Introduction
Gestational trophoblastic diseases (GTDs) are a
group of diseases characterized by abnormal cellular proliferation
of atypical trophoblasts and include hydatidiform moles, invasive
moles, choriocarcinomas, placental site trophoblastic tumors, and
epithelial trophoblastic tumors. A hydatidiform mole is an abnormal
pregnancy caused by genetic fertilization disorders, and it can be
classified as a complete hydatidiform mole (CHM) or a partial
hydatidiform mole (PHM). CHMs are androgenic in origin and occur
through fertilization of an ovum with an inactivated or eliminated
nucleus, with two sperms (dispermy) or with a haploid sperm
followed by duplication of its chromosomes (1,2).
PHMs are biparental and are generally dispermic triploids (3,4).
Hydatidiform moles, especially CHMs, have a high potential to
develop into cancer, with invasive moles developing in 15–24% of
CHM cases (5–7). The risk associated with
choriocarcinoma is 2,000–4,000 times higher in hydatidiform moles
than in normal pregnancy or abortion (2).
Previous studies on hydatidiform moles have examined
the genes or proteins involved using tissues of hydatidiform moles.
Investigation into the function of trophoblasts of hydatidiform
moles has not been performed and is largely limited by the short
life spans of primary cultured trophoblasts in vitro. A cell
line established from a hydatidiform mole would allow for
investigation into the function of molar trophoblasts. The cell
line CHM1 was established from CHM and has been used only for
genomic studies making use of characteristics of a single haplotype
(8,9). Because a hydatidiform mole is a type
of pregnancy, it is difficult to maintain a primary culture of
growing non-cancerous trophoblasts for more than a few weeks. There
are, however, previous studies that have established cell lines
from normal human or animal cells using a transfection technique
involving human telomerase reverse transcriptase (hTERT), as well
as mutant forms of CDK (CDK4R24C) and cyclin D1 (10–12). The gain in telomerase activity
obtained through expression of hTERT was found to be insufficient
to make a cell line derived from normal cells immortal (13). The aim of this study was to
establish cell lines from CHMs and to characterize the cells for
future studies on hydatidiform moles and gestational trophoblastic
neoplasia.
Materials and methods
Tissue collection and processing
This study was approved by the Ethics Committee of
Nagoya University Graduate School of Medicine. CHM tissues were
obtained from patients who had undergone evacuations at 8–10 weeks
of gestation (n=6). CHM diagnoses were confirmed by pathological
examination. Intraoral cells were obtained by OmniSwab (GE
Healthcare, Chicago, IL, USA) from two of the CHM patients and
their husbands for short tandem repeat (STR) analysis. Informed
consent was obtained from the patients and their husbands for the
use of the molar tissues and their intraoral cells.
Cell line and antibodies
Human choriocarcinoma cell line Jar was purchased
from the American Type Culture Collection (Manassas, VA, USA) and
was grown in RPMI-1640 medium (Sigma-Aldrich, St. Louis, MO, USA)
supplemented with 10% FCS, penicillin (100 U/ml), streptomycin (100
µg/ml), and 2 mM glutamine. Cultures were incubated at 37°C in 5%
CO2. Anti-hCG antibody (Ab) (N1534, rabbit polyclonal),
anti-human placental lactogen (hPL) Ab (A0137, rabbit polyclonal),
and anti-human cytokeratin 7 (CK7) Ab (ready-to-use N-series, mouse
monoclonal) were purchased from Dako (Glostrup, Denmark).
Anti-CK8/18 Ab (MM-1700-01, mouse monoclonal) was purchased from
ImmunoBioScience (Mukilteo, WA, USA) and anti-vimentin Ab
(ready-to-use N-series, mouse monoclonal) was purchased from
Nichirei Biosciences Inc. (Tokyo, Japan).
Human CHM explant culture
Molar explant culture was established using molar
tissues obtained from evacuations (8–10 weeks, n=6). Molar tissues
were washed with phosphate-buffered saline (PBS) and aseptically
dissected to remove blood and decidual tissues, and then only molar
vesicles were collected. After teasing apart small fragments of
molar vesicles, molar fragments were placed in collagen type
I-coated dishes (BD Biosciences, Franklin Lakes, NJ, USA) and
incubated in Dulbecco's modified Eagle's medium (DMEM) (Wako,
Osaka, Japan) supplemented with 10% FCS, penicillin (100 U/ml),
streptomycin (100 µg/ml), and 5% amphotericin B at 37°C in a 5%
CO2 atmosphere. Detached cells and molar fragments were
removed after incubation for 24 h, and medium was changed every 48
h until use in examinations.
Immunocytochemistry
Molar explants, which were cultured for 3 days, and
cells were used for immunocytochemistry. Dishes were washed gently
with cold PBS, and cells were fixed with 4% paraformaldehyde for 30
min at room temperature and methanol for 10 min at −20°C. After
blocking with 5% skim milk in PBS for 20 min at room temperature,
cells were immunostained using anti-hCG Ab (1:50), anti-hPL Ab
(1:1,000), anti-CK8/18 Ab (1:50), anti-vimentin Ab, and anti-human
CK7 Ab in the dilutions recommended by the manufacturer. For
negative controls, the primary antibody was replaced with PBS.
Vector construction and retroviral
infection
Construction of lentiviral vector plasmids
CSII-CMV-hTERT, -CDK4R24C, -cyclin D1, -p53C234 and -TetOff, were
described previously (12,14).
CDK4R24C is a mutant (p16INK4a-resistant) form of CDK4, and p53C234
encodes the carboxy-terminal 234 residues of p53 and functions as a
dominant-negative mutant. Similarly, entry vectors containing cDNAs
for hTERT, cyclin D1, and CDK4R24C were recombined with the
lentiviral vector CSII-TRE-Tight-RfA, in which the elongation
factor promoter in CSII-EF-RfA (a gift from Hiroyuki Miyoshi; Riken
BRC, Tsukuba, Japan) was replaced with the tetracycline-responsive
promoter from pTRE-Tight (Clontech, Mountain View, CA, USA) to
generate CSII-TRE-Tight-cyclin D1, -CDK4R24C and -p53C234.
CSII-TRE-Tight-MYC-2A-HRAS was constructed by recombining the
wild-type MYC and HRAS cDNA segments separated by the sequences
encoding the autonomous 'self-cleaving' 2A peptides derived from
foot-and-mouse disease virus (FMDV) (15) with CSII-TRE-Tight-RfA. Production
and infection of recombinant lentiviruses was performed as
described previously (12,16).
Establishment of immortalized human molar
cells
We used the primary-cultured molar cells from two
CHM patients (10 weeks) for immortalization. The primary cells from
hydatidiform mole tissue no. 1 (Mole1) were infected in 25
cm2 flasks with a combination of CSII-CMV-hTERT and
-CDK4R24C (#8); CSII-CMV-hTERT, -CDK4R24C, -cyclin D1 and -p53C234
(#2C); or CSII-CMV-hTERT, -TetOff, CSII-TRE-Tight-CDK4R24C and
-cyclin D1 (#3B) at multiplicity of infection values of >5.
Another batch of primary cells from hydatidiform mole tissue no. 3
(Mole3) was infected in a 12-well dish with a combination of
CSII-CMV-hTERT, -CDK4R24C, -cyclin D1, -TetOff,
CSII-TRE-Tight-p53C234 and -MYC-2A-HRAS (#1B). Among several
colonies growing in the dish, fibroblastic colonies were manually
removed, and colonies with epithelial morphology were expanded from
the culture and further characterized. HMol cell lines were
cultured in Epi-Life KG2 (Kurabo Industries Ltd., Osaka, Japan) and
DMEM at initial and final ratios of 4:1 and 1:1, respectively.
Epi-Life KG2 was used after adding insulin (10 µg/ml), human
epithelial growth factor (hEGF, 0.1 ng/ml), hydrocortisone (0.5
µg/ml), gentamycin (50 µg/ml), amphotericin B (50
ng/ml), and 0.4% v/v bovine pituitary extract (BPE).
Short tandem repeat analysis
Genomic DNA was extracted from HMol cell lines,
molar tissues (Mole1 and Mole3) and intraoral cells of the two
patients and their husbands. DNA (1 ng) was amplified using a
commercially available kit (AmpFlSTR Identifilter™ PCR
amplification kit; Applied Biosystems, Foster City, CA, USA) with
15 STR markers and a gender determination marker used for personal
identification and paternity tests. The amplified fragments were
loaded on a PRISM Genetic Analyzer 310 and were automatically
genotyped with GeneMapper ID software v3.2 (Applied
Biosystems).
Chromosome analysis
Chromosomes from Mole3 and four HMol cell lines
(HMol1-2C, HMol1-3B, HMol1-8 and HMol3-1B) were analyzed by G-band
and karyotyping. The analysis used fresh tissue from Mole3 and HMol
cell lines, which were cultured more than four months after gene
transfection.
Cell proliferation assay
For cell proliferation assay, 5×103 cells
were plated in 100 µl medium in 96-well plates. Cell
viability was determined by modified tetrazolium salt (MTS) assay
using the Cell Titer 96 Aqueous One Solution Proliferation assay
kit (Promega, Fitchburg, WI, USA) according to the manufacturer's
instructions. Data were obtained from three independent experiments
with eight samples each.
Transwell migration and invasion
assays
Assay of the migration and invasion abilities of Jar
cells and HMol cells was performed as previously reported (17). Transwells (Corning Inc., Corning,
NY, USA) with a filter of 6.5-mm diameter and 8.0-µm pore
size were used. Cell numbers were adjusted to 3.0×105/ml
in serum-free medium. A 200-µl sample was added in
triplicate to the upper wells, and 800 µl of serum-free
medium with 1% fibronectin was added to the lower wells. Assays
were performed after 24 h of incubation. Invasion assay was
performed under the same conditions as those for migration assay,
except that wells were coated with Matrigel (Collaborative
Biomedical Products, Bedford, MA, USA). The number of cells was
counted under a microscope at ×200 magnification. Data were
obtained from three independent experiments and are expressed as
the mean ± standard deviation (SD).
Assay for hCG and hPL in the culture
medium
To detect the presence of hCG and hPL in the culture
medium, 5×104 cells were plated in a 24-well chamber and
incubated for 24 h. The medium was replaced with 600 µl of
serum-free DMEM, and cells were cultured for 48 h. To examine the
effect of forskolin treatment on hCG secretion, serum-free medium
containing 20 µM of forskolin (Sigma-Aldrich) was used.
Conditioned media were collected, and levels of hCG and hPL were
quantified in triplicate with an enzyme immunoassay (EIA) using an
hCG-β-CTP Ab and a latex agglutination immunoassay (both from SRL
Inc., Tokyo, Japan), respectively. Data are expressed as the mean ±
SD.
Fluorescent staining
Cells were cultured in DMEM for 24 h before the
medium was removed and replaced with serum-free DMEM with or
without 100 µM forskolin. After incubation for 48 h, cells
were fixed with 4% paraformaldehyde for 30 min at room temperature
and permeabilized with PBS containing 0.2% Triton-X. After blocking
with PBS containing 7% fetal bovine serum (FBS) for 20 min at room
temperature, cells were incubated with a fluorescein
isothiocyanate-labeled antibody against phalloidin (R-415;
Invitrogen, Waltham, MA, USA) at a dilution of 1/500 and Hoechst
33342 (Bio-Rad Laboratories, Hercules, CA, USA) at a dilution of
1/100 for 1 h. After washing with PBS, cells were analyzed using a
Biorevo BZ-8000 fluorescence microscope (Keyence, Osaka,
Japan).
Results
Primary culture of molar explants and
immunocytochemical expression of trophoblastic markers in cultured
molar cells
We first investigated whether molar trophoblasts
could be cultured and isolated from hydatidiform molar tissues
using the same method as that used to isolate EVTs from human
chorionic villi (18). We found
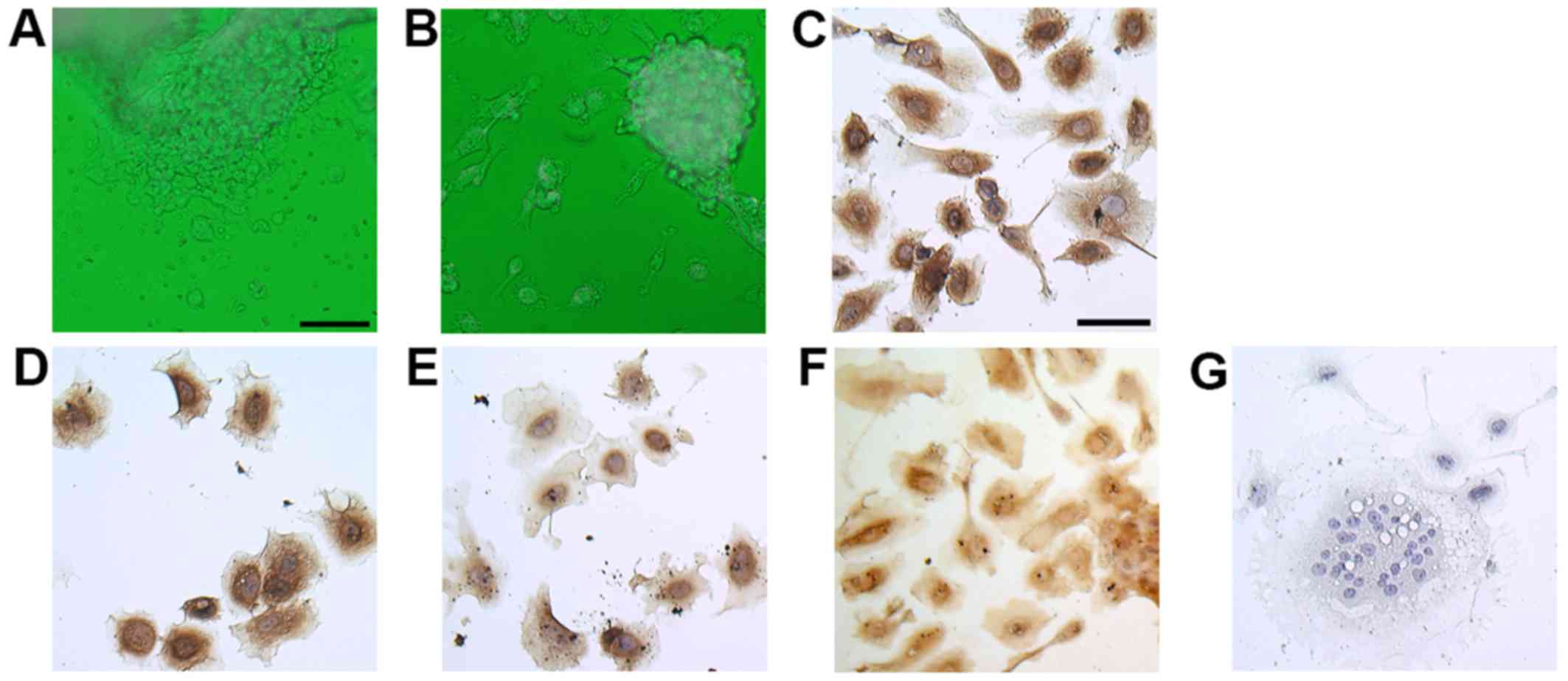
that round cells started growing from the explanted molar tissue
tips after 24 h of incubation (Fig.
1A) and that cells were able to grow for 7 days on
collagen-coated dishes (Fig. 1B).
To confirm the trophoblastic character of the cells, we performed
immunocytochemistry using isolated molar cells after 3 days of
culture. These cells exhibited positive immunoreactivity against
the trophoblast markers CK7 and CK8/18 (Fig. 1C and D) (17–19). Hydatidiform moles secrete more hCG
than normal pregnancies do. hPL is produced mainly by
syncytiotrophoblasts and can be used as a marker of intermediate
trophoblasts (20). Both hCG and
hPL were expressed in isolated molar cells (Fig. 1E and F). Most cells were
mononuclear cells, but some were multinuclear like
syncytiotrophoblasts (Fig.
1G).
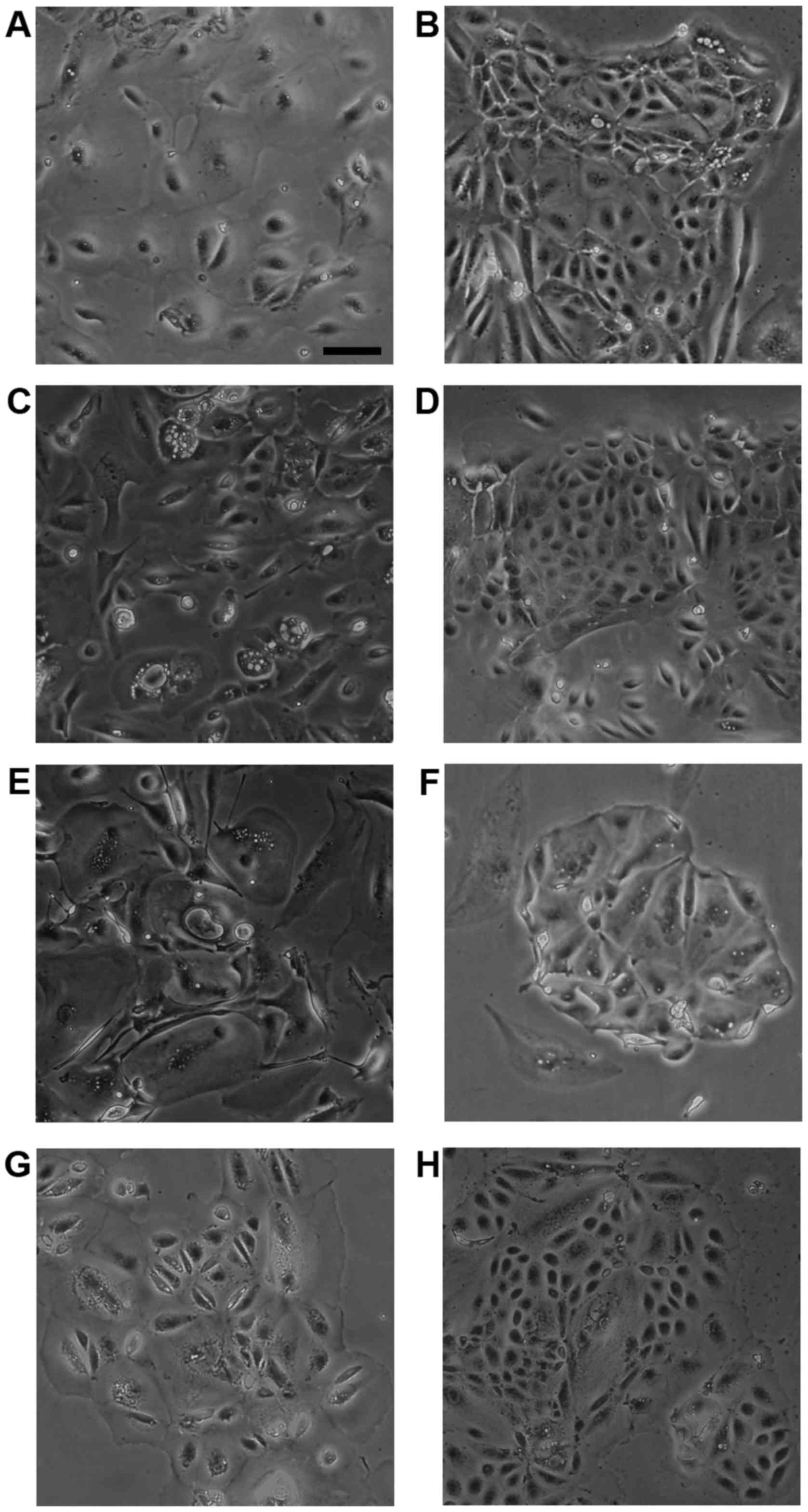
Morphological analysis of HMol cells
Cell lines were established from isolated molar
cells derived from Mole1 (three lines) and Mole3 (one line) tissues
following transduction of different sets of lentiviruses, including
CSII-CMV-hTERT. These independent cell lines were named HMol1-2C,
HMol1-3B, HMol1-8 and HMol3-1B. To create epithelial cell lines, we
removed fibroblastic colonies manually and cultured the cells with
medium containing Epi-Life KG2 and growth factors such as insulin,
hEGF, and hydrocortisone. HMol1-2C, HMol1-3B and HMol3-1B cells
exhibited two cell patterns with small cells and large cells,
similar to cytotrophoblasts and syncytiotrophoblasts (Fig. 2B, D and H). In contrast, HMol1-8
cells were round and bigger than those of the other three cell
lines (Fig. 2F), and it took
longer to grow and remove HMol1-8 cells from dishes using trypsin
compared to those of the other three cell lines. We assumed
therefore that HMol1-8 cells may exhibit different characteristics
than the others.
STR analysis
To confirm the genetic origins of the HMol cell
lines, we performed STR analysis using DNA from the four cell
lines, the two patients, their partners, and the CHM tissues (Mole1
and Mole3). The results showed that the two molar tissues each
contained only one paternal allele at most loci. Although eight
loci from one mother (Mother1) and nine loci from the other mother
(Mother3) showed the same alleles as Mole1 and Mole3, respectively,
it was clear that the two molar tissues were not biparental,
suggesting that they arose from a single sperm fertilizing an empty
ovum (Table I). Results from
HMol1-3B and HMol1-2C were identical to that of Mole1, and HMol3-1B
was genetically identical to Mole3. However, HMol1-8 contained the
same alleles as the maternal cells from which Mole1 was derived.
These results indicated that HMol1-3B, HMol1-2C and HMol3-1B
originated from CHMs, but HMol1-8 was derived from the maternal
cells of Mole1. The results of STR and cell morphology analyses
suggested that HMol1-8 was likely established from decidual
cells.
 | Table IShort tandem repeat analysis of DNA
from molar tissues and established cell lines. |
Table I
Short tandem repeat analysis of DNA
from molar tissues and established cell lines.
Karyotype analysis
Table II shows
the results of chromosome number analysis of 100 cells from each
cell line. The analysis revealed that chromosome numbers in
HMol1-2C, HMol1-3B, and HMol3-1B primarily ranged from 80 to 88
(Table II), and karyotype
analysis showed that most chromosomes had four bands in these three
cell line. The karyotype of Mole3 was 46, XX (data not shown), and
karyotype analysis of Mole1 was not performed. Lawler et al
performed genetic studies of 149 CHMs, and the results showed that
128 were diploid, 1 triploid, 1 haploid, and 19 unknown (21). These results suggest that the
three cell lines may have been established from tetraploid cells
after duplication of diploid cells, with loss and recombination of
some chromosomes. On the other hand, 81% of HMol1-8 cells had a
karyotype of 48, XX, with trisomies 2 and 5 noted in most cells. We
assumed that HMol1-8 cells originated from diploid (46, XX) cells
and that the chromosomal alterations were induced during gene
transduction and culture.
 | Table IIKaryotype analysis of the newly
established cell lines. |
Table II
Karyotype analysis of the newly
established cell lines.
| HMol1-2C | No. of
chromosomes | ≤77 | 78 | 79 | 80 | 81 | 82 | 83 | 84 | 85 | 86 | 87 | 88–99 | ≤100 | Total |
| No. of cells | 14 | 2 | 6 | 8 | 11 | 11 | 16 | 10 | 3 | 5 | 1 | 4 | 9 | 100 |
| HMol1-3B | No. of
chromosomes | ≤79 | 80 | 81 | 82 | 83 | 84 | 85 | 86 | 87 | 88 | 89 | 90–99 | ≤100 | Total |
| No. of cells | 6 | 1 | 0 | 4 | 5 | 15 | 13 | 14 | 15 | 8 | 4 | 7 | 9 | 100 |
| HMol1-8 | No. of
chromosomes | ≤46 | 47 | 48 | 49 | 50 | 51 | 94–98 | Total | | | | | | |
| No. of cells | 2 | 3 | 81 | 2 | 1 | 0 | 11 | 100 | | | | | | |
| HMol3-1B | No. of
chromosomes | ≤79 | 80–81 | 82 | 83 | 84 | 85 | 86 | 87 | 88–100 | 168± | Total | | | |
| No. of cells | 7 | 0 | 9 | 12 | 15 | 20 | 13 | 3 | 2 | 9 | 90 | | | |
Immunocytochemical analysis of HMol1-2C,
HMol1-3B, HMol1-8 and HMol3-1B
Next, we performed immunocytochemistry to confirm
HMol1-2C, HMol1-3B and HMol3-1B as trophoblastic cells. We used the
choriocarcinoma cell line Jar as a representative trophoblastic
cell line for comparison with the three HMol cell lines. All three
HMol cell lines showed positive staining for CK7, hCG and hPL but
were negative for vimentin, similar to Jar staining patterns
(Fig. 3). The results of
HMol1-2C, HMol1-3B, and HMol3-1B staining are consistent with the
characteristics of trophoblastic cells. Immunocytochemistry of
HMol1-8 showed that the round cells were very weakly positive for
CK7 and positive for hCG, hPL, and vimentin. Although cytokeratin
and vimentin are used as markers for epithelial cells and
mesenchymal cells, decidual cells are reported to be positive for
vimentin as well (22). These
results suggest that HMol1-8 cells have the characteristics of
decidual cells.
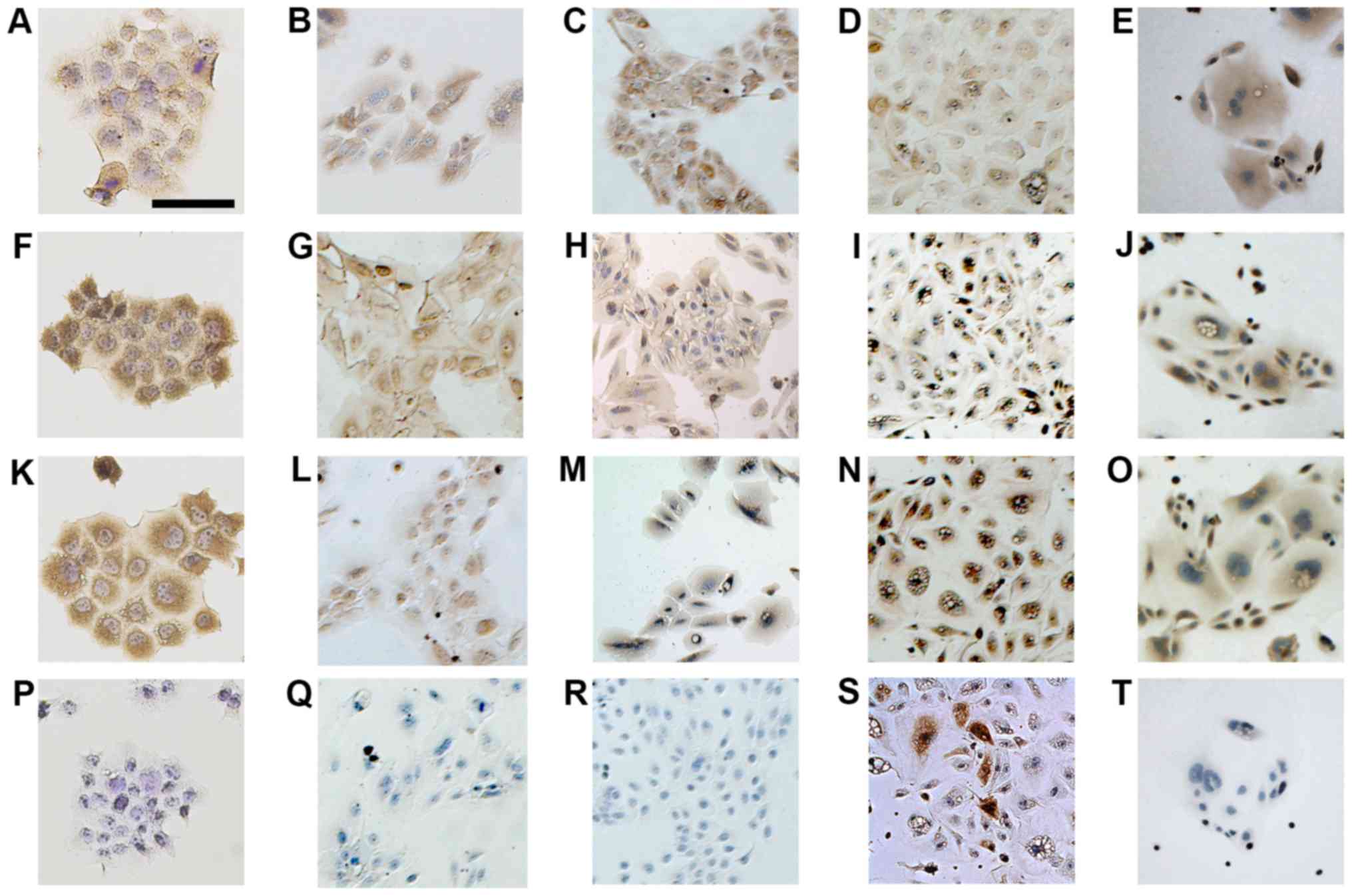 | Figure 3Immunocytochemistry of cell lines
established from primary cultures of complete hydatidiform moles
compared with that of a choriocarcinoma cell line, Jar.
Immunostaining of Jar with (A) CK7, (F) human chorionic
gonadotropin (hCG), (K) human placental lactogen (hPL), and (P)
vimentin. Jar was positive for CK7, hCG and hPL and negative for
vimentin. Immunostaining of (B, G, L and Q) HMol1-2C, (C, H, M and
R) HMol1-3B, (D, I, N and S) HMol1-8 and (E, J, O and T) HMol3-1B,
using antibodies against (B-E) CK7, (G-J) hCG, (L-O) hPL and (Q-T)
vimentin. Magnification, ×100; scale bar, 100 µm. |
Cell proliferation
Cell proliferation of the four established cell
lines was examined by MTS assay. Cell growth was fastest in the
HMol1-3B cells, and HMol1-2C and HMol3-1B cells grew nearly at the
same speeds. HMol1-8 cells grew very slowly, with only a 17.0%
increase after 72 h of incubation (Fig. 4A).
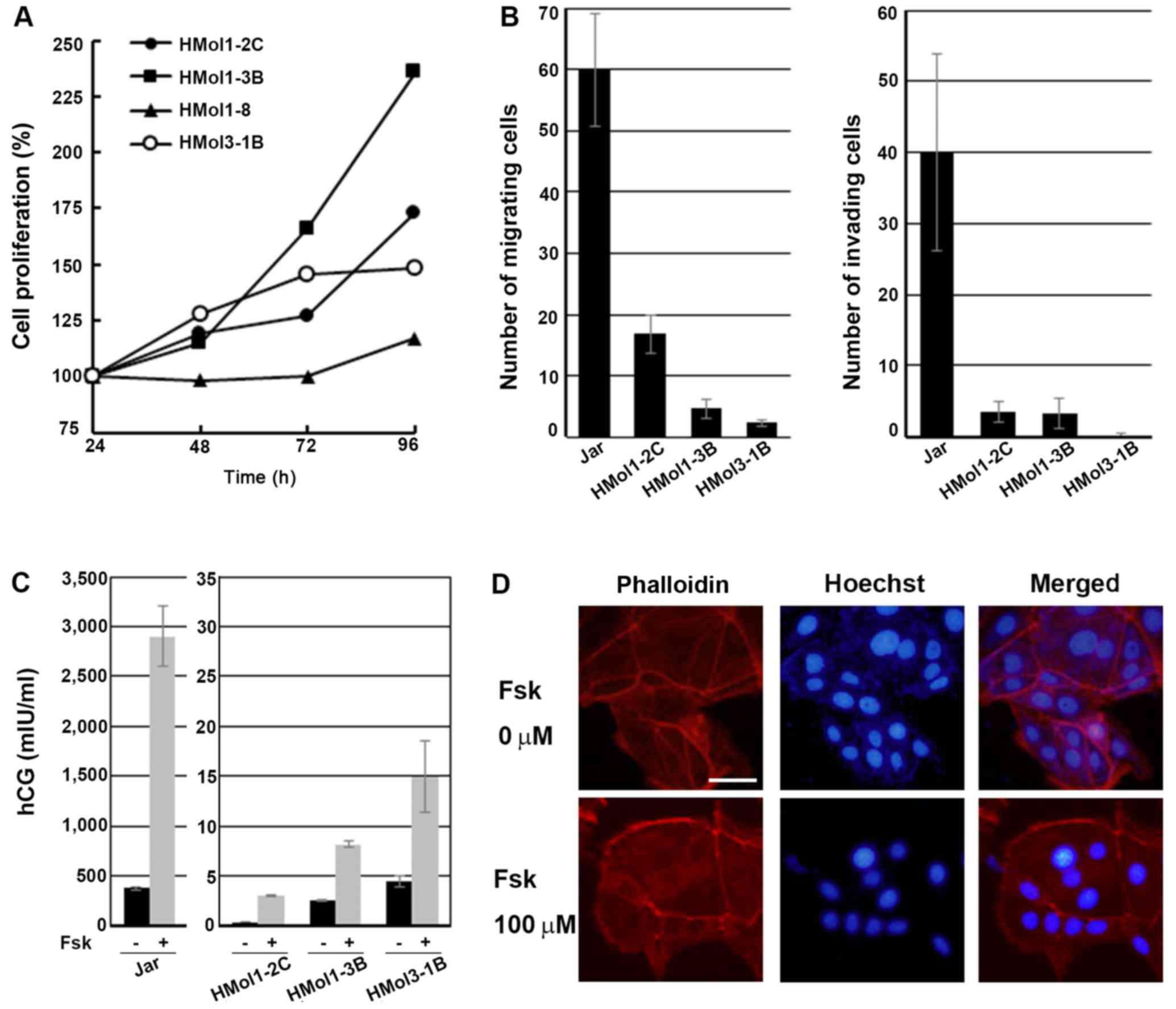 | Figure 4Assays to determine cell
proliferation, migration, invasion, human chorionic gonadotropin
(hCG) secretion, and the effect of forskolin treatment in
established cell lines. (A) Graphical depiction of the relative
absorbance readings after modified tetrazolium salt (MTS) assays,
demonstrating that all established cell lines were immortal and
that cell proliferation of HMol1-8 was much lower than those of
HMol1-2C, HMol1-3B and HMol3-1B. Mean values of three different
experiments performed in eight wells are shown. (B) Graphical
depiction of data obtained from migration assays (left panel, n=3)
and Matrigel invasion assays (right panel, n=3) of Jar, HMol1-2C,
HMol1-3B and HMol3-1B, demonstrating that the three established
molar cell lines exhibited much weaker migration and invasion
abilities compared to those of Jar. Data were obtained from three
independent experiments. Each bar represents the mean distance of
the control ± SD. (C) Graphical depiction of data obtained from hCG
assay of conditioned media of Jar, HMol1-2C, HMol1-3B and HMol3-1B
with and without forskolin treatment. Data were obtained from three
independent experiments, demonstrating increases in hCG secretion
following forskolin treatment in HMol1-2C, HMol1-3B and HMol3-1B,
as well as Jar cells. Each bar represents the mean hCG
concentration (mIU/ml) ± SD. (D) Immunofluorescent double-staining
of HMol3-1B cells for nuclei (Hoechst) and the actin cytoskeleton
(phalloidin), demonstrating the differentiation into
multi-nucleated cells after incubation with 100 µM forskolin
for 48 h. Scale bar, 50 µm. |
Migration and invasion assays
We next examined whether the three cell lines
originating from CHMs have the same migration and invasion
abilities as Jar cells (18,23). While cells from the three cell
lines did migrate, the migrated cell numbers were much lower than
those of Jar cells. Migration ability was strongest in HMol1-2C and
weakest in HMol3-1B (Fig. 4B,
left panel). Invasion assay showed that HMol1-2C and HMol1-3B cells
possessed invasion abilities, but these were much weaker than those
of the Jar cells (Fig. 4B, right
panel). HMol3-1B cells underwent very little invasion under the
same condition as those of the other cell lines. We assumed that
the migration and invasion abilities of the molar cell lines were
weaker than those of Jar cells because hydatidiform moles are a
type of pregnancy while Jar cells are established from
choriocarcinoma.
Differentiation ability of HMol cells
after forskolin treatment
The characteristics of trophoblastic cells include
proliferation, migration, invasion and hormone production, and
these depend on the type and differentiation of the trophoblasts
involved. Immunocytochemistry showed that HMol1-2C, HMol1-3B and
HMol3-1B cells expressed hCG and hPL (Fig. 3). To identify additional
characteristics of the three cell lines, we measured secretion
levels of hCG and hPL and examined the effects on the cells of
forskolin treatment, which is used for differentiation of
trophoblasts in vitro. Levels of hPL in conditioned media
were <0.05 mg/ml in all three molar cell lines and in Jar cells.
Secretion of hCG was stimulated by forskolin treatment in all three
molar cell lines as well as in Jar cells, although basal and
induced levels of hCG in molar cell lines were much lower than
those in Jar (Fig. 4C).
Interestingly, the molar cell line with the highest migration
ability (HMol1-2C) had the lowest level of hCG secretion. An
increase in hCG secretion suggests that forskolin treatment may
induce the molar cell lines to differentiate into
syncytiotrophoblastic cells. Immunofluorescent double-staining for
nuclei (Hoechst) and the actin cytoskeleton (phalloidin) was
performed to visualize the number of nuclei in each cell as well as
cell borders (Fig. 4D). We used
HMol3-1B cells to examine the effect of forskolin on cell fusion
because among the three lines, this cell line showed the highest
hCG secretion with forskolin treatment. HMol3-1B cells became
multi-nucleated after incubation for 48 h with 100 µM
forskolin (Fig. 4D). These
results suggest that treatment with forskolin upregulates
differentiation and fusion of HMol3-1B cells.
Discussion
In the present study, three cell lines (HMol1-2C,
HMol1-3B and HMol3-1B) were established and confirmed to originate
from CHM trophoblasts. STR analysis revealed that HMol1-2C,
HMol1-3B and HMol3-1B were genetically identical to their
corresponding Mole1 and Mole3 progenitors. HMol1-2C, HMol1-3B and
HMol3-1B cells were thus considered to have characteristics of
cytotrophoblasts rather than differentiated trophoblasts such as
syncytiotrophoblasts and EVTs. The three cell lines showed low
levels of hCG secretion and low cell invasion abilities; hCG is
mainly produced by syncytiotrophoblasts and EVTs have invasion
ability. The observed increases in cell fusion and hCG secretion
following forskolin treatment is consistent with the
characteristics of cytotrophoblasts, which can differentiate into
syncytiotrophoblasts. In the primary culture of molar tissues,
small, round cells first derived from the tips of molar explants,
and then some spindle-shaped, EVT-like cells appeared around day 7.
Some multi-nucleated, syncytiotrophoblast-like cells were found
during primary culture. After establishment of HMol cell lines, the
number of multi-nucleated cells diminished, and more small cells
were noted in later passages. These results suggest that the major
constituents of the three cell lines may exhibit characteristics of
cytotrophoblasts.
We established HMol1-8, which may have originated
from decidual cells. STR analysis revealed that HMol1-8 was derived
from maternal cells. Decidual and squamous cells of the uterine
cervix and the vagina were considered to be contaminants. Decidual
cells may inadvertently be included with the molar vesicles since
the tips of the villi are attached to the decidua basalis with
fibrinoids. Surface cells of the uterine cervix and the vagina may
act as contaminants during operations. The HMol1-8 cell line
consisted of large round cells that showed positive
immunoreactivity for vimentin. Squamous cells are epithelial cells
that are positive for cytokeratin, and decidual cells express
vimentin because they are differentiated from endometrial stromal
cells (24). These results
suggest that HMol1-8 originated from decidual cells. However, we
did not examine the expression or secretion of prolactin (25–27), insulin-like growth factor binding
protein (25,27,28), or tissue factor (27,29) which are reported markers of
decidual cells. Further studies are needed to confirm the origin
and characteristics of the HMol1-8 cell line.
To immortalize molar trophoblastic cells, we
transduced cells with hTERT, CDK4R24C and cyclin D1, and we
transduced some cells with p53C234 and MYC-2A-HRAS using
recombinant lentiviral vectors. In previous studies, HTR-8/SVneo
and B6Tert were established by transfection with SV40 large T
antigen and hTERT while maintaining the characteristics of the
parental cells, which were derived from first-trimester human EVTs
and normal placental-origin cytotrophoblasts, respectively
(30–32). Transduction with hTERT, CDK4R24C
and cyclin D1 has been suggested to be an effective method for
immortalizing normal cells with differentiation capacities while
maintaining the original phenotype of the primary cells according
to previous studies (10,11,14,33–35). However, introduction of hTERT,
CDK4R24C and cyclin D1 was not sufficient to immortalize
non-luteinized granulosa cells, and adding p53C234 was effective at
lengthening the lifespan of the cells (14). To immortalize cells effectively,
we transfected several genes in various combinations, and we
successfully established three CHM cell lines.
In this study, karyotypes of all three cell lines
established from CHMs were almost tetraploid, caused by duplication
of diploid progenitors. Formally, there are two major possibilities
for the tetraploidization of the three cell lines: i) original
diploid molar cells became tetraploid during primary culture or ii)
diploid primary molar trophoblasts became tetraploid after
induction of genes for immortalization. Karyotype analysis of 403
CHM samples revealed that 15 were post-zygotic tetraploids, which
are likely to have developed by somatic endoreduplication of
androgenic diploid cells (36).
Previous studies on the immortalization of non-cancerous cell lines
via the introduction of hTERT, CDK4R24C, cyclin D1 and TetOff
showed intact karyotypes (10–12), except for two lines. One of these
exhibited tetraploidy in 70% of cells and another showed a
chromosomal deletion of chromosome 22 (33). In this study, tetraploidy may have
been caused by the characteristics of the CHMs rather than being an
effect of the introduction of additional genes such as p53C234, MYC
and HRAS, as tetraploidy was observed in all CHM cell lines but not
in HMol1-8. These results may suggest the possibility of
tetraploidization during primary culture.
There are some limitations to this study. First, the
molar cell lines secretes hCG at low levels although hCG secretion
is the major characteristic of hydatidiform moles. This is because
differentiated cells such as syncytiotrophoblasts are more
difficult to grow continuously compared with undifferentiated cells
such as cytotrophoblasts, and therefore, the cell lines became
cyncytiotrophoblastic after passing many times. Endogenous hCG
production involves an autocrine regulation of invasion in
trophoblasts (37). However, this
study showed that forskolin treatment is a good method for inducing
molar cell lines to increase hCG production and differentiate into
syncytiotrophoblastic cells. Moreover, these cell lines may be
suitable models for identifying genes that exhibit increased
malignant behaviors, such as invasion or cell proliferation,
together with an increase in hCG production, which is also the
major characteristic of choriocarcinoma. Secondly, the molar cell
lines were almost tetraploid although most CHMs are diploid.
Histological morphology analysis did not reveal any differences
between diploid and tetraploid CHMs (36), supporting the conclusion that the
three CHM cell lines maintained the characteristics of the original
molar cells.
In conclusion, we successfully established three
cell lines from complete hydatidiform moles by introduction of
hTERT, CDK4R24C and cyclin D1 with or without p53C234 and
MYC-2A-HRAS. The genetic origins of each cell line were identical
with those of the original CHM tissues, and the cell lines
exhibited characteristics of trophoblastic cells, which are similar
to those of undifferentiated cytotrophoblasts.
Acknowledgments
We thank Hiroyuki Miyoshi (RIKEN Tsukuba Institute)
for providing the lentiviral vectors and packaging constructs. This
study was supported by grant-in-aid no. 23592445 (to E.Y.) from the
Japanese Ministry of Education, Culture, Sports, Science and
Technology and by the National Cancer Center Research and
Development Fund (23B-1). We would like to thank Editage
(www.editage.jp) for English language editing.
References
|
1
|
Kajii T and Ohama K: Androgenetic origin
of hydatidiform mole. Nature. 268:633–634. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wake N, Seki T, Fujita H, Okubo H, Sakai
K, Okuyama K, Hayashi H, Shiina Y, Sato H, Kuroda M, et al:
Malignant potential of homozygous and heterozygous complete moles.
Cancer Res. 44:1226–1230. 1984.PubMed/NCBI
|
|
3
|
Vejerslev LO, Dissing J, Hansen HE and
Poulsen H: Hydatidiform mole: Genetic origin in polyploid
conceptuses. Hum Genet. 76:11–19. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ohama K, Ueda K, Okamoto E, Takenaka M and
Fujiwara A: Cytogenetic and clinicopathologic studies of partial
moles. Obstet Gynecol. 68:259–262. 1986.PubMed/NCBI
|
|
5
|
Kaneki E, Kobayashi H, Hirakawa T, Matsuda
T, Kato H and Wake N: Incidence of postmolar gestational
trophoblastic disease in androgenetic moles and the morphological
features associated with low risk postmolar gestational
trophoblastic disease. Cancer Sci. 101:1717–1721. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Seckl MJ, Sebire NJ and Berkowitz RS:
Gestational trophoblastic disease. Lancet. 376:717–729. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kan M, Yamamoto E, Niimi K, Tamakoshi K,
Sekiya Y, Nishino K, Ino K and Kikkawa F: Gestational trophoblastic
neoplasia and pregnancy outcome after routine second curettage for
hydatidiform mole: A retrospective observational study. J Reprod
Med. 61:373–379. 2016.
|
|
8
|
Taillon-Miller P, Bauer-Sardiña I, Zakeri
H, Hillier L, Mutch DG and Kwok PY: The homozygous complete
hydatidiform mole: A unique resource for genome studies. Genomics.
46:307–310. 1997. View Article : Google Scholar
|
|
9
|
Steinberg KM, Schneider VA, Graves-Lindsay
TA, Fulton RS, Agarwala R, Huddleston J, Shiryev SA, Morgulis A,
Surti U, Warren WC, et al: Single haplotype assembly of the human
genome from a hydatidiform mole. Genome Res. 24:2066–2076. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kuroda K, Kiyono T, Eitsuka T, Isogai H,
Takahashi K, Donai K, Isogai E and Fukuda T: Establishment of cell
lines derived from the genus Macaca through controlled expression
of cell cycle regulators. J Cell Biochem. 116:205–211. 2015.
View Article : Google Scholar
|
|
11
|
Donai K, Kiyono T, Eitsuka T, Guo Y,
Kuroda K, Sone H, Isogai E and Fukuda T: Bovine and porcine
fibroblasts can be immortalized with intact karyotype by the
expression of mutant cyclin dependent kinase 4, cyclin D, and
telomerase. J Biotechnol. 176:50–57. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sasaki R, Narisawa-Saito M, Yugawa T,
Fujita M, Tashiro H, Katabuchi H and Kiyono T: Oncogenic
transformation of human ovarian surface epithelial cells with
defined cellular oncogenes. Carcinogenesis. 30:423–431. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Okamoto T, Aoyama T, Nakayama T, Nakamata
T, Hosaka T, Nishijo K, Nakamura T, Kiyono T and Toguchida J:
Clonal heterogeneity in differentiation potential of immortalized
human mesenchymal stem cells. Biochem Biophys Res Commun.
295:354–361. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bayasula IA, Iwase A, Kiyono T, Takikawa
S, Goto M, Nakamura T, Nagatomo Y, Nakahara T, Kotani T, Kobayashi
H, et al: Establishment of a human nonluteinized granulosa cell
line that transitions from the gonadotropin-independent to the
gonadotropin-dependent status. Endocrinology. 153:2851–2860. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Carey BW, Markoulaki S, Hanna J, Saha K,
Gao Q, Mitalipova M and Jaenisch R: Reprogramming of murine and
human somatic cells using a single polycistronic vector. Proc Natl
Acad Sci USA. 106:157–162. 2009. View Article : Google Scholar :
|
|
16
|
Miyoshi H, Blömer U, Takahashi M, Gage FH
and Verma IM: Development of a self-inactivating lentivirus vector.
J Virol. 72:8150–8157. 1998.PubMed/NCBI
|
|
17
|
Yamamoto E, Ito T, Abe A, Sido F, Ino K,
Itakura A, Mizutani S, Dovat S, Nomura S and Kikkawa F: Ikaros is
expressed in human extravillous trophoblasts and involved in their
migration and invasion. Mol Hum Reprod. 11:825–831. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yamamoto E, Ino K, Miyoshi E, Inamori K,
Abe A, Sumigama S, Iwase A, Kajiyama H, Shibata K, Nawa A, et al:
N-acetylglucosaminyltransferase V regulates extravillous
trophoblast invasion through glycosylation of alpha5beta1 integrin.
Endocrinology. 150:990–999. 2009. View Article : Google Scholar
|
|
19
|
Austgulen R, Chedwick L, Vogt Isaksen C,
Vatten L and Craven C: Trophoblast apoptosis in human placenta at
term as detected by expression of a cytokeratin 18 degradation
product of caspase. Arch Pathol Lab Med. 126:1480–1486.
2002.PubMed/NCBI
|
|
20
|
Sasagawa M, Yamazaki T, Endo M, Kanazawa K
and Takeuchi S: Immunohistochemical localization of HLA antigens
and placental proteins (alpha hCG, beta hCG CTP, hPL and SP1 in
villous and extravillous trophoblast in normal human pregnancy: A
distinctive pathway of differentiation of extravillous trophoblast.
Placenta. 8:515–528. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lawler SD, Fisher RA and Dent J: A
prospective genetic study of complete and partial hydatidiform
moles. Am J Obstet Gynecol. 164:1270–1277. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Devergne O, Coulomb-L'Herminé A, Capel F,
Moussa M and Capron F: Expression of Epstein-Barr virus-induced
gene 3, an interleukin-12 p40-related molecule, throughout human
pregnancy: Involvement of syncytiotrophoblasts and extravillous
trophoblasts. Am J Pathol. 159:1763–1776. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Niimi K, Yamamoto E, Fujiwara S, Shinjo K,
Kotani T, Umezu T, Kajiyama H, Shibata K, Ino K and Kikkawa F: High
expression of N-acetylglucosaminyltransferase IVa promotes invasion
of choriocarcinoma. Br J Cancer. 107:1969–1977. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhu H, Hou CC, Luo LF, Hu YJ and Yang WX:
Endometrial stromal cells and decidualized stromal cells: Origins,
transformation and functions. Gene. 551:1–14. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Richards RG, Brar AK, Frank GR, Hartman SM
and Jikihara H: Fibroblast cells from term human decidua closely
resemble endometrial stromal cells: Induction of prolactin and
insulin-like growth factor binding protein-1 expression. Biol
Reprod. 52:609–615. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Telgmann R and Gellersen B: Marker genes
of decidualization: Activation of the decidual prolactin gene. Hum
Reprod Update. 4:472–479. 1998. View Article : Google Scholar
|
|
27
|
Dunn CL, Kelly RW and Critchley HO:
Decidualization of the human endometrial stromal cell: An enigmatic
transformation. Reprod Biomed Online. 7:151–161. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim JJ, Jaffe RC and Fazleabas AT:
Insulin-like growth factor binding protein-1 expression in baboon
endometrial stromal cells: Regulation by filamentous actin and
requirement for de novo protein synthesis. Endocrinology.
140:997–1004. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lockwood CJ, Krikun G, Caze R, Rahman M,
Buchwalder LF and Schatz F: Decidual cell-expressed tissue factor
in human pregnancy and its involvement in hemostasis and
preeclampsia-related angiogenesis. Ann NY Acad Sci. 1127:67–72.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Graham CH, Hawley TS, Hawley RG,
MacDougall JR, Kerbel RS, Khoo N and Lala PK: Establishment and
characterization of first trimester human trophoblast cells with
extended lifespan. Exp Cell Res. 206:204–211. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang YL, Qiu W, Feng HC, Li YX, Zhuang LZ,
Wang Z, Liu Y, Zhou JQ, Zhang DH and Tsao GS: Immortalization of
normal human cytotrophoblast cells by reconstitution of telomeric
reverse transcriptase activity. Mol Hum Reprod. 12:451–460. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li RH and Zhuang LZ: The effects of growth
factors on human normal placental cytotrophoblast cell
proliferation. Hum Reprod. 12:830–834. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Katayama M, Kiyono T, Horie K, Hirayama T,
Eitsuka T, Kuroda K, Donai K, Hidema S, Nishimori K and Fukuda T:
Establishment of an immortalized cell line derived from the prairie
vole via lentivirus-mediated transduction of mutant
cyclin-dependent kinase 4, cyclin D, and telomerase reverse
transcriptase. Exp Anim. 65:87–96. 2016. View Article : Google Scholar :
|
|
34
|
Kuroda K, Kiyono T, Isogai E, Masuda M,
Narita M, Okuno K and Koyanagi Y: Fukuda T. Immortalization of
fetal bovine colon epithelial cells by expression of human cyclin
D1, mutant cyclin dependent kinase 4, and telomerase reverse
transcriptase: An in vitro model for bacterial infection. PLoS One.
10:e01434732015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Inagawa Y, Yamada K, Yugawa T, Ohno S,
Hiraoka N, Esaki M, Shibata T, Aoki K, Saya H and Kiyono T: A human
cancer xenograft model utilizing normal pancreatic duct epithelial
cells conditionally transformed with defined oncogenes.
Carcinogenesis. 35:1840–1846. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sundvall L, Lund H, Niemann I, Jensen UB,
Bolund L and Sunde L: Tetraploidy in hydatidiform moles. Hum
Reprod. 28:2010–2020. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zygmunt M, Hahn D, Münstedt K, Bischof P
and Lang U: Invasion of cytotrophoblastic JEG-3 cells is stimulated
by hCG in vitro. Placenta. 19:587–593. 1998. View Article : Google Scholar : PubMed/NCBI
|