Introduction
Nasopharyngeal carcinoma (NPC) is one of the most
common malignant tumors in South China, and its morbidity and
mortality are ranked first globally. The etiology of NPC is
multifactorial. Accumulated epidemiological and etiological
evidence indicate that NPC develops from a complex interaction
between genetic factors, exposure to chemical carcinogens and
latent Epstein-Barr virus (EBV) infection (1).
In recent years, studies have indicated that genetic
changes in the classical oncogenes and tumor suppressor genes are
rare in NPC, and the epigenetic changes in tumor suppressor genes,
particlarly DNA methylation, play an important role in its
development. NPC exhibits a high frequency of genetic CpG island
methylation of the tumor. Moreover, tumor-associated gene
methylation changes in NPC cells is involved in many functional
processes, including cell cycle regulation, DNA repair, apoptosis,
tumor invasion and metastasis (1–3).
However, to date, and to the best of our knowledge, the epigenetic
changes of the MutS homolog human 3 (MSH3) gene in NPC have not yet
been investigated.
Mismatch repair genes maintain the stability of the
genome by correction of base mismatch during DNA recombination and
replication, and this eliminates the potential for carcinogenic
changes by mutant cell growth via induction of cellular apoptosis.
Thus, the mismatch repair of genes in the maintenance of genomic
integrity, the repair of damage caused by cancer-specific factors
and the anticancer process itself all play a very important role
(4).
MSH3 is a member of the mismatch repair genes, which
is located on chromosome 5q11–13. The MSH3 gene consists of 24
exons and 23 introns and is expressed in a variety of human
tissues. MutSβ is a heterodimer of MSH2 and MSH3, and is primarily
responsible for the repair of insertion-deletion loops (IDLs) of
1–15 nucleotides, as well as DNA loop and DNA double-strand breaks,
which can trigger apoptosis of cells with high levels of DNA
damage, and can also regulate the sensitivity of tumor cells to
radiotherapy and chemotherapeutic drugs (5–10).
It has been suggested that human MSH3 (hMSH3)
deficiency can drive hamartomatous polyposis syndrome tumorigenesis
(11). MSH3 deficiency in human
colonic epithelial cells results in elevated microsatellite
instability at selected tetranucleotide repeats, and in the
formation of DNA double-strand breaks, as well as significant
proteomic changes, although it lacks oncogenic transformation
(12). MSH3 knockout mice develop
cancers only in later life (13);
however, a double knockout of MSH3 and MSH6 renders mice far more
susceptible to cancer than the single knockout of either gene
(14). Therefore, MSH3 is
considered a tumor suppressor gene.
Previous studies have considered that gene mutation
and homozygous deletion are the main mechanisms of tumor suppressor
gene transcription inactivation. However, recent studies have
confirmed that promoter methylation is the third mechanism of the
tumor suppression of gene transcription inactivation, and in some
cases the only mechanism of tumor suppressor gene inactivation
(15). MSH3 protein expression
has been shown to be downregulated in colorectal (16), gastric (17), bladder (18), prostate (19), ovarian (20) and other cancers, although each
cancer type presents with its own inactivation mechanism. Mutations
have rarely been found in MSH3 (21), and the loss of heterozygosity
(LOH) and gene promoter GpG island methylation are the predominant
modes of inactivation of MSH3 (16–18,22).
In this study, to screen novel epigenetic
inactivation of tumor suppressor genes in NPC, we used a
genome-wide screening of genes that were downregulated by promoter
hypermethylation. In our previous study, the mRNA expression levels
were frequently absent or downregulated in MSH3-methylated NPC
primary tumor biopsies as compared to normal nasopharyngeal
epithelial (NNE) tissues (unpublished data). However,
MSH3-unmethylated cases exhibited upregulated or parallel mRNA
expression levels. Thus, MSH3 may be a target gene with its
expression suppressed by promoter hypermethylation in NPC.
In this study, we detected the methylation status,
and the mRNA and protein expression levels in NPC primary tumor
biopsies. We further analyzed the correlation between promoter
methylation and mRNA expression. The above-mentioned evidence
supports our hypothesis that MSH3 is epigenetically inactivated in
NPC by promoter hypermethylation.
Materials and methods
Primary tumor biopsies and NNE
tissues
A total of 54 NPC primary tumor biopsies was
collected from the Department of Otolaryngology Head and Neck
Surgery, Hangzhou First People's Hospital (Hangzhou, China), and 16
NNE tissues were obtained by tonsillectomy as normal controls,
after obtaining written informed consent from the donors. Diagnoses
were established by experienced pathologists according to the World
Health Organization (WHO) classification. Biopsy samples were
stored in liquid nitrogen prior to DNA or RNA extraction or
paraffin sectioning. This study was approved by the local Ethics
Committee of Hangzhou First People's Hospital, Hangzhou, China
(approval ID: 201202501).
Semi-quantitative reverse
transcription-PCR (RT-PCR)
The preparation of total RNA, first-strand synthesis
of cDNA and RT-PCR was performed as previously described (23). All primer sequences, annealing
temperatures, cycling conditions and expected PCR product sizes are
listed in Table I. β-actin was
amplified from the same cDNA sample as an internal control. The
amplified PCR products were visualized following electrophoresis on
2% agarose gels and semi-quantitative analysis was performed using
Quantity One v 4.4.0 software (Bio-Rad Laboratories, Inc, Hercules,
CA, USA).
 | Table IPrimer sequence of MSH3 and β-actin
genes for the semi-quantitative reverse transcription PCR (RT-PCR)
assay. |
Table I
Primer sequence of MSH3 and β-actin
genes for the semi-quantitative reverse transcription PCR (RT-PCR)
assay.
| Primer | Primer sequence
5′→3′ | Product size | Annealing
temperature |
|---|
| MSH3 primer | | | |
| Sense |
5′-GATGGCATTTTCACAAGGATGGG-3′ | 244 bp | 57°C |
| Antisense |
5′-CTGGCGGATAATGGGTGACAAAC-3′ | | |
| β-actin primer | | | |
| Sense |
5′-ACACTGTGCCCATCTACGAGG-3′ | 621 bp | 58°C |
| Antisense |
5′-AGGGGCCGGACTCGTCATACT-3′ | | |
Immunohistochemical staining
The immunohistochemical staining procedure was
carried out as previously described (24). Briefly, human tissue sections were
stained for the expression of MSH3 [EPR4334(2); ab111107; 1:500] (Abcam, Cambridge,
MA, USA) and detected by streptavidin-biotin-horseradish peroxidase
complex formation. Tumor sections that were stained by
isotype-matched immunoglobulin G instead of primary antibodies were
used as a negative control.
The intensity of staining of the tissues was scored
from 0–3 (i.e., absent, mild, moderate and intense) and the
percentage of positive staining of cells was scored from 0–3 (i.e.,
0, 0–5%; 1, 6–25%; 2, 26–50%; and 3, 51–100%) The MSH3 protein
expression score was calculated by multiplying these two scores
(i.e., as indicated by the codes: −, 0; +, 1 to 2; ++, 3 to 5; and
+++, 6 to 9).
Sodium bisulphite modification of genomic
DNA and methylation-specific PCR (MSP)
The procedure for the sodium bisulfite modification
of DNA was performed as previously described (23,25). Bisulfite-modified DNA was
amplified using MSP with primer sets that specifically detected
methylated or unmethylated alleles. All primer sequences, annealing
temperatures, cycling conditions and expected PCR product sizes are
listed in Table II, PCR products
were separated on 2% agarose gels.
 | Table IIPrimer sequence of MSH3 genes for the
methylation-specific PCR. |
Table II
Primer sequence of MSH3 genes for the
methylation-specific PCR.
| Primer | Sequence 5′→3′ | Product size | Annealing
temperature |
|---|
| Methylated
primer | | | |
| Sense |
5′-GGAGGATTTTCGAGTTCGTTC-3′ | 174 bp | 57.5°C |
| Antisense |
5′-CGACCGCAATTCCCAAACG-3′ | | |
| Unmethylated
primer | | | |
| Sense |
5′-GGAGGATTTTTGAGTTTGTTT-3′ | 175 bp | 55.5°C |
| Antisense |
5′-ACAACCACAATTCCCAAACA-3′ | | |
Statistical analysis
The statistical software package SPSS 17.0 (SPSS,
Inc., Chicago, IL, USA) was used in this study to biometrically
assess the data. MSH3 expression levels in primary tumors versus
NNE tumors and MSH3 methylated versus unmethylated tumors were
analyzed by Mann-Whitney's U test. The correlations between MSH3
mRNA or protein expression levels and the clinicopathological
characteristics were analyzed by the Student's t-test or the rank
sum test. A P-value <0.05 was considered statistically
significant.
Results
Primary tumors frequently lack MSH3 mRNA
and protein expression
To evaluate the mRNA expression levels of MSH3 in
primary NPC biopsies, semi-quantitative RT-PCR was carried out.
MSH3 mRNA expression was detected in all NNE tissues (Fig. 1: N1, N2 and N3 used as samples);
however, MSH3 expression was frequently absent or downregulated in
the 54 NPC biopsy specimens (Fig.
1: T1, T2 and T3 used as samples). In addition, to evaluate the
protein expression of MSH3 in the NNE tissues and NPC biopsy
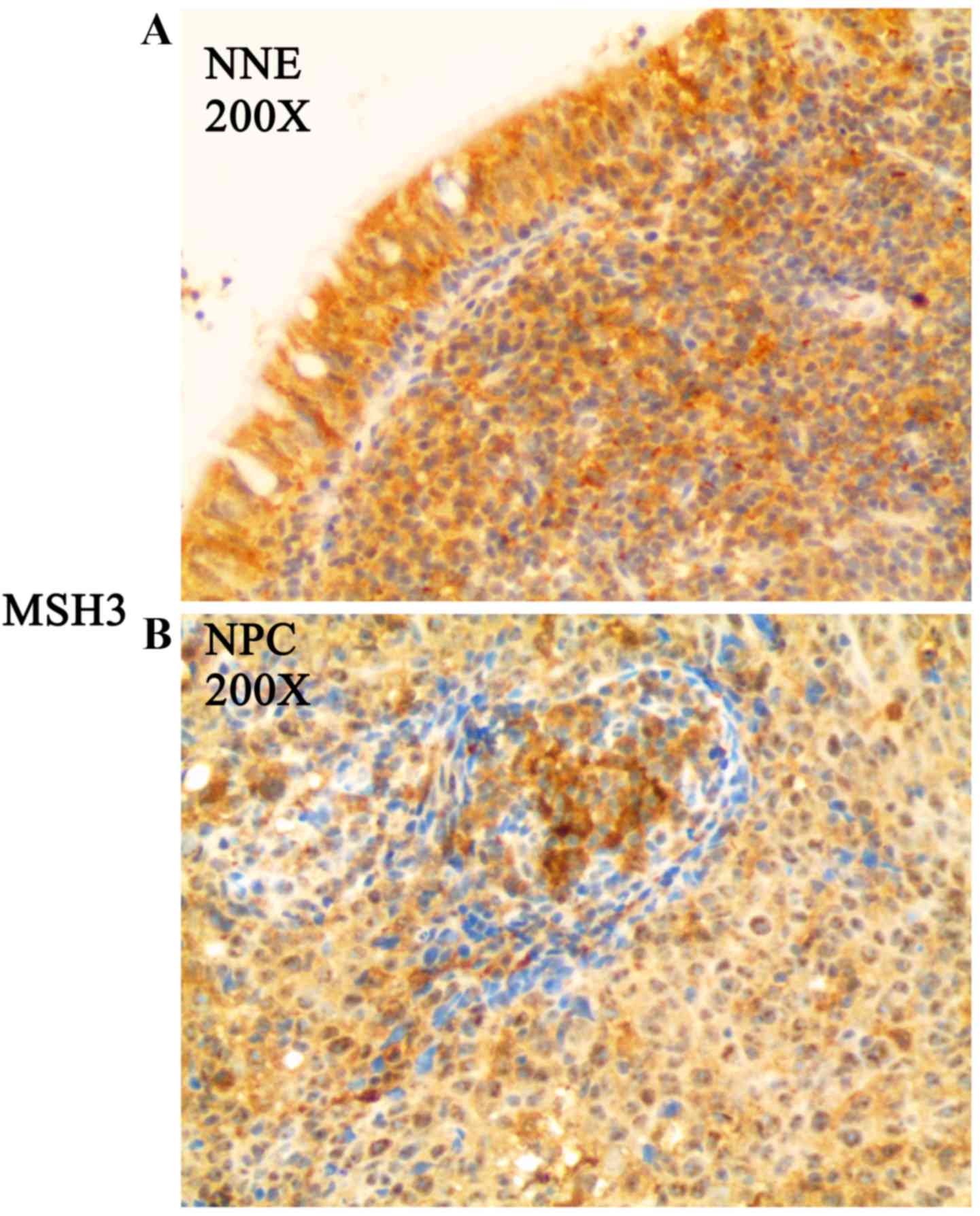
specimens, immunohistochemical staining was performed. MSH3 protein
expression was detected in all NNE tissues (Fig. 2A and Table III), while MSH3 protein
expression was frequently absent or downregulated in the 54 NPC
biopsy specimens (Fig. 2B and
Table III).
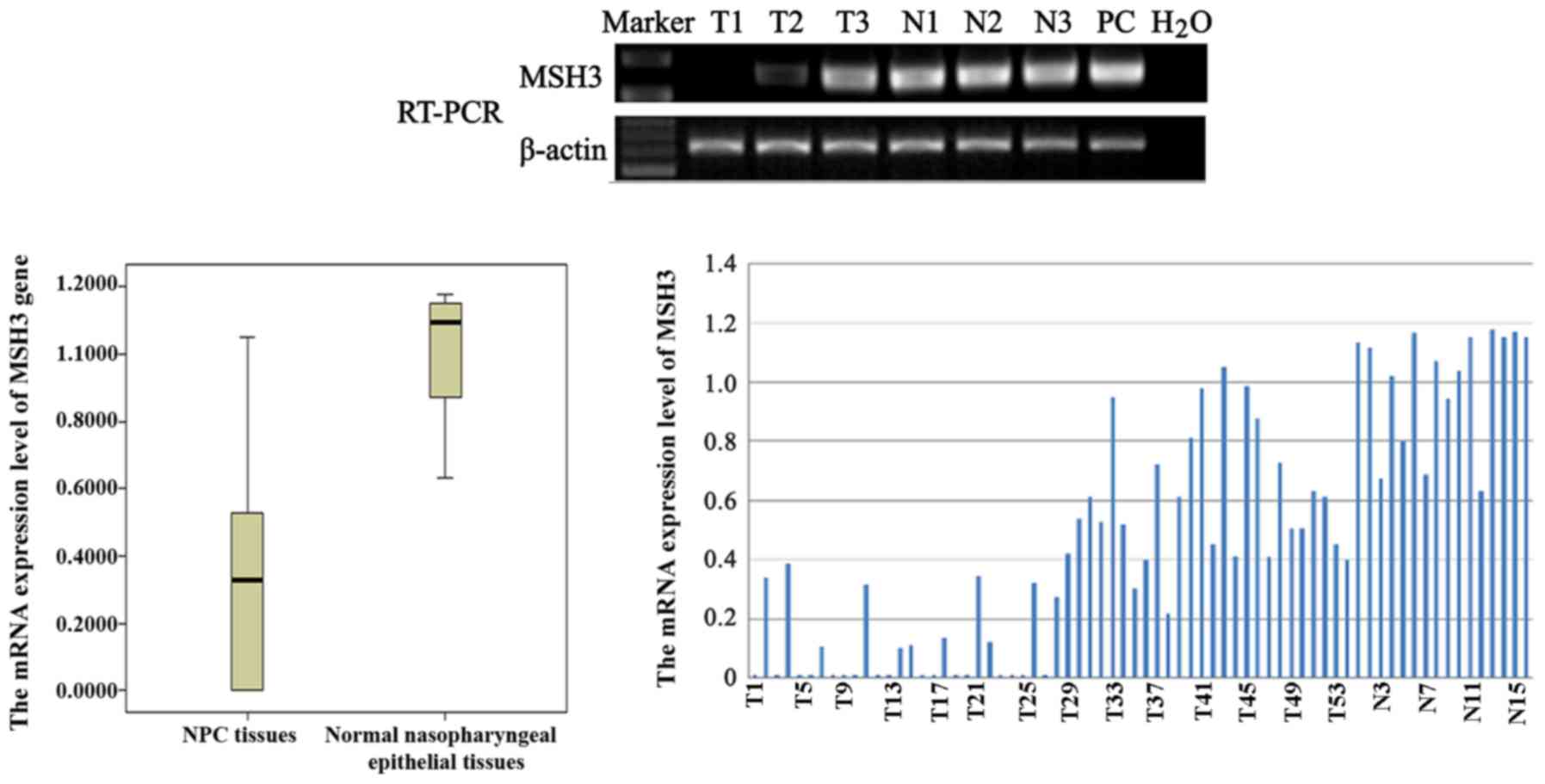 | Figure 1RT-PCR analysis of the mRNA expression
of MutS homolog human 3 (MSH3), and the mRNA expression levels of
MSH3 in nasopharyngeal carcinoma (NPC) biopsies (i.e., T1, T2, and
T3) and normal nasopharyngeal epithelial (i.e., N1, N2, and N3)
samples. The data are representative of 2 independent experiments.
In addition, β-actin, normal colorectal tissue and water were used
as an internal control, positive controls (PC) and blank control,
respectively. |
 | Table IIIThe protein expression levels of MSH3
in NNE tissues and NPC biopsies. |
Table III
The protein expression levels of MSH3
in NNE tissues and NPC biopsies.
| Biopsies | − | + | ++ | +++ | z | P-value |
|---|
| NPC | 16 | 14 | 13 | 11 | 3.986 | 0.0003 |
| NNE | 0 | 0 | 3 | 13 | | |
Hypermethylation of MSH3 in NPC primary
tumors
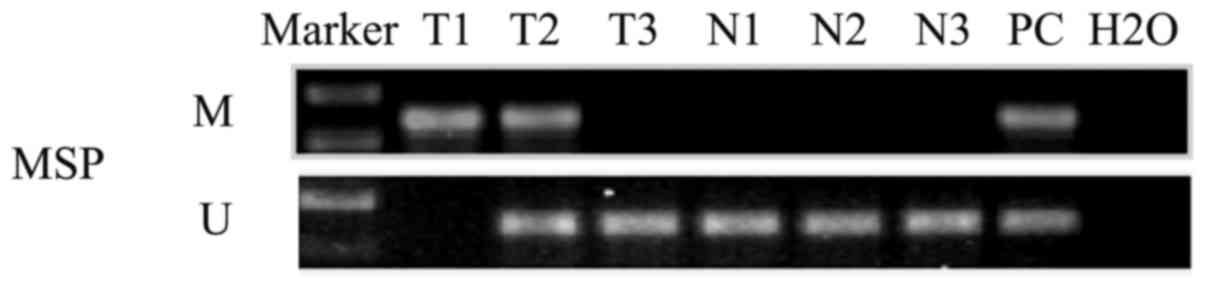
To investigate the promoter methylation status of
MSH3, MSP was performed. The promoter methylation of MSH3 was
detected in 50% (27/54) of the primary tumors (Fig. 4: T1, T2 and T3 used as samples);
however, this was not found in any of the 16 cases of normal
nasopharyngeal primary epithelial tissues (Fig. 3: N1, N2 and N3).
Inactivation of MSH3 correlates with its
promoter hypermethylation
To verify whether the mRNA inactivation of MSH3 is
related to promoter methylation in NPC, we detected the methylation
status of the MSH3 gene by MSP. The promoter methylation of MSH3
was detected in 50% (27/54) of primary tumors, among which 16 cases
(i.e., T1,T5,T6,T8,T9, T10,T12,T13,T16,T17,T19,T20,T23,T24,T25,T27;
Fig. 1) were MSH3-silenced.
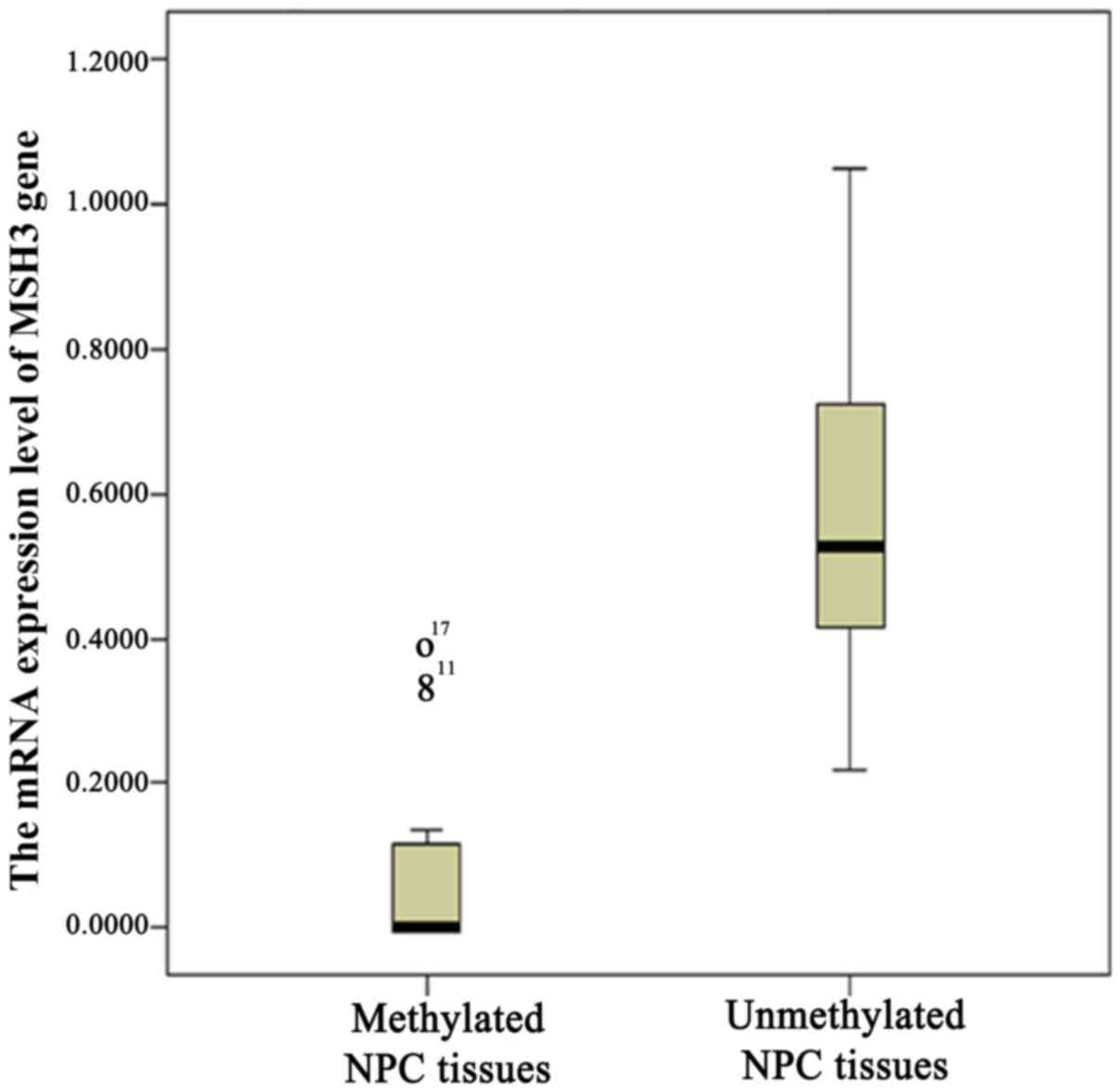
Combined with the previous RT-PCR results, a decreased level of
MSH3 expression was observed among the MSH3-methylated NPC cases as
compared to the unmethylated cases (P<0.05, Mann-Whitney's U
test; Fig. 4).
Clinicopathological significance of MSH3
gene expression
We found that the MSH3 mRNA and protein expression
levels were markedly associated with the variable T stage
(P<0.05); however, this did not correlate with the age or sex of
the patients, the stage, NM classification, or the
histopathological subtype (P>0.05; Tables IV and V).
 | Table IVCorrelation between MSH3 mRNA
expression and the clinical data of the patients with NPC. |
Table IV
Correlation between MSH3 mRNA
expression and the clinical data of the patients with NPC.
| Clinical data | n (case) | mRNA
expression | P-value |
|---|
| Sex | | | P=0.935 |
| Male | 39 | 0.3346±0.2950 | |
| Female | 15 | 0.3426±0.3744 | |
| Age, years | | | P=0.890 |
| <50 | 33 | 0.3417±0.2918 | |
| ≥50 | 21 | 0.3293±0.3566 | |
| Tumor size
stage | | | P=0.033 |
| T1T2 | 35 | 0.4077±0.3462 | |
| T3T4 | 19 | 0.2164±0.2118 | |
| Lymph node
metastasis | | | P=0.636 |
| Absence | 18 | 0.3659±0.2968 | |
| Presence | 36 | 0.3223±0.3273 | |
| Tumor stage | | | P=0.505 |
| III | 19 | 0.3761±0.3153 | |
| IIIIV | 35 | 0.3155±0.3178 | |
| Histological
subtypes | | | P=0.724 |
| Non-keratinizing
carcinoma | 48 | 0.3156±0.4024 | |
| Keratinizing
squamous cell carcinoma | 6 | 0.3342±0.3563 | |
 | Table VCorrelation between MSH3 protein
expression and the clinical data of the patients with NPC. |
Table V
Correlation between MSH3 protein
expression and the clinical data of the patients with NPC.
| Clinical data | n (case) | − | + | ++ | +++ | P-value |
|---|
| Sex | | | | | | P=0.928 |
| Male | 39 | 10 | 12 | 11 | 6 | |
| Female | 15 | 6 | 2 | 2 | 5 | |
| Age, years | | | | | | P=0.509 |
| <50 | 33 | 8 | 9 | 10 | 6 | |
| ≥50 | 21 | 8 | 5 | 3 | 5 | |
| Tumor size
stage | | | | | | P=0.047 |
| T1T2 | 35 | 7 | 11 | 6 | 11 | |
| T3T4 | 19 | 9 | 3 | 7 | 0 | |
| Lymph node
metastasis | | | | | | P=0.292 |
| Absence | 18 | 2 | 7 | 6 | 3 | |
| Presence | 36 | 14 | 7 | 7 | 8 | |
| Tumor stage | | | | | | P=0.245 |
| III | 19 | 2 | 9 | 3 | 5 | |
| IIIIV | 35 | 14 | 5 | 10 | 6 | |
| Histological
subtypes | | | | | | P=0.424 |
| Non-keratinizing
carcinoma | 48 | 24 | 1 | 15 | 8 | |
| Keratinizing
squamous cell carcinoma | 6 | 3 | 0 | 2 | 1 | |
Discussion
Tumor cells frequently exhibit deficiencies in the
signaling or repair of DNA damage. These deficiencies probably
contribute to the pathogenesis of many diseases; however, a type of
genetic stability is required, which can be obtained by
overexpressing specific DNA repair genes, in order to produce
primary tumor cells that are sufficiently genetically stable to be
able to invade and give rise to distant metastases. They also
present an opportunity to target the tumor. The dual roles of the
DNA repair pathway in cancer highlights how an understanding of DNA
repair processes can be used in the development of novel cancer
treatments. Thus, the role and expression of the regulatory
mechanism of the DNA mismatch repair gene, MSH3, in NPC is of
particular importance and is largely unresolved.
In the present study, we demonstrated that the mRNA
and protein expression levels of MSH3 in the NPC tissues were
downregulated, which indicated that MSH3 played a major role as a
tumor suppressor gene in the development of NPC. Our MSP results
revealed that MSH3 promoter methylation was detected in 50% of the
primary tumors, but was not detected in the NNE tumors. Therefore,
MSH3 methylation was a frequent and tumor-specific process in NPC.
The MSP and semi-quantitative RT-PCR data exhibited a correlation
between the mRNA expression levels and the methylation status in
NPC primary tumors. In this context, methylation inactivation
appears to be a major mechanism in the loss of MSH3 expression.
Thus, MSH3 gene methylation has a similar role in gastric cancer,
bladder cancer and NPC, and yet plays differential roles in
colorectal cancer, ovarian cancer and oral squamous cell carcinoma,
in which LOH was the major regulatory mechanism for the
inactivation of MSH3 gene expression (16–18,22).
Vymetalkova et al found the significant
overexpression of the MSH3 gene in colon tumors as compared to
adjacent mucosal tissues (26).
By contrast, it has been reported that the high frequency of LOH,
as well as aberrant protein expression, indicate an involvement of
impaired MSH3 in the low level of microsatellite instability in
colorectal cancer (16). Kawakami
et al revealed that decreased hMSH3 protein expression
levels in bladder cancer may play a significant role in the
progression of bladder tumors. An inverse correlation between hMSH3
protein expression levels with the pathological grade was also
found (18). However, in the
present study, we demonstrated that MSH3 mRNA and protein
expression levels in the NPC tissues were significantly and
inversely associated with the variable T stage. This observation
suggested that MSH3 mRNA and protein expression could be considered
an indicator of local invasion by the primary tumor. Thus, the role
of MSH3 as a tumor suppressor gene in NPC is similar to that of
colorectal and bladder cancer, although the specific mechanisms of
functional gene expression of MSH3 in different cancers vary
considerably.
Conde et al discovered that the variant in
MSH3, referred to as Ala1045Thr, was associated with a decreased
risk of breast cancer (27). The
results presented in the studies by Jafary et al and Hirata
et al demonstrated that MSH3 codon 222 and MSH3 codon 1036
polymorphisms may represent an increased risk factor for sporadic
prostate cancer (28,29). Michiels et al reported that
MSH3 single-nucleotide polymorphisms may be associated with an
increased risk of lung cancer (30). Koessler et al reported a
single nucleotide polymorphism of rs863221, located in MSH3 that
was associated with disease-specific survival in patients with
colorectal cancer (31). Dong
et al discovered that MSH3 single-nucleotide polymorphisms
correlated with overall survival in patients with pancreatic cancer
(32). However, the majority of
the cases were followed-up for >5 years, and thus the 5-year
survival rate of the patients was not analyzed. In the future, the
role of MSH3 in the pathogenesis, prognosis and risk estimation of
NPC needs to be further clarified after the relevant follow-up data
of the past 5 years is analyzed statistically.
Previous evidence has suggested that MSH3-deficient
as compared to proficient colorectal cancer cells exhibit an
increased sensitivity to the irinotecan metabolite, SN-38, and to
oxaliplatin, but not to 5-FU (9).
Another study reported an attempt to identify an association
between MSH2 and MSH3 genetic variants and development of
radiosensitivity in breast cancer patients (10). Thus, the sensitivity of
nasopharyngeal cancer patients to radiotherapy and chemotherapy is
related to MSH3, which need to be confirmed by further
experiments.
In addition, methylation-mediated inactivation is
potentially a reversible phenomenon (33). As the MSH3 gene product has an
anticancer effect, turning this process around and upregulating
MSH3 by using demethylating agent treatment may probably prevent or
reverse the malignant phenotype, and might therefore translate into
a novel demethylating agent and therapeutic target in NPC.
In this study, it is a pity that we have only used
tissue samples and have not performed any in vitro
experiments on cell lines. We aim to further analyze the mRNA
expression of the MSH3 gene in NPC cell lines before and after
treatment with the methyltransferase inhibitor,
5-aza-2-deoxycytidine, in future studies. We aim to provide
evidence to further verify that MSH3 is epigenetically inactivated
in NPC by promoter hypermethylation. The present study revealed
that MSH3 is frequently inactivated by its promoter methylation and
its mRNA and protein expression correlate with the primary tumor
stage in NPC, suggesting that MSH3 mRNA and protein expression can
be considered as such, an indicator of primary tumor local
invasion, and would have a potential value in clinical
applications. MSH3 may probably translate to a novel demethylating
agent and therapeutic target in NPC. Whether the MSH3 gene can be
used as a predictive indicator of the sensitivity of patients with
NPC to radiotherapy and chemotherapy should be confirmed by further
experiments.
Acknowledgments
This study was supported by grants from the Medical
and Health Backbone Platform Foundation Project of Zhejiang
Province, China (grant no. 2012RCB038).
References
|
1
|
Tao Q and Chan AT: Nasopharyngeal
carcinoma: molecular pathogenesis and therapeutic developments.
Expert Rev Mol Med. 9:1–24. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lung HL, Cheung AK, Ko JM, Cheng Y,
Stanbridge EJ and Lung ML: Deciphering the molecular genetic basis
of NPC through functional approaches. Semin Cancer Biol. 22:87–95.
2012. View Article : Google Scholar
|
|
3
|
Lo KW, Chung GT and To KF: Deciphering the
molecular genetic basis of NPC through molecular, cytogenetic, and
epigenetic approaches. Semin Cancer Biol. 22:79–86. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fishel R: The selection for mismatch
repair defects in hereditary nonpolyposis colorectal cancer:
revising the mutator hypothesis. Cancer Res. 61:7369–7374.
2001.PubMed/NCBI
|
|
5
|
Jensen LE, Jauert PA and Kirkpatrick DT:
The large loop repair and mismatch repair pathways of Saccharomyces
cerevisiae act on distinct substrates during meiosis. Genetics.
170:1033–1043. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gupta S, Gellert M and Yang W: Mechanism
of mismatch recognition revealed by human MutSβ bound to unpaired
DNA loops. Nat Struct Mol Biol. 19:72–78. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
van Oers JM, Edwards Y, Chahwan R, Zhang
W, Smith C, Pechuan X, Schaetzlein S, Jin B, Wang Y, Bergman A, et
al: The MutSβ complex is a modulator of p53-driven tumorigenesis
through its functions in both DNA double-strand break repair and
mismatch repair. Oncogene. 33:3939–3946. 2014. View Article : Google Scholar
|
|
8
|
Owen BA, H Lang W and McMurray CT: The
nucleotide binding dynamics of human MSH2–MSH3 are lesion
dependent. Nat Struct Mol Biol. 16:550–557. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Park JM, Huang S, Tougeron D and Sinicrope
FA: MSH3 mismatch repair protein regulates sensitivity to cytotoxic
drugs and a histone deacetylase inhibitor in human colon carcinoma
cells. PLoS One. 8:e653692013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mangoni M, Bisanzi S, Carozzi F, Sani C,
Biti G, Livi L, Barletta E, Costantini AS and Gorini G: Association
between genetic polymorphisms in the XRCC1, XRCC3, XPD, GSTM1,
GSTT1, MSH2, MLH1, MSH3, and MGMT genes and radiosensitivity in
breast cancer patients. Int J Radiat Oncol Biol Phys. 81:52–58.
2011. View Article : Google Scholar
|
|
11
|
Huang SC, Lee JK, Smith EJ, Doctolero RT,
Tajima A, Beck SE, Weidner N and Carethers JM: Evidence for an
hMSH3 defect in familial hamartomatous polyps. Cancer. 117:492–500.
2011. View Article : Google Scholar
|
|
12
|
Campregher C, Schmid G, Ferk F, Knasmüller
S, Khare V, Kortüm B, Dammann K, Lang M, Scharl T, Spittler A, et
al: MSH3-deficiency initiates EMAST without oncogenic
transformation of human colon epithelial cells. PLoS One.
7:e505412012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
de Wind N, Dekker M, Claij N, Jansen L,
van Klink Y, Radman M, Riggins G, van der Valk M, van't Wout K and
te Riele H: HNPCC-like cancer predisposition in mice through
simultaneous loss of Msh3 and Msh6 mismatch-repair protein
functions. Nat Genet. 23:359–362. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Edelmann W, Umar A, Yang K, Heyer J,
Kucherlapati M, Lia M, Kneitz B, Avdievich E, Fan K, Wong E, et al:
The DNA mismatch repair genes Msh3 and Msh6 cooperate in intestinal
tumor suppression. Cancer Res. 60:803–807. 2000.PubMed/NCBI
|
|
15
|
Jones PA and Laird PW: Cancer epigenetics
comes of age. Nat Genet. 21:163–167. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Plaschke J, Preußler M, Ziegler A and
Schackert HK: Aberrant protein expression and frequent allelic loss
of MSH3 in colorectal cancer with low-level microsatellite
instability. Int J Colorectal Dis. 27:911–919. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim HG, Lee S, Kim DY, Ryu SY, Joo JK, Kim
JC, Lee KH and Lee JH: Aberrant methylation of DNA mismatch repair
genes in elderly patients with sporadic gastric carcinoma: a
comparison with younger patients. J Surg Oncol. 101:28–35. 2010.
View Article : Google Scholar
|
|
18
|
Kawakami T, Shiina H, Igawa M, Deguchi M,
Nakajima K, Ogishima T, Tokizane T, Urakami S, Enokida H, Miura K,
et al: Inactivation of the hMSH3 mismatch repair gene in bladder
cancer. Biochem Biophys Res Commun. 325:934–942. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sun X, Chen C, Vessella RL and Dong JT:
Microsatellite instability and mismatch repair target gene
mutations in cell lines and xenografts of prostate cancer.
Prostate. 66:660–666. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xiao X, Melton DW and Gourley C: Mismatch
repair deficiency in ovarian cancer - molecular characteristics and
clinical implications. Gynecol Oncol. 132:506–512. 2014. View Article : Google Scholar
|
|
21
|
Heinen CD: Genotype to phenotype:
analyzing the effects of inherited mutations in colorectal cancer
families. Mutat Res. 693:32–45. 2010. View Article : Google Scholar :
|
|
22
|
Nunn J, Nagini S, Risk JM, Prime W,
Maloney P, Liloglou T, Jones AS, Rogers SR, Gosney JR, Woolgar J
and Field JK: Allelic imbalance at the DNA mismatch repair loci,
hMSH2, hMLH1, hPMS1, hPMS2 and hMSH3, in squamous cell carcinoma of
the head and neck. Oral Oncol. 39:115–129. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang Z, Sun D, Van N, Tang A, Hu L and
Huang G: Inactivation of RASSF2A by promoter methylation correlates
with lymph node metastasis in nasopharyngeal carcinoma. Int J
Cancer. 120:32–38. 2007. View Article : Google Scholar
|
|
24
|
Huang XM, Dai CB, Mou ZL, Wang LJ, Wen WP,
Lin SG, Xu G and Li HB: Overproduction of cyclin D1 is dependent on
activated mTORC1 signal in nasopharyngeal carcinoma: implication
for therapy. Cancer Lett. 279:47–56. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Olek A, Oswald J and Walter J: A modified
and improved method for bisulphite based cytosine methylation
analysis. Nucleic Acids Res. 24:5064–5066. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vymetalkova VP, Slyskova J, Korenkova V,
Bielik L, Langerova L, Prochazka P, Rejhova A, Schwarzova L,
Pardini B, Naccarati A and Vodicka P: Molecular characteristics of
mismatch repair genes in sporadic colorectal tumors in Czech
patients. BMC Med Genet. 15:172014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Conde J, Silva SN, Azevedo AP, Teixeira V,
Pina JE, Rueff J and Gaspar JF: Association of common variants in
mismatch repair genes and breast cancer susceptibility: a multigene
study. BMC Cancer. 9:3442009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jafary F, Salehi M, Sedghi M, Nouri N,
Jafary F, Sadeghi F, Motamedi S and Talebi M: Association between
mismatch repair gene MSH3 codons 1036 and 222 polymorphisms and
sporadic prostate cancer in the Iranian population. Asian Pac J
Cancer Prev. 13:6055–6057. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hirata H, Hinoda Y, Kawamoto K, Kikuno N,
Suehiro Y, Okayama N, Tanaka Y and Dahiya R: Mismatch repair gene
MSH3 polymorphism is associated with the risk of sporadic prostate
cancer. J Urol. 179:2020–2024. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Michiels S, Danoy P, Dessen P, Bera A,
Boulet T, Bouchardy C, Lathrop M, Sarasin A and Benhamou S:
Polymorphism discovery in 62 DNA repair genes and haplotype
associations with risks for lung and head and neck cancers.
Carcinogenesis. 28:1731–1739. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Koessler T, Azzato EM, Perkins B, Macinnis
RJ, Greenberg D, Easton DF and Pharoah PD: Common germline
variation in mismatch repair genes and survival after a diagnosis
of colorectal cancer. Int J Cancer. 124:1887–1891. 2009. View Article : Google Scholar
|
|
32
|
Dong X, Li Y, Hess KR, Abbruzzese JL and
Li D: DNA mismatch repair gene polymorphisms affect survival in
pancreatic cancer. Oncologist. 16:61–70. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Verma M, Maruvada P and Srivastava S:
Epigenetics and cancer. Crit Rev Clin Lab Sci. 41:585–607. 2004.
View Article : Google Scholar : PubMed/NCBI
|