|
1
|
Kim YK, Kim YS, Yoo KJ, Lee HJ, Lee DR,
Yeo CY and Baek KH: The expression of Usp42 during embryogenesis
and spermatogenesis in mouse. Gene Expr Patterns. 7:143–148. 2007.
View Article : Google Scholar
|
|
2
|
Quesada V, Díaz-Perales A,
Gutiérrez-Fernández A, Garabaya C, Cal S and López-Otín C: Cloning
and enzymatic analysis of 22 novel human ubiquitin-specific
proteases. Biochem Biophys Res Commun. 314:54–62. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Baek SH, Park KC, Lee JI II, Kim KI II,
Yoo YJ, Tanaka K, Baker RT and Chung CH: A novel family of
ubiquitin-specific proteases in chick skeletal muscle with distinct
N- and C-terminal extensions. Biochem J. 334:677–684. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
D'Andrea A and Pellman D: Deubiquitinating
enzymes: a new class of biological regulators. Crit Rev Biochem Mol
Biol. 33:337–352. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wing SS: Deubiquitinating enzymes - the
importance of driving in reverse along the ubiquitin-proteasome
pathway. Int J Biochem Cell Biol. 35:590–605. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Everett RD, Meredith M, Orr A, Cross A,
Kathoria M and Parkinson J: A novel ubiquitin-specific protease is
dynamically associated with the PML nuclear domain and binds to a
herpesvirus regulatory protein. EMBO J. 16:1519–1530. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang W, Tian QB, Li QK, Wang JM, Wang CN,
Liu T, Liu DW and Wang MW: Lysine 92 amino acid residue of USP46, a
gene associated with 'behavioral despair' in mice, influences the
deubiquitinating enzyme activity. PLoS One. 6:e262972011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dang LC, Melandri FD and Stein RL: Kinetic
and mechanistic studies on the hydrolysis of ubiquitin C-terminal
7-amido-4-methylcoumarin by deubiquitinating enzymes. Biochemistry.
37:1868–1879. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yin ST, Huang H, Zhang YH, Zhou ZR, Song
AX, Hong FS and Hu HY: A fluorescence assay for elucidating the
substrate specificities of deubiquitinating enzymes. Biochem
Biophys Res Commun. 416:76–79. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Timms KM, Ansari-Lari MA, Morris W, Brown
SN and Gibbs RA: The genomic organization of isopeptidase T-3
(ISOT-3), a new member of the ubiquitin specific protease family
(UBP). Gene. 217:101–106. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Stein RL, Chen Z and Melandri F: Kinetic
studies of isopeptidase T: modulation of peptidase activity by
ubiquitin. Biochemistry. 34:12616–12623. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lacombe T and Gabriel JM: Further
characterization of the putative human isopeptidase T catalytic
site. FEBS Lett. 531:469–474. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Reyes-Turcu FE, Horton JR, Mullally JE,
Heroux A, Cheng X and Wilkinson KD: The ubiquitin binding domain
ZnF UBP recognizes the C-terminal diglycine motif of unanchored
ubiquitin. Cell. 124:1197–1208. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
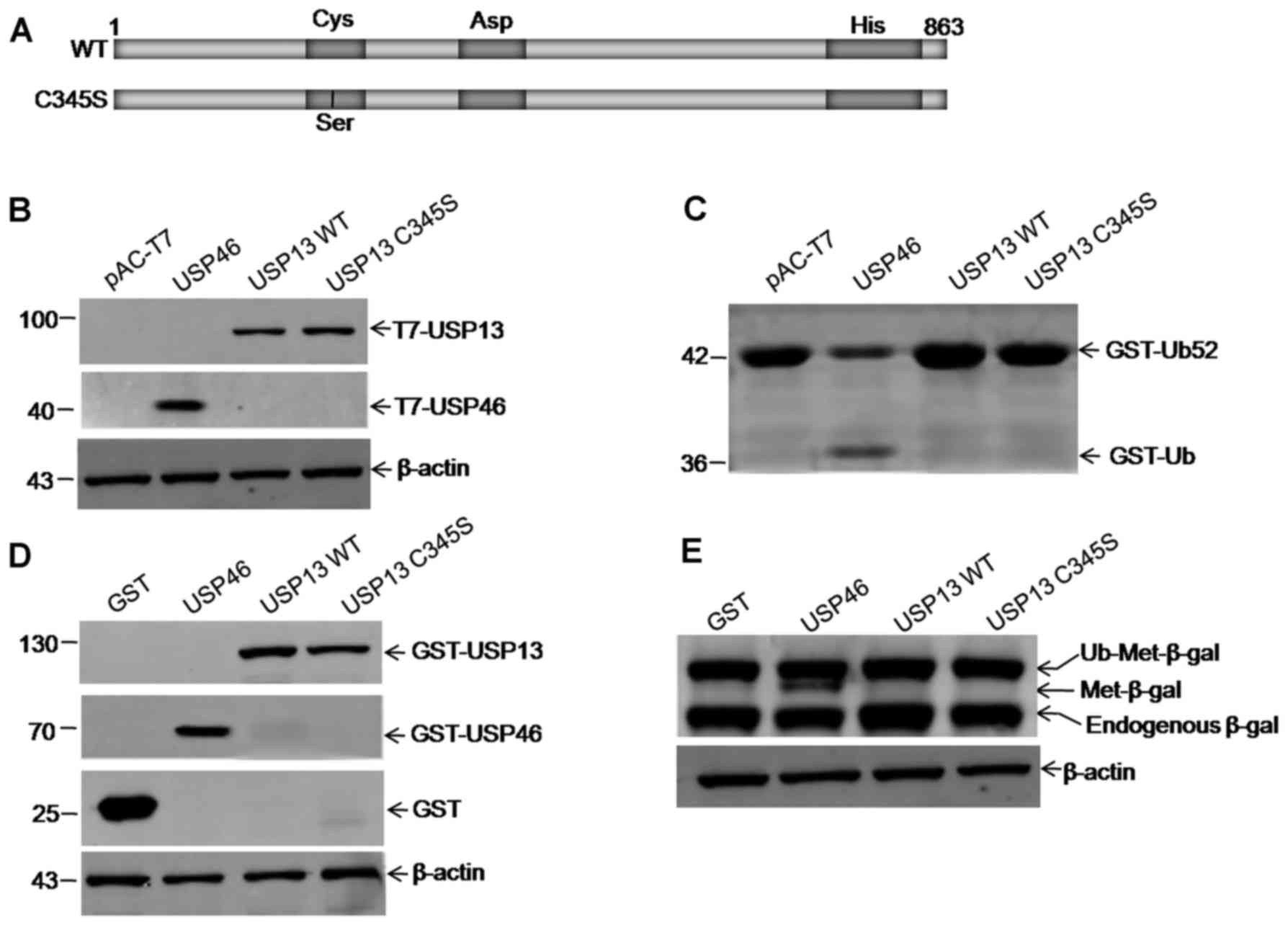
Zhang YH, Zhou CJ, Zhou ZR, Song AX and Hu
HY: Domain analysis reveals that a deubiquitinating enzyme USP13
performs non-activating catalysis for Lys63-linked polyubiquitin.
PLoS One. 6:e293622011. View Article : Google Scholar
|
|
15
|
Bonnet J, Romier C, Tora L and Devys D:
Zinc-finger UBPs: regulators of deubiquitylation. Trends Biochem
Sci. 33:369–375. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Catic A, Fiebiger E, Korbel GA, Blom D,
Galardy PJ and Ploegh HL: Screen for ISG15-crossreactive
deubiquitinases. PLoS One. 2:e6792007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu Y, Soetandyo N, Lee JG, Liu L, Xu Y,
Clemons WM Jr and Ye Y: USP13 antagonizes gp78 to maintain
functionality of a chaperone in ER-associated degradation. eLife.
3:e013692014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Scortegagna M, Subtil T, Qi J, Kim H, Zhao
W, Gu W, Kluger H and Ronai ZA: USP13 enzyme regulates Siah2 ligase
stability and activity via noncatalytic ubiquitin-binding domains.
J Biol Chem. 286:27333–27341. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yeh HM, Yu CY, Yang HC, Ko SH, Liao CL and
Lin YL: Ubiquitin-specific protease 13 regulates IFN signaling by
stabilizing STAT1. J Immunol. 191:3328–3336. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen M, Gutierrez GJ and Ronai ZA:
Ubiquitin-recognition protein Ufd1 couples the endoplasmic
reticulum (ER) stress response to cell cycle control. Proc Natl
Acad Sci USA. 108:9119–9124. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao X, Fiske B, Kawakami A, Li J and
Fisher DE: Regulation of MITF stability by the USP13
deubiquitinase. Nat Commun. 2:4142011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L,
Cai Y, Norberg HV, Zhang T, Furuya T, et al: Beclin1 controls the
levels of p53 by regulating the deubiquitination activity of USP10
and USP13. Cell. 147:223–234. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang J, Zhang P, Wei Y, Piao HL, Wang W,
Maddika S, Wang M, Chen D, Sun Y, Hung MC, et al: Deubiquitylation
and stabilization of PTEN by USP13. Nat Cell Biol. 15:1486–1494.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Geng J, Huang X, Li Y, Xu X, Li S, Jiang
D, Liang J, Jiang D, Wang C and Dai H: Down-regulation of USP13
mediates phenotype transformation of fibroblasts in idiopathic
pulmonary fibrosis. Respir Res. 16:1242015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu YL, Zheng J, Tang LJ, Han W, Wang JM,
Liu DW and Tian QB: The deubiquitinating enzyme activity of USP22
is necessary for regulating HeLa cell growth. Gene. 572:49–56.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tian QB, Okano A, Nakayama K, Miyazawa S,
Endo S and Suzuki T: A novel ubiquitin-specific protease, synUSP,
is localized at the post-synaptic density and post-synaptic lipid
raft. J Neurochem. 87:665–675. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tang LJ, Li Y, Liu YL, Wang JM, Liu DW and
Tian QB: USP12 regulates cell cycle progression by involving c-Myc,
cyclin D2 and BMI-1. Gene. 578:92–99. 2016. View Article : Google Scholar
|
|
28
|
Bradford MM: A rapid and sensitive method
for the quantitation of microgram quantities of protein utilizing
the principle of protein-dye binding. Anal Biochem. 72:248–254.
1976. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tang W: Quantitative analysis of plasma
membrane proteome using two-dimensional difference gel
electrophoresis. Methods Mol Biol. 876:67–82. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
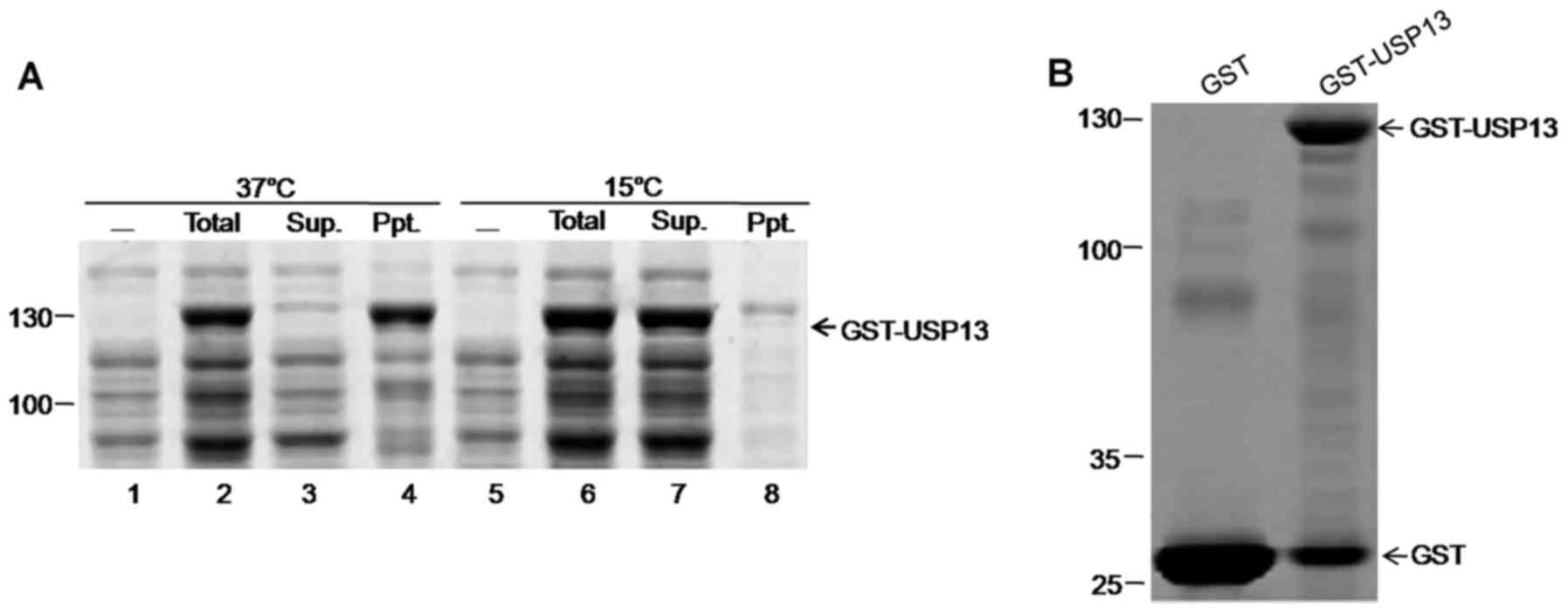
Shirano Y and Shibata D: Low temperature
cultivation of Escherichia coli carrying a rice lipoxygenase L-2
cDNA produces a soluble and active enzyme at a high level. FEBS
Lett. 271:128–130. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kataeva I, Chang J, Xu H, Luan CH, Zhou J,
Uversky VN, Lin D, Horanyi P, Liu ZJ, Ljungdahl LG, et al:
Improving solubility of Shewanella oneidensis MR-1 and Clostridium
thermocellum JW-20 proteins expressed into Esherichia coli. J
Proteome Res. 4:1942–1951. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Volontè F, Marinelli F, Gastaldo L, Sacchi
S, Pilone MS, Pollegioni L and Molla G: Optimization of
glutaryl-7-aminocephalosporanic acid acylase expression in E coli.
Protein Expr Purif. 61:131–137. 2008. View Article : Google Scholar
|
|
33
|
Chou CP: Engineering cell physiology to
enhance recombinant protein production in Escherichia coli. Appl
Microbiol Biotechnol. 76:521–532. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Larsen CN, Krantz BA and Wilkinson KD:
Substrate specificity of deubiquitinating enzymes: ubiquitin
C-terminal hydrolases. Biochemistry. 37:3358–3368. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kolkman A, Dirksen EH, Slijper M and Heck
AJ: Double standards in quantitative proteomics: direct comparative
assessment of difference in gel electrophoresis and metabolic
stable isotope labeling. Mol Cell Proteomics. 4:255–266. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Levy C, Khaled M and Fisher DE: MITF:
master regulator of melanocyte development and melanoma oncogene.
Trends Mol Med. 12:406–414. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
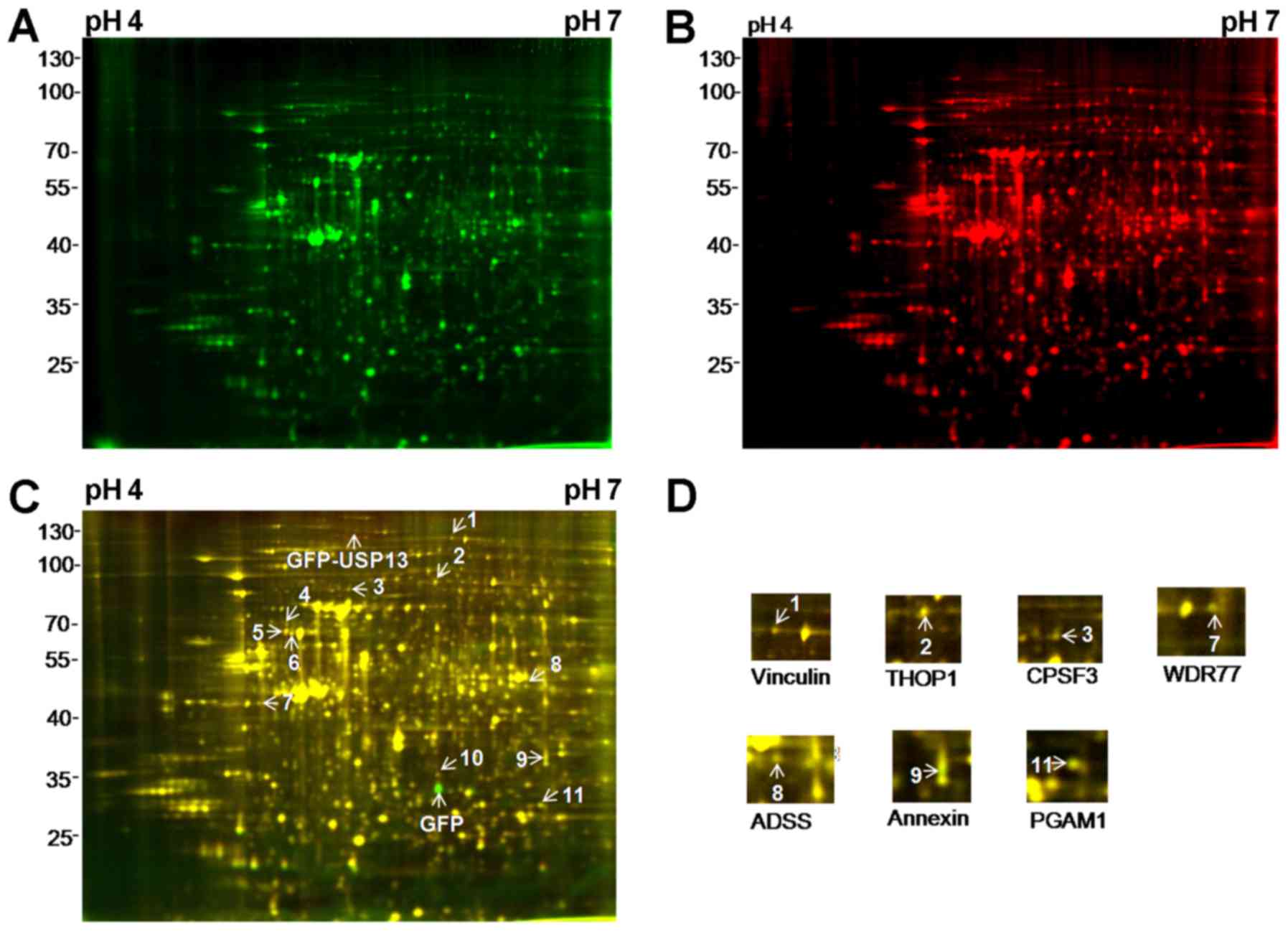
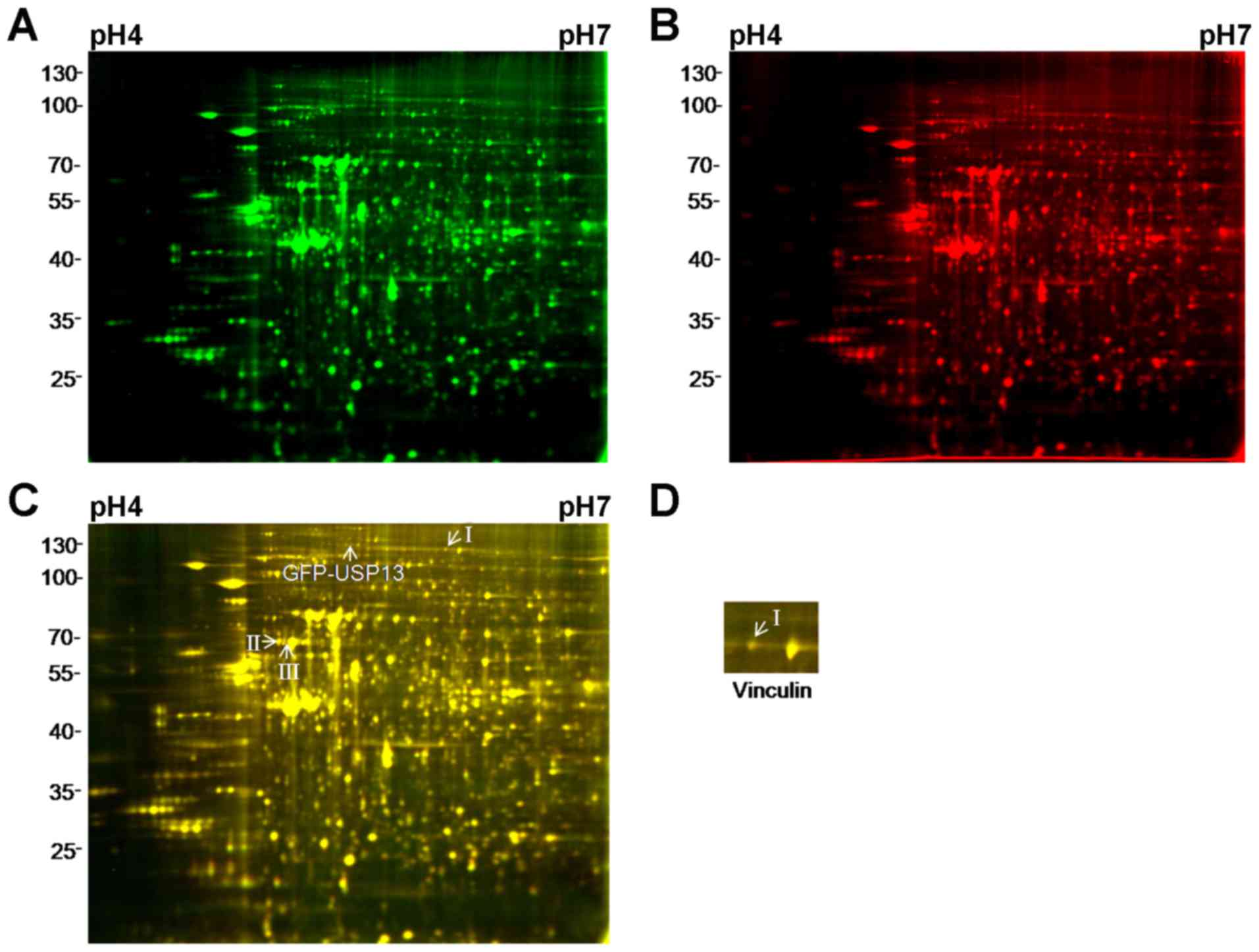
Ziegler WH, Liddington RC and Critchley
DR: The structure and regulation of vinculin. Trends Cell Biol.
16:453–460. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Das C, Hoang QQ, Kreinbring CA, Luchansky
SJ, Meray RK, Ray SS, Lansbury PT, Ringe D and Petsko GA:
Structural basis for conformational plasticity of the Parkinson's
disease-associated ubiquitin hydrolase UCH-L1. Proc Natl Acad Sci
USA. 103:4675–4680. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Frisan T, Coppotelli G, Dryselius R and
Masucci MG: Ubiquitin C-terminal hydrolase-L1 interacts with
adhesion complexes and promotes cell migration, survival, and
anchorage independent growth. FASEB J. 26:5060–5070. 2012.
View Article : Google Scholar : PubMed/NCBI
|