-induced secretion of LGI3
from human keratinocytes promoted cell survival (5). We also demonstrated that LGI3
increased keratinocyte migration and melanocyte pigmentation
(6,7).
We have also previously reported that LGI3 is
upregulated in the adipose tissues of ob/ob mice and high fat
diet-fed mice (4,8). We further demonstrated that LGI3 was
downregulated during adipocyte differentiation, and suppressed
adipogenesis through its receptor, a disintegrin and
metalloproteinase domain-containing protein 23 (ADAM23) (4). LGI3 was shown to downregulate
adiponectin, an anti-inflammatory adipokine (8). We also demonstrated that LGI3
increased the levels of pro-inflammatory proteins, including tumor
necrosis factor-α (TNF-α) in macrophages (4). LGI3 and TNF-α are upregulated
mutually through a key inflammatory transcription factor, nuclear
factor-κB (NF-κB) (9). We
proposed that LGI3 may be a pro-inflammatory adipokine that
functionally interacts with other adipokines and cytokines in
metabolic inflammation (4,8,9).
Our previous findings supported the hypothesis that
LGI3 may be a multifunctional cytokine and may participate in the
cytokine network in metabolic inflammation. LGI3 and its receptor,
ADAM23, may transduce intracellular signals through Akt, focal
adhesion kinase (FAK), mouse double minute 2 homolog (MDM2), p53,
β-catenin and glycogen synthase kinase (GSK)3β (3,5,7).
In order to gain insight into the functional network
of LGI3, in this study, we conducted experimental and integrative
analyses of the gene products regulated by LGI3 and identified a
cluster of protein interaction network that may represent an
LGI3-regulated cytokine network.
Materials and methods
Animals and cell culture
All the animal protocols were approved by the
Institutional Animal Care and Use Committee. LGI3 knockout mice
were generated by Macrogen, Inc. (Seoul, Korea) (8). LGI3 knockout mice and wild-type
littermates (n=3) were used in this study. White adipose tissues
(WATs; epididymal fat) and plasma were obtained from 10-week-old
LGI3 knockout mice and wild-type littermates. WAT was isolated
after blood collection and after the mice were sacrificed. The
culture of 3T3-L1 (ATCC CL-173) and RAW 264.7 (ATCC TIB-71) cells
(ATCC, Manassas, VA, USA), and the differentiation of 3T3-L1 cells
was carried out as previously described (4). Briefly, 3T3-L1 cells were cultured
in Dulbecco's modified Eagle's medium (DMEM) containing 10% calf
serum and antibiotics (100 U/ml penicillin and 100 U/ml
streptomycin) at 37°C in 5% CO2. Two days after reaching
confluence, the cells were cultured in DMEM containing 1
µg/ml insulin, 1 µM dexamethasone (Dex), 0.5 mM
3-isobutyl-1-methylxanthine (IBMX), 10% fetal bovine serum (FBS)
and antibiotics for 3 days. The cells were then maintained in DMEM
containing 1 µg/ml insulin, 10% FBS and antibiotics for 4
days. The medium was changed every 2 days until the cells were
harvested. Differentiation was confirmed by Oil Red O staining
(data not shown). RAW 264.7 cells were maintained in DMEM
supplemented with 10% FBS. The cells were treated with LGI3 (10
ng/ml) or Dulbecco's phosphate-buffered saline (DPBS, control) for
1 or 24 h as indicated in the figure legends.
Preparation of recombinant LGI3 and
protein analysis
The preparation of recombinant LGI3 protein and
lysates of cells and tissues were carried out as previously
described (3). Briefly,
LGI3-His6 protein was expressed in E. coli
BL21(DE3) using pET28a(+) expression vector (Novagen, Madison, WI,
USA) and chaperone system (pGro7) (Takara, Otsu, Japan). The
protein was purified by Talon metal affinity resin (Clontech,
Mountain View, CA, USA). Protein array analysis was performed using
adipokine and cytokine array kits (R&D Systems, Inc.,
Minneapolis, MN, USA; product nos. ARY013 for 38 adipokines and
ARY006 for 40 cytokines) according to the manufacturer's
instructions. Briefly, the array membrane was incubated with tissue
extracts in RIPA buffer (Sigma, St. Louis, MO, USA) and bound
proteins were visualized by detection antibody cocktail and
streptavidin-horseradish peroxidase using chemiluminescence
detection reagent. Phosphoprotein array analysis was conducted
using PathScan array kits (intracellular signaling array and Akt
signaling antibody array kits; Cell Signaling Technology, Danvers,
MA, USA) according to the manufacturer's instructions.
Quantitative PCR
Quantitative PCR was performed as previously
described (4). Total RNA was
extracted using RNeasy lipid tissue mini kit (Qiagen, Hilden,
Germany). Thermal cycling using Applied Biosystems StepOne system
was, 10 min at 25°C, 120 min at 37°C, 10 min at 95°C and 40 cycles
of 15 sec at 95°C and 1 min at 60°C. The assay-on-demand gene
expression products (Applied Biosystems, Foster City, CA, USA) were
used to measure the mRNA expression levels of the following genes:
CD11c, Mm00498698_m1; CD68, Mm03047340_m1; F4/80, Mm00802529_m1;
interleukin (IL)-6, Mm00446190_m1; inducible nitric oxide synthase
(iNOS, Mm00440502_m1); monocyte chemoattractant protein 1 (MCP-1),
Mm00441242_ m1); NADPH oxidase (NOX-2, Mm01287743_m1); p22phox,
Mm00514478_m1; p47phox, Mm00447921_m1; p67phox, Mm00726636_s1;
TNF-α, Mm00443260_g1; and 18S rRNA (endogenous control),
Hs99999901_s1.
Functional enrichment and protein-protein
interaction network analyses
For functional enrichment of a group of regulated
genes, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and
Gene Ontology (GO) enrichment analyses were performed using the
Database for Annotation, Visualization and Integration Discovery
(DAVID) (http://david.abcc.ncifcrf.gov), as previously
described (10). The results were
ranked by their p-values and 20 entries with the lowest p-values
were presented. The protein-protein interaction network was
constructed using information provided by the Search Tool for the
Retrieval of Interacting Genes (STRING) (http://string-db.org/) (11). Cluster analysis and GO enrichment
analysis of subnetwork clusters were performed using ClusterONE
application in Cytoscape 3.3.0, as previously described (12).
Statistical analysis
Data are expressed as the means ± standard error of
the mean (SEM). Statistical significance between 2 groups was
assessed using the Student's t-test. The results were considered
statistically significant with a p-value <0.05.
Results
Effect of LGI3 knockout on adipokine and
cytokine profiles
Homozygous mice with disruption of the LGI3 gene
were viable and fertile, and exhibited no gross abnormalities under
normal growth conditions (8).
These mice exhibited increased levels of adiponectin in WAT and
plasma, as described in our previous study (8). We analyzed cytokine and adipokine
profiles using protein arrays in WAT and plasma of the wild-type
and LGI3 knockout mice. The results revealed that the levels of
multiple cytokines and adipokines were increased or decreased in
LGI3 knockout mice (Fig. 1). The
factors with increased expression were adiponectin, insulin-like
growth factor-binding protein-1 (IGFBP-1), C-reactive protein
(CRP), endocan, pre-adipocyte factor-1 (Pref-1), serpin E1, C5/C5a,
macrophage colony-stimulating factor (M-CSF) (Fig. 1, a–h). The factors with decreased
expression were insulin-like growth factor-1 (IGF-1), insulin-like
growth factor binding protein-5 (IGFBP-5), B lymphocyte
chemoattractant (BLC), granulocyte colony-stimulating factor
(G-CSF), monocyte chemotactic protein 5 (MCP-5), macrophage
inflammatory protein-2 (MIP-2) and tissue inhibitor of
metalloproteinase-1 (TIMP-1) (Fig. 1,
i–o). An increase in the levels of adiponectin was observed
both in WAT and plasma, as previously observed (8), and alterations in the levels of
other factors were observed only in plasma (Fig. 1).
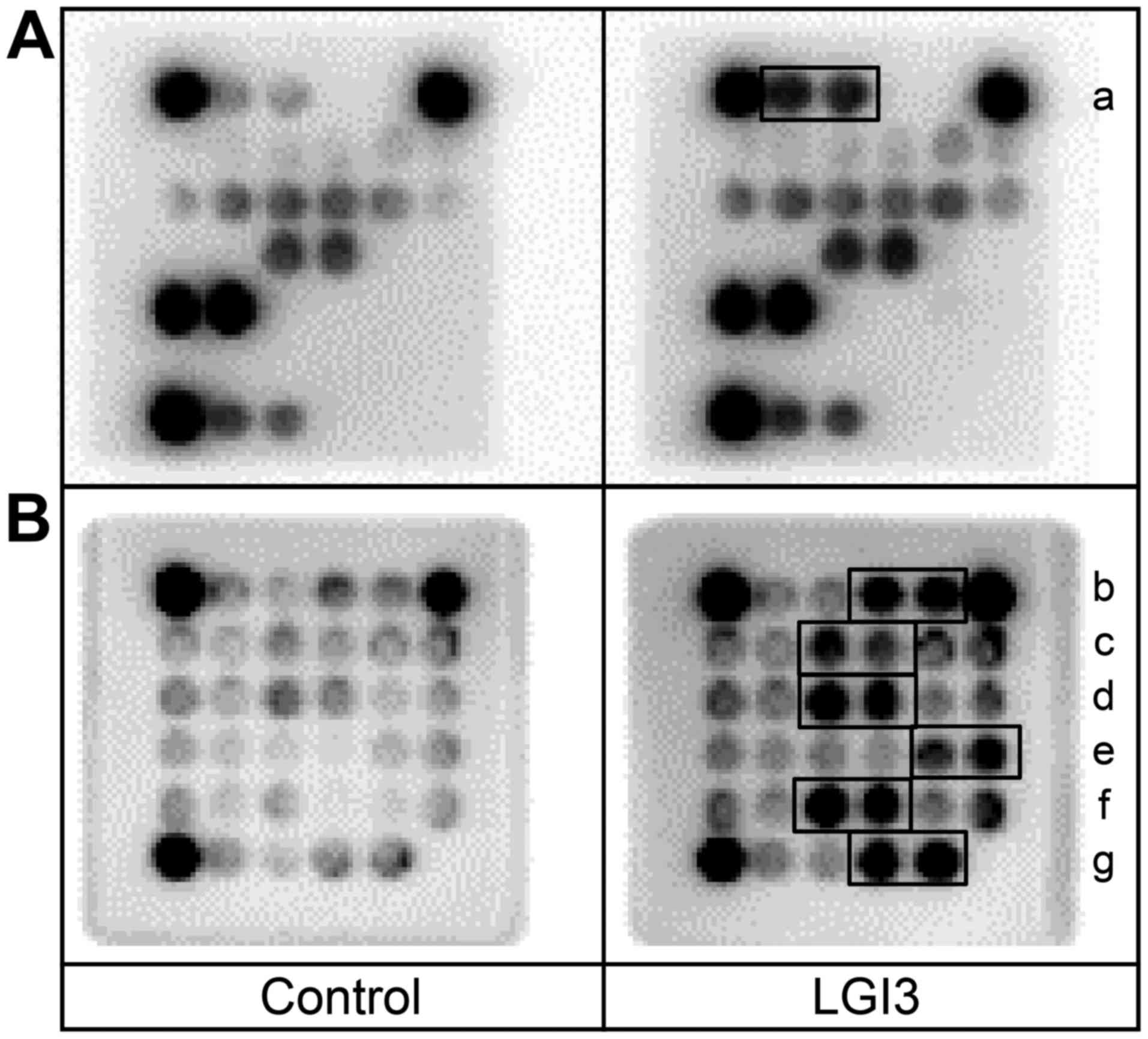
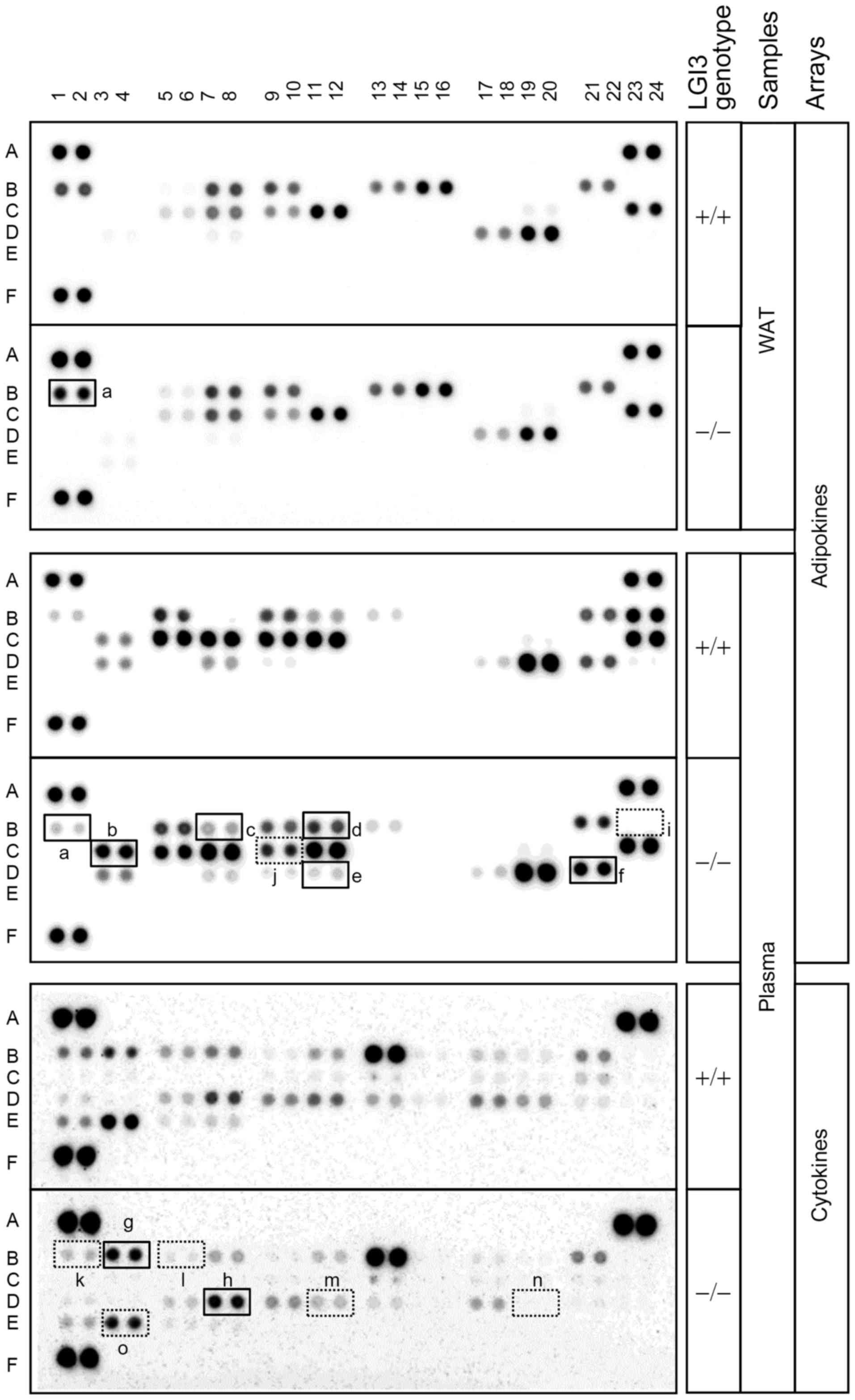 | Figure 1Effect of leucine-rich glioma
inactivated 3 (LGI3) knockout on adipokine and cytokine profiles.
White adipose tissue (WAT) and plasma from the wild-type (+/+) and
homozygous LGI3 knockout (−/−) mice were analyzed by adipokine and
cytokine arrays. Solid line box, increased proteins in knockout
mice; dotted line box, decreased protein in knockout mice. a,
adiponectin; b, IGFBP-1; c, C-reactive protein; d, endocan; e,
pref-1; f, serpin E1; g, C5/C5a; h, M-CSF; i, IGF-1; j, IGFBP-5; k,
BLC; l, G-CSF; m, MCP-5; n, MIP-2; o, TIMP-1. Numbers and capital
letters indicate array coordinates. |
Effect of LGI3 treatment on the
phosphorylation of signaling proteins
As shown in our previous study, LGI3 and its
receptor ADAM23 was predominantly expressed at the pre-adipocyte
stage in 3T3-L1 cells (4). Thus,
LGI3 may transduce intracellular signals primarily in
pre-adipocytes. To explore proteins that convey LGI3 signaling, we
conducted phosphoprotein array analysis using the extracts from
3T3-L1 pre-adipocytes treated with LGI3 protein. The results
revealed increases in the phosphorylation levels of extracellular
signal-regulated kinase (Erk)1/2 (Thr202/Tyr204), Akt (Ser473),
AMP-activated protein kinase (AMPK; Thr172), GSK3α (Ser21), Bad
(Ser112), phosphatase and tensin homolog (PTEN) (Ser380) and
eukaryotic translation initiation factor 4E-binding protein 1
(4E-BP1) (Thr37/46) in the LGI3-treated cells (Fig. 2, a–g).
Effect of LGI3 treatment on the
expression of inflammatory genes
As shown in our previous study, treatment with LGI3
protein increased the level of TNF-α in 3T3-L1 pre-adipocytes, and
those of iNOS, cyclooxygenase (Cox)-2 and TNF-α in RAW 264.7
macrophages (4,9). We thus hypothesized that LGI3 may
upregulate inflammatory genes in adipose tissues from obese mice
(4). We thus examined the effects
of LGI3 treatment on expression of various inflammatory genes in
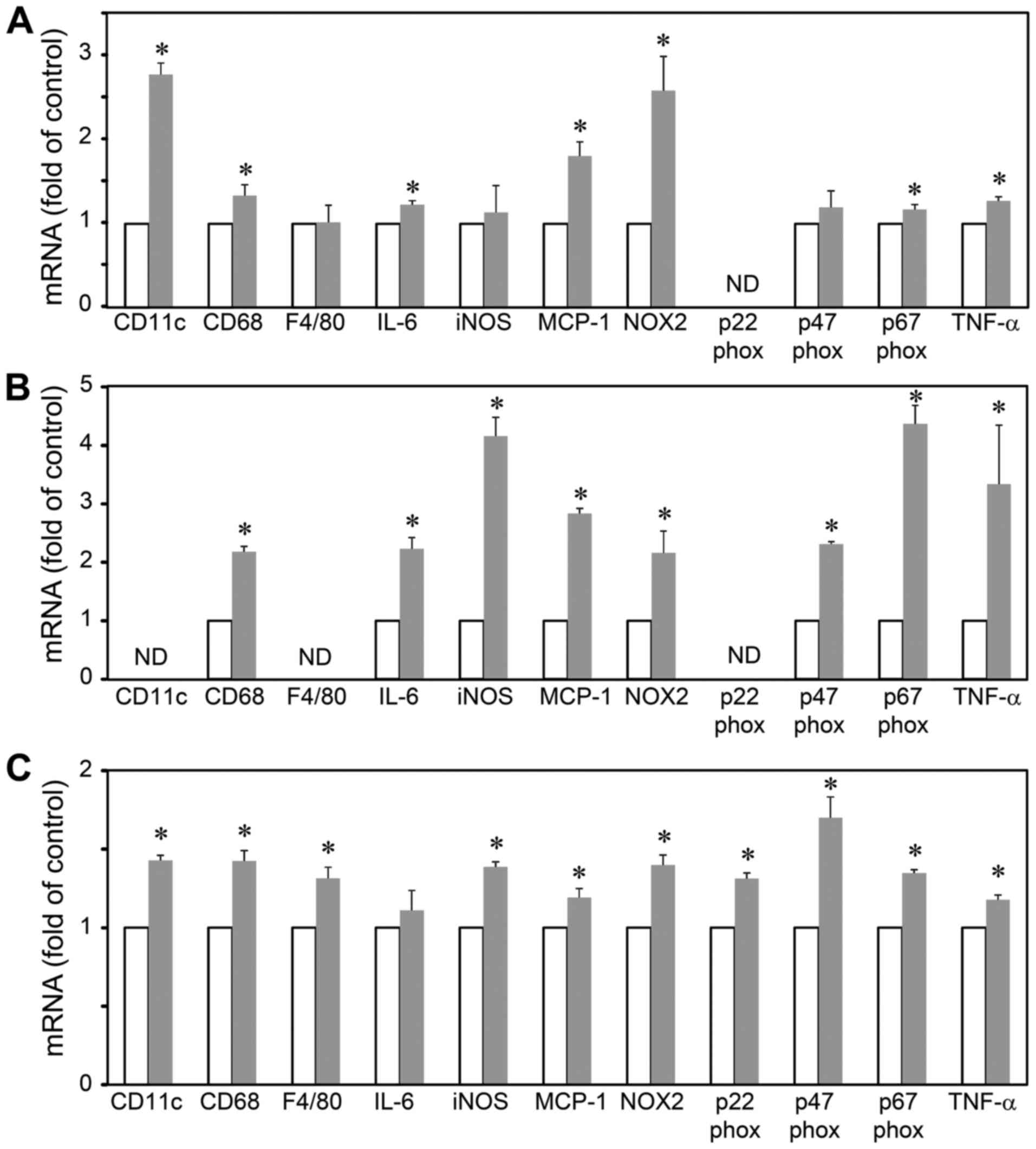
pre-adipocytes, adipocytes and macrophages (Fig. 3). The results revealed that LGI3
increased the levels of CD11c, CD68, IL-6, MCP-1, NOX-2, p67phox
and TNF-α in the 3T3-L1 pre-adipocytes (Fig. 3A). In the 3T3-L1 adipocytes, LGI3
increased the expression of CD68, IL-6, iNOS, MCP-1, NOX-2,
p47phox, p67phox and TNF-α (Fig.
3B). The LGI3-treated RAW 264.7 cells exhibited an increased
expression of CD11c, CD68, F4/80, iNOS, MCP-1, NOX-2, p22phox,
p47phox, p67phox and TNF-α (Fig.
3C).
Integrative functional enrichment
analysis of LGI3-regulated genes
The results of adipokine and cytokine arrays,
phosphoprotein arrays and quantitative PCR revealed 23 gene
products upregulated and 11 gene products that were downregulated
by LGI3 in this study (Table I
and Figs. 1Figure 2–3). The proteins that were increased or
decreased in LGI3 knockout mice were presumed to be the proteins
downregulated or upregulated by LGI3, respectively. The findings of
our previous studies indicated that 14 genes were regulated
positively or negatively by LGI3 (Table I) (2–7).
We conducted functional enrichment analysis for LGI3-regulated gene
products using integrative database of functional annotations
(Table II). GO terms of
upregulated genes with the highest significance were associated
with various types of cancer, inflammatory responses, apoptosis and
response to wounding (Table
IIA). Downregulated genes were categorized with the highest
significance into the terms of cell differentiation, growth and
homeostasis (Table IIB).
Functional annotation clustering analysis of all LGI3-regulated
genes revealed that the 5 functional clusters with the highest
scores, included response to endogenous and hormone stimuli,
regulation of cell proliferation, response to wounding,
inflammatory response and regulation of apoptosis (Table III).
 | Table ISummary of LGI3-regulated gene
products from the present study and our previous studies. |
Table I
Summary of LGI3-regulated gene
products from the present study and our previous studies.
| Upregulated
genes | Downregulated
genes |
|---|
| Adipokine/cytokine
arraysa | Adipokine/cytokine
arraysa |
| BLC (CXCL13) | Adiponectin
(ADIPOQ) (8) |
| G-CSF (CSF3) | C5/C5a (C5,
HC) |
| IGF-1 (IGF1) | C-reactive protein
(CRP) |
| IGFBP-5
(IGFBP5) | Endocan
(ESM1) |
| MCP-5 (CCL12) | IGFBP-1
(IGFBP1) |
| MIP-2 (CXCL2) | M-CSF (CSF1) |
| TIMP-1
(TIMP1) | Pref-1 (DLK1) |
| Serpin E1
(SERPINE1) |
| PathScan | PathScanb |
| Akt (AKT1)
(3) | 4E-BP1
(EIF4EBP1) |
| AMPKα
(PRKAA1) | Bad (BAD) |
| Erk1/2
(MAPK3/1) | GSK3α (GSK3A) |
| PTEN (PTEN) | |
| Quantitative
PCR | Previous
studies |
| CD11c (ITGAX) | C/EBPα (CEBPA)
(4) |
| CD68 (CD68) | FABP4 (FABP4)
(4) |
| F4/80 (EMR1) | GSK3β (GSK3B)
(7) |
| IL-6 (IL6) | LPL (LPL) (4) |
| iNOS (NOS2)
(4) | p53 (TP53)
(5) |
| MCP-1 (CCL2)
(9) | PPARγ (PPARG)
(4) |
| NOX-2 (CYBB) | Syntaxin 1
(STX1A)c (2) |
| p22phox
(CYBA) | |
| p47phox
(NCF1) | |
| p67phox
(NCF2) | |
| TNF-α (TNF)
(4,9) | |
| Previous
studies | |
| β-catenin (CTNNB1)
(7) | |
| Cox-2 (PTGS2) | |
| FAK (PTK2)
(3) | |
| MDM2 (MDM2)
(5) | |
| MITF (MIFT)
(6) | |
| NF-κB (NFKB1)
(9) | |
| PI3K (PIK3CA)
(3) | |
 | Table IIFunctional enrichment analysis of the
gene products regulated by LGI3. |
Table II
Functional enrichment analysis of the
gene products regulated by LGI3.
| A, Gene products
regulated positively by LGI3 |
|---|
|
|---|
| Category | Term | P-value |
|---|
| KEGG_PATHWAY | mmu05200:Pathways
in cancer | 1.24E-12 |
| KEGG_PATHWAY | mmu05215:Prostate
cancer | 3.67E-12 |
| KEGG_PATHWAY | mmu04062:Chemokine
signaling pathway | 2.03E-10 |
| KEGG_PATHWAY |
mmu05218:Melanoma | 6.76E-10 |
| KEGG_PATHWAY |
mmu05213:Endometrial cancer | 2.56E-09 |
| KEGG_PATHWAY | mmu04621:NOD-like
receptor signaling pathway | 9.11E-09 |
| KEGG_PATHWAY | mmu04510:Focal
adhesion | 1.58E-07 |
| GOTERM_BP_FAT |
GO:0006954-inflammatory response | 2.74E-07 |
| KEGG_PATHWAY |
mmu05214:Glioma | 3.74E-07 |
| GOTERM_CC_FAT |
GO:0005829-cytosol | 6.18E-07 |
| GOTERM_BP_FAT |
GO:0042981-regulation of apoptosis | 9.91E-07 |
| KEGG_PATHWAY | mmu04370:VEGF
signaling pathway | 1.05E-06 |
| KEGG_PATHWAY | mmu05220:Chronic
myeloid leukemia | 1.05E-06 |
| GOTERM_BP_FAT |
GO:0043067-regulation of programmed cell
death | 1.10E-06 |
| GOTERM_BP_FAT |
GO:0010941-regulation of cell death | 1.15E-06 |
| KEGG_PATHWAY | mmu05222:Small cell
lung cancer | 2.04E-06 |
| KEGG_PATHWAY | mmu05210:Colorectal
cancer | 2.19E-06 |
| KEGG_PATHWAY | mmu04012:ErbB
signaling pathway | 2.34E-06 |
| KEGG_PATHWAY | mmu04150:mTOR
signaling pathway | 4.22E-06 |
| GOTERM_BP_FAT | GO:0009611-response
to wounding | 4.97E-06 |
| B, Gene products
regulated negatively by LGI3 |
|---|
|
|---|
| Category | Term | P-value |
|---|
| GOTERM_BP_FAT |
GO:0010743-regulation of foam cell
differentiation | 1.63E-07 |
| GOTERM_BP_FAT |
GO:0040008-regulation of growth | 2.40E-05 |
| GOTERM_BP_FAT |
GO:0001558-regulation of cell growth | 4.64E-05 |
| GOTERM_BP_FAT | GO:0010558-negative
regulation of macromolecule biosynthetic process | 8.43E-05 |
| GOTERM_BP_FAT | GO:0031327-negative
regulation of cellular biosynthetic process | 8.97E-05 |
| GOTERM_BP_FAT | GO:0009890-negative
regulation of biosynthetic process | 1.01E-04 |
| INTERPRO |
IPR000867:Insulin-like growth
factor-binding protein, IGFBP | 1.43E-04 |
| KEGG_PATHWAY | bta03320:PPAR
signaling pathway | 1.89E-04 |
| GOTERM_BP_FAT | GO:0010605-negative
regulation of macromolecule metabolic process | 1.95E-04 |
| GOTERM_BP_FAT | GO:0050873-brown
fat cell differentiation | 2.59E-04 |
| GOTERM_MF_FAT |
GO:0005520-insulin-like growth factor
binding | 2.89E-04 |
| SMART | SM00121:IB -insulin
growth factor-binding protein homologues | 3.64E-04 |
| GOTERM_BP_FAT |
GO:0042127-regulation of cell
proliferation | 3.81E-04 |
| GOTERM_CC_FAT |
GO:0005576-extracellular region | 5.76E-04 |
| GOTERM_BP_FAT | GO:0045444-fat cell
differentiation | 6.34E-04 |
| GOTERM_BP_FAT |
GO:0042592-homeostatic process | 9.81E-04 |
| GOTERM_MF_FAT | GO:0019838-growth
factor binding | 1.90E-03 |
| GOTERM_MF_FAT | GO:0046983-protein
dimerization activity | 2.46E-03 |
| GOTERM_BP_FAT | GO:0048878-chemical
homeostasis | 2.81E-03 |
| GOTERM_BP_FAT | GO:0045596-negative
regulation of cell differentiation | 4.73E-03 |
 | Table IIIFunctional annotation clustering
analysis of LGI3-regulated gene products. |
Table III
Functional annotation clustering
analysis of LGI3-regulated gene products.
| Category | Term | Count | P-value |
|---|
| Annotation cluster
1 | Enrichment score:
10.13 | | |
| GOTERM_BP_FAT | GO:0009719-response
to endogenous stimulus | 19 | 3.63E-13 |
| GOTERM_BP_FAT | GO:0009725-response
to hormone stimulus | 18 | 7.76E-13 |
| GOTERM_BP_FAT | GO:0048545-response
to steroid hormone stimulus | 14 | 1.85E-11 |
| GOTERM_BP_FAT | GO:0010033-response
to organic substance | 20 | 1.22E-10 |
| GOTERM_BP_FAT | GO:0051384-response
to glucocorticoid stimulus | 9 | 1.27E-08 |
| GOTERM_BP_FAT | GO:0031960-response
to corticosteroid stimulus | 9 | 1.95E-08 |
| Annotation cluster
2 | Enrichment score:
9.36 | | |
| GOTERM_BP_FAT |
GO:0042127-regulation of cell
proliferation | 19 | 4.52E-12 |
| GOTERM_BP_FAT | GO:0048545-response
to steroid hormone stimulus | 14 | 1.85E-11 |
| GOTERM_BP_FAT | GO:0008284-positive
regulation of cell proliferation | 11 | 9.64E-07 |
| Annotation cluster
3 | Enrichment score:
8.51 | | |
| GOTERM_BP_FAT | GO:0009611-response
to wounding | 16 | 1.06E-11 |
| GOTERM_BP_FAT |
GO:0006954-inflammatory response | 11 | 4.10E-09 |
| GOTERM_BP_FAT | GO:0006952-defense
response | 13 | 1.15E-08 |
| GOTERM_BP_FAT | GO:0006955-immune
response | 12 | 1.84E-07 |
| Annotation cluster
4 | Enrichment score:
7.49 | | |
| GOTERM_BP_FAT |
GO:0042981-regulation of apoptosis | 17 | 7.48E-10 |
| GOTERM_BP_FAT |
GO:0043067-regulation of programmed cell
death | 17 | 9.07E-10 |
| GOTERM_BP_FAT |
GO:0010941-regulation of cell death | 17 | 9.67E-10 |
| GOTERM_BP_FAT | GO:0043066-negative
regulation of apoptosis | 13 | 1.49E-09 |
| GOTERM_BP_FAT | GO:0043069-negative
regulation of programmed cell death | 13 | 1.77E-09 |
| GOTERM_BP_FAT | GO:0060548-negative
regulation of cell death | 13 | 1.83E-09 |
| GOTERM_BP_FAT | GO:0043065-positive
regulation of apoptosis | 9 | 2.13E-05 |
| GOTERM_BP_FAT | GO:0043068-positive
regulation of programmed cell death | 9 | 2.22E-05 |
| GOTERM_BP_FAT | GO:0010942-positive
regulation of cell death | 9 | 2.37E-05 |
| Annotation cluster
5 | Enrichment score:
4.81 | | |
| GOTERM_BP_FAT |
GO:0040007-growth | 10 | 6.21E-08 |
| GOTERM_BP_FAT |
GO:0031099-regeneration | 7 | 3.77E-06 |
| GOTERM_BP_FAT | GO:0042060-wound
healing | 6 | 4.02E-04 |
| GOTERM_BP_FAT | GO:0042246-tissue
regeneration | 4 | 5.73E-04 |
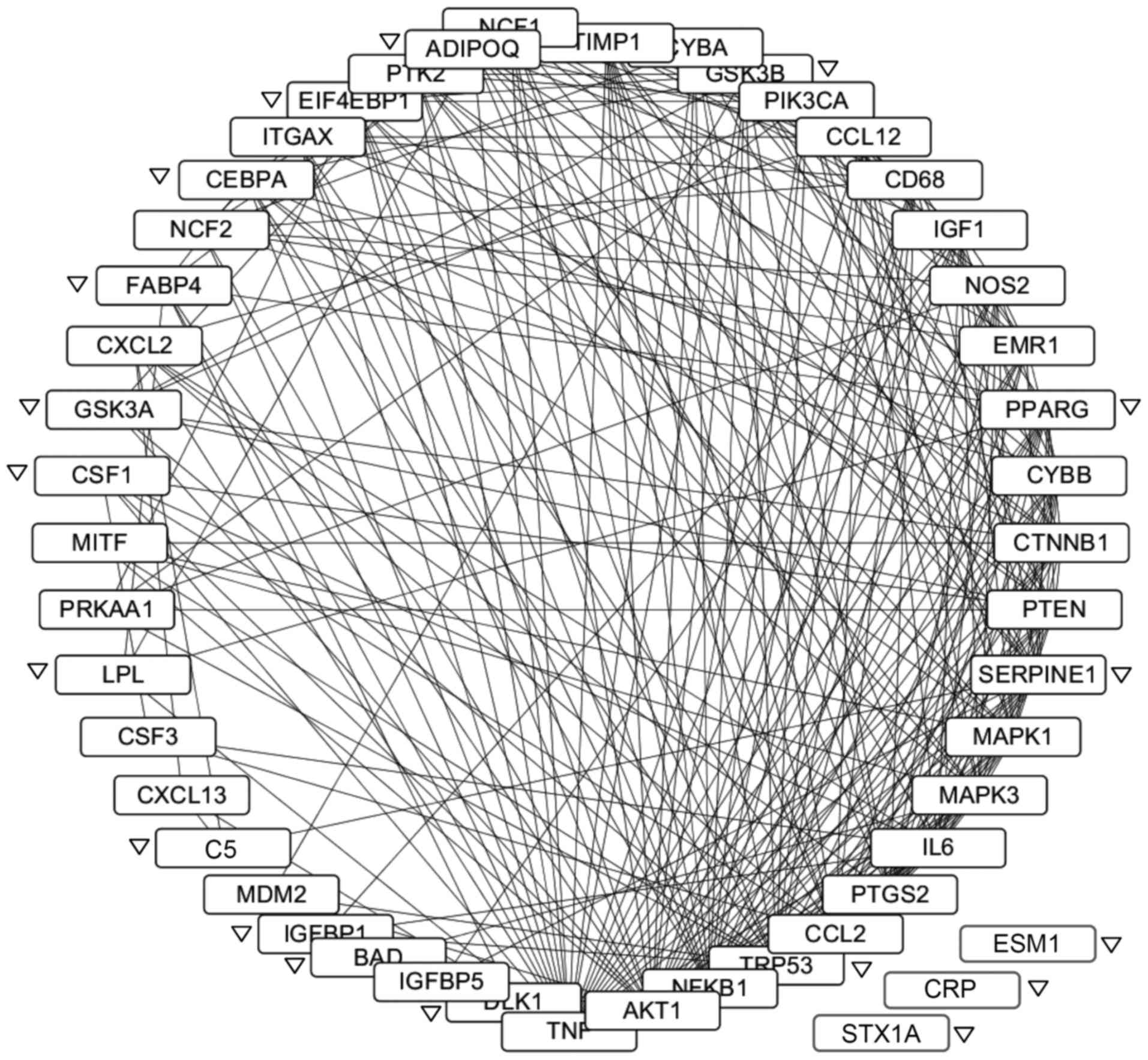
Protein-protein interaction network of
LGI3-regulated gene products
The protein-protein interaction network of
LGI3-regulated genes was constructed (Fig. 4). All gene products except for
endocan, CRP and syntaxin 1A formed an interaction cluster. A total
of 270 interactions were observed involving 45 gene products.
TNF-α, Akt, NF-κB, p53 and CCL2 (MCP-1) were the gene products with
the highest degree of interactions. All the genes upregulated by
LGI3 and 83% (15/18) of the downregulated genes participated in the
network. Four subnetworks were identified by ClusterONE (p<0.01,
data not shown). Functional enrichment analysis indicated that the
subnetworks were associated with developmental process, the
regulation of apoptosis, response to hormone stimuli and response
to stress (data not shown).
Discussion
LGI3 has been postulated to participate in the
adipokine network through the regulation of adiponectin and TNF-α
in obesity-associated metabolic inflammation (8,9).
Among the multiple molecular mass forms of LGI3 (75-, 60- and
35-kDa), 60-kDa LGI3 has been shown to be selectively increased in
obese adipose tissues and has been suggested to be a major
adipokine form (4,8). LGI3 knockout mice exhibit a
selective ablation of 60-kDa LGI3 (8). The alteration of various adipokines
and cytokines in LGI3 knockout mice supports the widespread
functional interaction of adipokines and cytokines with LGI3
(Fig. 1). The increased levels of
adipokines and cytokines in LGI3 knockout mice may represent
compensatory upregulation under LGI3 deficiency or may be the
factors that are negatively regulated by LGI3 in wild-type mice
(Fig. 1, a–h). Downregulated
factors in LGI3 knockout mice suggested that LGI3 may cooperate
with these factors by positive regulation. These results revealed
the adipokine and cytokine network regulated by LGI3 that may play
a role in fine-tuning responses to metabolic perturbation and other
stimuli.
Intracellular signaling pathways of LGI3 were
explored in our previous studies. In neuronal cells, LGI3-promoted
neurite outgrowth was mediated by Akt and FAK (3). MDM2 and p53 were shown to be
involved in LGI3-promoted survival under UVB irradiation in
keratinocytes (5). Keratinocyte
migration induced by LGI3 was shown to employ β-catenin and GSK3β
(7). The present study on
pre-adipocytes demonstrated that LGI3 treatment regulated a
multitude of signaling proteins (Erk1/2, AMPK, GSK3α, Bad, PTEN and
4E-BP1, as well as previously observed Akt) (Fig. 2, a–g) (3,7).
These results suggested that the intracellular signaling of LGI3
may consist of common and unique signaling components in different
target cell types. The suppressive effect of LGI3 on adipogenesis
was shown to be mediated by its receptor, ADAM23 (4). ADAM22, other putative LGI3 receptor
abundantly expressed in brain, was not expressed in adipose tissues
and 3T3-L1 cells (4). Thus, LGI3
and ADAM23 may transduce intracellular signaling through these
proteins (Fig. 2, a–g) in
pre-adipocytes in adipose tissues.
We previously proposed that LGI3 may be a
pro-inflammatory adipokine based on its upregulating effect on
COX-2, iNOS, MCP-1, TNF-α and NF-κB (4,9).
In this study, LGI3 increased the expression of multiple
inflammatory genes differentially in pre-adipocytes, adipocytes and
macrophages that are the predominant cell types in adipose tissues
(Fig. 3). All of the
LGI3-regulated inflammatory gene products have been previously
reported to be associated positively with obesity-associated
metabolic disorders (13–23). In this study, following treatment
with LGI3, the significant upregulation of CD68, MCP-1, NOX-2,
p67phox and TNF-α was observed in 3 cell types. F4/80 and p22phox
were only upregulated in RAW 264.7 macrophages (Fig. 3C). As LGI3 was previously shown to
be secreted predominantly by pre-adipocytes and macrophages
(4), these results suggested that
LGI3 may act as an autocrine and paracrine adipokine that conveys
pro-inflammatory stimuli on target cells.
The findings of the present study and those of
previous studies demonstrated that an array of proteins were
regulated by LGI3 in different cell types (Table I). As expected, functional
enrichment analysis revealed that LGI3-regulated gene products were
associated with inflammatory responses and related path-ways, such
as chemokines, apoptosis, focal adhesion (Table IIA) and with cell growth and
differentiation (Table IIB). It
is noted that GO terms of the genes upregulated by LGI3 were
associated with various cancers including prostate, melanoma,
endometrial and glioma cancers (Table IIA). A previous study
demonstrated that LGI3 was expressed at high levels in glioma,
neuroblastoma, melanoma, colon and breast cancer cells (24). Among four LGI family members, LGI3
was the only member expressed at very high levels in gliomas,
melanomas and neuroblastoma cells (24). Cytokine networks play regulatory
roles in cancer through inflammatory immune responses to tumors
(25,26) or chronic and carcinogenic
inflammation (25). TNF-α and
adiponectin have been described to be risk factors and potential
prognostic biomarkers in various types of cancer (26,27). Since LGI3 has been shown to
regulate adiponectin and TNF-α (8,9),
LGI3 may also be associated with cytokine networks in cancer.
Furthermore, functional annotation clustering analysis supported
the notion that LGI3 may act as a pleiotropic cytokine in various
biological processes such as hormonal stimuli, proliferation,
wounding and inflammatory responses and apoptosis (Table III).
It is remarkable that a majority of LGI3-regulated
gene products formed a cluster of protein-protein interaction
network (Fig. 4). The
upregulation of LGI3 in obese adipose tissues has been proposed to
contribute to perturbation of cytokine network in metabolic
inflammation (4,8). The LGI3-regulated protein
interaction network may account for the complex mechanisms in
dysregulation of adipose tissue homeostasis in obesity. This notion
was supported by the proteins with the highest degree of
interactions in the network (TNF-α, Akt, NF-κB, p53, CCL2, IL-6,
Erk2, serpin E1, PTEN, PPARγ and IGF-1) that were previously
reported to be involved in obesity-associated metabolic disorders
(15,16,28–34). Since NF-κB, a crucial inflammatory
transcription factor, was shown to mediate mutual upregulation of
LGI3 and TNF-α in adipose tissues (9), TNF-α and NF-κB may be the major
players that mediate the effect of LGI3 on this network.
In conclusion, the present study presented an
integrative insight into LGI3-regulated gene products identified in
this study and our previous studies. High degree interactions
within the network of LGI3-regulated gene products suggested that
LGI3 may be profoundly involved in a wide variety of biological
processes regulated by cytokines. These findings supported our
hypothesis that LGI3 is an adipokine that participates in cytokine
networks involved in metabolic inflammation and related
disorders.
Acknowledgments
This study was supported by Basic Science Research
Program through the National Research Foundation of Korea (NRF)
funded by the Ministry of Education (grant no.
NRF-2015R1D1A1A01056981).
References
|
1
|
Lee SE, Lee AY, Park WJ, Jun DH, Kwon NS,
Baek KJ, Kim YG and Yun HY: Mouse LGI3 gene: expression in brain
and promoter analysis. Gene. 372:8–17. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Park WJ, Lee SE, Kwon NS, Baek KJ, Kim DS
and Yun HY: Leucine-rich glioma inactivated 3 associates with
syntaxin 1. Neurosci Lett. 444:240–244. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Park WJ, Lim YY, Kwon NS, Baek KJ, Kim DS
and Yun HY: Leucine-rich glioma inactivated 3 induces neurite
outgrowth through Akt and focal adhesion kinase. Neurochem Res.
35:789–796. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim HA, Park WJ, Jeong HS, Lee HE, Lee SH,
Kwon NS, Baek KJ, Kim DS and Yun HY: Leucine-rich glioma
inactivated 3 regulates adipogenesis through ADAM23. Biochim
Biophys Acta. 1821:914–922. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee SH, Jeong YM, Kim SY, Jeong HS, Park
KC, Baek KJ, Kwon NS, Yun HY and Kim DS: Ultraviolet B-induced LGI3
secretion protects human keratinocytes. Exp Dermatol. 21:716–718.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jeong HS, Jeong YM, Kim J, Lee SH, Choi
HR, Park KC, Kim BJ, Baek KJ, Kwon NS, Yun HY, et al: Leucine-rich
glioma inactivated 3 is a melanogenic cytokine in human skin. Exp
Dermatol. 23:600–602. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jeong YM, Park WJ, Kim MK, Baek KJ, Kwon
NS, Yun HY and Kim DS: Leucine-rich glioma inactivated 3 promotes
HaCaT keratinocyte migration. Wound Repair Regen. 21:634–640. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim HA, Kwon NS, Baek KJ, Kim DS and Yun
HY: Leucine-rich glioma inactivated 3 associates negatively with
adiponectin. Cytokine. 62:206–209. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim HA, Kwon NS, Baek KJ, Kim DS and Yun
HY: Leucine-rich glioma inactivated 3 and tumor necrosis factor-α
regulate mutually through NF-κB. Cytokine. 72:220–223. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar
|
|
11
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar
|
|
12
|
Lopes CT, Franz M, Kazi F, Donaldson SL,
Morris Q and Bader GD: Cytoscape web: an interactive web-based
network browser. Bioinformatics. 26:2347–2348. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Antonopoulos AS, Margaritis M, Coutinho P,
Shirodaria C, Psarros C, Herdman L, Sanna F, De Silva R, Petrou M,
Sayeed R, et al: Adiponectin as a link between type 2 diabetes and
vascular NADPH oxidase activity in the human arterial wall: the
regulatory role of perivascular adipose tissue. Diabetes.
64:2207–2219. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Du J, Fan LM, Mai A and Li JM: Crucial
roles of Nox2-derived oxidative stress in deteriorating the
function of insulin receptors and endothelium in dietary obesity of
middle-aged mice. Br J Pharmacol. 170:1064–1077. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fernandez-Twinn DS, Blackmore HL, Siggens
L, Giussani DA, Cross CM, Foo R and Ozanne SE: The programming of
cardiac hypertrophy in the offspring by maternal obesity is
associated with hyperinsulinemia, AKT, ERK, and mTOR activation.
Endocrinology. 153:5961–5971. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hotamisligil GS, Shargill NS and
Spiegelman BM: Adipose expression of tumor necrosis factor-alpha:
direct role in obesity-linked insulin resistance. Science.
259:87–91. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kanda H, Tateya S, Tamori Y, Kotani K,
Hiasa K, Kitazawa R, Kitazawa S, Miyachi H, Maeda S, Egashira K, et
al: MCP-1 contributes to macrophage infiltration into adipose
tissue, insulin resistance, and hepatic steatosis in obesity. J
Clin Invest. 116:1494–1505. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kapur S, Marcotte B and Marette A:
Mechanism of adipose tissue iNOS induction in endotoxemia. Am J
Physiol. 276:E635–E641. 1999.PubMed/NCBI
|
|
19
|
Pietiläinen KH, Kannisto K,
Korsheninnikova E, Rissanen A, Kaprio J, Ehrenborg E, Hamsten A and
Yki-Järvinen H: Acquired obesity increases CD68 and tumor necrosis
factor-alpha and decreases adiponectin gene expression in adipose
tissue: a study in monozygotic twins. J Clin Endocrinol Metab.
91:2776–2781. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ronis MJ, Sharma N, Vantrease J,
Borengasser SJ, Ferguson M, Mercer KE, Cleves MA, Gomez-Acevedo H
and Badger TM: Female mice lacking p47phox have altered
adipose tissue gene expression and are protected against high
fat-induced obesity. Physiol Genomics. 45:351–366. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sindhu S, Thomas R, Shihab P, Sriraman D,
Behbehani K and Ahmad R: Obesity is a positive modulator of IL-6R
and IL-6 expression in the subcutaneous adipose tissue:
significance for metabolic inflammation. PLoS One. 10:e01334942015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Uchida K, Satoh M, Inoue G, Onuma K,
Miyagi M, Iwabuchi K and Takaso M: CD11c(+) macrophages and levels
of TNF-α and MMP-3 are increased in synovial and adipose tissues of
osteoarthritic mice with hyperlipidaemia. Clin Exp Immunol.
180:551–559. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Weisberg SP, McCann D, Desai M, Rosenbaum
M, Leibel RL and Ferrante AW Jr: Obesity is associated with
macrophage accumulation in adipose tissue. J Clin Invest.
112:1796–1808. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rossi MR, Huntoon K and Cowell JK:
Differential expression of the LGI and SLIT families of genes in
human cancer cells. Gene. 356:85–90. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
West NR, McCuaig S, Franchini F and Powrie
F: Emerging cytokine networks in colorectal cancer. Nat Rev
Immunol. 15:615–629. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lippitz BE: Cytokine patterns in patients
with cancer: a systematic review. Lancet Oncol. 14:e218–e228. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dalamaga M, Diakopoulos KN and Mantzoros
CS: The role of adiponectin in cancer: a review of current
evidence. Endocr Rev. 33:547–594. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Choi JH, Banks AS, Estall JL, Kajimura S,
Boström P, Laznik D, Ruas JL, Chalmers MJ, Kamenecka TM, Blüher M,
et al: Anti-diabetic drugs inhibit obesity-linked phosphorylation
of PPARgamma by Cdk5. Nature. 466:451–456. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang XF and Chen JZ: Obesity, the
PI3K/Akt signal pathway and colon cancer. Obes Rev. 10:610–616.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kaur P, Reis MD, Couchman GR, Forjuoh SN,
Greene JF and Asea A: SERPINE 1 links obesity and diabetes: a pilot
study. J Proteomics Bioinform. 3:191–199. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kumar PA, Chitra PS, Lu C, Sobhanaditya J
and Menon R: Growth hormone (GH) differentially regulates NF-kB
activity in preadipocytes and macrophages: implications for GH's
role in adipose tissue homeostasis in obesity. J Physiol Biochem.
70:433–440. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mauer J, Chaurasia B, Goldau J, Vogt MC,
Ruud J, Nguyen KD, Theurich S, Hausen AC, Schmitz J, Brönneke HS,
et al: Signaling by IL-6 promotes alternative activation of
macrophages to limit endotoxemia and obesity-associated resistance
to insulin. Nat Immunol. 15:423–430. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pal A, Barber TM, Van de Bunt M, Rudge SA,
Zhang Q, Lachlan KL, Cooper NS, Linden H, Levy JC, Wakelam MJ, et
al: PTEN mutations as a cause of constitutive insulin sensitivity
and obesity. N Engl J Med. 367:1002–1011. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yokoyama M, Okada S, Nakagomi A, Moriya J,
Shimizu I, Nojima A, Yoshida Y, Ichimiya H, Kamimura N, Kobayashi
Y, et al: Inhibition of endothelial p53 improves metabolic
abnor-malities related to dietary obesity. Cell Reports.
7:1691–1703. 2014. View Article : Google Scholar
|