Introduction
Tumorigenesis has been a serious threat to human
health and survival for decades and among the malignant cancers,
osteosarcoma (OS) is one of the most common tumors, particularly in
adolescents (1). The occurrence
of OS denotes a complex mechanism where multiple factors from
environmental and genetic origins may largely contribute to the
complexity (2). During the
development of OS, immature bones are formed and normal bones are
jeopardized (3). In addition,
long bones are at a greater risk of the onset of OS, particularly
those in the knees (4). The
survival rates of patients with OS are particularly low despite
significant improvements being made, including combinatorial
chemotherapy and surgery (5,6).
The mechanisms responsible for the development of OS are very
complex. Therefore, the in-depth understanding of the occurrence
and development of OS poses a new challenge to biological
researchers.
The p21-activated kinases (PAKs) belong to the
serine/threonine kinases which can be activated by GTPases
(7). PAKs are classified into 2
subgroups, namely group I PAKs (PAK1-3) and group II PAKs (PAK4-6),
while PAK7 is a different type of PAK compared with group I and
group II (8). PAKs are implicated
in massive biological processes, particularly in neoplasms
(7). The expression of PAK7 is
usually found to be upregulated in diverse types of cancer,
including OS, lung cancer, colorectal cancer and pancreatic cancer
(9–13). Therefore, the oncogene PAK7
may be implicated in the progression of numerous types of
cancer.
A class of non-coding RNAs with a short length known
as microRNAs (miRNAs of miRs) (approximately 22 nucleotides in
length) are usually evolutionarily conserved and play pivotal roles
in various biological processes (14). Complementary binding to the
3′-untranslated region (3′-UTR) of target mRNAs can either silence
the translation or promote their degradation (15). The involvement of miRs has been
reported in numerous types of cancer, such as lung cancer,
hepatocellular carcinoma (HCC), breast cancer and colon cancer
(16–21). The role of miRs in tumorigenesis
is complex, partly due to the fact that miRs can either function as
tumor suppressors or oncogenes (17–19,22–24). For example, He et al
suggested that miR-34 inhibits the development of OS in a
p53-dependent manner (25).
However, Li et al demonstrated that miR-296-5p induced the
proliferation of gastric cancer cells by targeting caudal-related
homeobox 1 (CRH1) (26). The
expression and epigenetics of miRs are usually altered in tumorous
tissues (19,27). These findings suggest that miRs
are intensively implicated in tumorigenesis. However, even though
numerous studies have been carried out on miRs and cancer, the
association between miR-492 and OS remains poorly understood.
In this study, we found that miR-492 was usually
downregulated in both OS cell lines and human specimens. The
ectopic expression of miR-492 significantly attenuated the adverse
effects on OS, inhibiting the proliferation, migration and invasion
of U-2 OS and MG-63 cells in vitro. Furthermore,
transfection with a miR-492 overexpression vector also decreased
the tumor volume in an implantation assay in vivo. We
further identified PAK7 as a potential miR-492 target in OS. The
inverse correlation between PAK7 and miR-492 was also evident in
tumorous tissues. Given the oncogenic role of PAK7, to the best of
our knowedge, we are the first to demonstrate that miR-492 can
serve as a candidate tumor suppressor by targeting PAK7 in OS. Our
findings provided important insight into the miR-targeted therapy
of OS.
Materials and methods
Cell culture and human specimens
The OS cell lines, Saos-2, HOS, 143B, U-2 OS, MG-63
and G292, as well as the control osteoblast cell line, hFOB, were
purchased from the American Type Culture Collection (ATCC;
Rockville, MD, USA). The ZK-58 cells were obtained from Shanghai
Institute of Biochemistry and Cell Biology (Shanghai, China). The
cells were cultured in RPMI-1640 medium obtained from Sigma-Aldrich
(Shanghai, China). The medium was further supplemented with 5%
fetal bovine serum (FBS) and penicillin (100 U/ml) (Sigma-Aldrich)
in a humidified atmosphere of 5% CO2 at 37°C. The OS
specimens were all surgical archives from patients (age, 41–79;
n=77) at the First Affiliated Hospital of Harbin Medical University
(Harbin, China) obtained between September 2013 and July 2015.
Paired normal adjacent tissues were resected as controls. All
specimens were kept in liquid nitrogen at −80°C following
resection. All patients provided written informed consent for their
use of their samples. The protocols of the experimental procedures
related to human samples were formally approved by the Human
Research Ethics Committee of the First Affiliated Hospital of
Harbin Medical University (no. 2013F0012).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from both cell lines and
human specimens using TRIzol reagent(Invitrogen Life Technologies,
Carlsbad, CA, USA) according to the manufacturer's instructions.
The cDNA (20 µg in total) obtained by reverse transcription
was generated by the SYBR Premix Taq™ Toolkit (Takara Bio, Inc.,
Otsu, Japan) according to the manufacturer's instructions. The
TaqMan microRNA qRT-PCR kit (Applied Biosystems, Foster
City, CA, USA) was used in present study. To measure PAK7
mRNA expression, the SYBR-Green PCR Master kit was utilized (Takara
Bio, Inc.). GAPDH was used as the control. Reactions were
performed with the ABI PRISM® 7000 Sequence Detection
system (Applied Biosystems) following the manufacturer's
instructions. miR-492 and PAK7 expression levels were
determined using the 2−ΔΔCt method. All experiments were
performed in 3 replicates. The primer sequences used were as
follows: miR-492, 5′-GGCTATGCTTGAGTACG-3′ (forward) and
5′-CTGAGTTAGCGTACGAGT-3′ (reverse); GAPDH,
5′-CTCGCCGCAGTGCATTCGT-3′ (forward) and
5′-ACGCTTCGCGATCGTGCGTGAT-3′ (reverse); and PAK7,
5′-GCTACGTAGACCCTGAT-3′ (forward) and 5′-CAGTCACTCCGTACGG-3′
(reverse).
Generation of stably transfected cell
lines
The lentiviral system to ectopically overexpress
miR-492 in U-2 OS and MG-63 was used in this study. The lentivirus
with miR-492 mimics (miR-492) and negative controls were
synthesized and purchased from Sigma-Aldrich. The Lipo fectamine™
2000 system (Invitrogen Life Technologies, Shanghai, China) was
used for viral transfection. At 24 h post-transfection, the
cultured medium was replaced with fresh medium. All plasmids were
experimentally verified by RT-qPCR.
Luciferase reporter assay
The 3′-UTR region for PAK7 was amplified by
PCR using the primer sequences as follows:
5′-ATGCCGTCGCCTCTTGTGTCTTC-3′ (forward) and
5′-GTACTGAGTCCTTCTGAGGC-3 (reverse). GAPDH was used as a
control. The 3′-UTR for PAK7 with predicted binding sites
for hsa-miR-492 was cloned into the Xbal downstream of
Renilla luciferase reporter plasmid phRL-TK (Promega,
Madison, WI, USA) to obtain the wild-type PAK7 luciferase
plasmids (PAK7 3′-UTR WT). A construction with PAK7
point mutation in 3′-UTR was similarly obtained (PAK7 3′-UTR
MUT). 293T cells (Shanghai Institute of Biochemistry and Cell
Biology) were loaded into a 96-well plate 24 h prior to
transfection and then co-transfected with PAK7 3′-UTR WT or
MUT vector. The β-gal control plasmid (Ambion, Carlsbad, CA,
USA), miR-492 or control vectors were transfected into the cells
using the Lipofectamine 2000 system (Invitrogen Life Technologies).
The Luciferase activities were measured using the Dual-Luciferase
reporter system (Promega) as relative luciferase units following
the manufacturer's instructions.
Colony formation assay
At 2 days following transfection, at total of 400
U-2 OS cells were seeded into a 12-well plate. The plate was
replenished with fresh medium at an interval of 2 days. After 2
weeks, the clones were washed with phosphate-buffered saline (PBS)
and fixed in 5% paraformaldehyde (PFA) for 15 min at 37°C. Giemsa
staining (no. G9641; Sigma-Aldrich) was then performed for 20 min
and the cells were washed with water.
Invasion assay
The upper chamber of a Transwell (8-mm pore size;
EMD Millipore, Billerica, MA, USA) was pre-coated with Matrigel
(Invitrogen Life Technologies) overnight. After 24 h, the U-2 OS
and MG-63 cells transfected with lentivirus were suspended and
loaded in the upper chamber (104 cells/well) in
RPMI-1640 medium (Sigma-Aldrich). The lower chambers were
supplemented with RPMI-1640 medium with additional 5% FBS. After a
further 24 h, the upper chamber was removed. The migrating cells
into the lower chamber were fixed with 5% PFA and stained with
crystal violet (Sigma-Aldrich). Cells from 5 non-overlapping views
were quantified using a Leica microscope fluorescence microscope
(DM IRB; Leica Microsystems GmbH, Wetzlar, Germany).
Migration assay
The Boyden chambers (BD Biosciences, San Jose, CA,
USA) were used in a 12-well plate. At 24 h following transfection,
105 cells from the RPMI-1640 medium were seeded in the
upper chamber (Sigma-Aldrich). RPMI-1640 with 15% FBS was used as
an attractant. Non-migrated cells were removed 24 h after the
experiment, loaded into 5% PFA and stained with crystal violet.
Proliferation assay
Alterations in cell proliferation were determined
using the Cell Counting Kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc., Shanghai, China). The U-2 OS and MG-63 cells
(104 cell/well) transfected with either lentivirus
control or miR-492 lentivirus were seeded on a 96-well plate for 5
days. A total of 20 µl MTT solution were added into the
culture (10 mg/ml) for 4 h at an interval of 1 day. The optical
density (OD) at 490 nm was determined using Spectramax M5
microplate monitor (Molecular Devices, LLC, Sunnyvale, CA, USA)
following the manufacturer's instructions.
Cancer cell implantation
In total, 105 U-2 OS cells transfected
with lentiviral miR-492 or lentiviral controls were injected
subcutaneously into BALB/c nude mice. In total, 12 mice (age, 4–5
weeks; average weight, 16.6 g; male: 6, female: 6) were used in the
current study. Mice were housed at 20°C, with 50–60% humidity and a
light-dark cycle of 12 h. Ad libitum access to food and
water was provided. Animal experiments were formally reviewed and
approved by the Ethics Committee for Animal Research at the First
Affiliated Hospital of Harbin Medical University. The tumor volume
was measured at an interval of 3 days for 30 days. By the end of
the implantation, all mice were sacrificed and Ki-67 immunostaining
was performed using the KI-67 kit (Sigma-Aldrich) according to the
manufacturer's instructions.
Western blot analysis
The U-2 OS and MG-63 cells were harvested with lysis
buffer (12% glycerol and 5% NP-40) obtained from Sigma-Aldrich. The
protein extracts (equally 100 µg for each) were dissolved in
10% SDS-PAGE and transferred onto nitrocellulose membranes (Bio-Rad
Laboratories, Hercules, CA, USA). The membranes were coated with
monoclonal anti-PAK7 antibody (cat. no. K3265) and anti-GAPDH
antibody (cat. no. G8795) (both from Sigma-Aldrich) overnight. The
HRP-conjugated secondary antibodies (1:1,000) were supplemented at
20°C for 2 h. The chemiluminescence film system (Amersham Pharmacia
Biotech, Shanghai, China) was used to visualize the
immunoblots.
Predicting miR-492 target
We used algorithms for target gene prediction
TargetScan (http://genes.mit.edu/targetscan), DIANA-microT
(http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=microT_CDS/index)
and miRDB (www.mirdb.org) as previously described
(28–30). Briefly, putative targets were
ranked by Z scores. The top ranked overlapping targets are selected
for experimental verification.
Statistical analysis
Statistical results were all analyzed using SPSS
software (version 16.0; SPSS, Inc., Chicago, IL, USA). All
experiments were carried out in triplicate. P-values <0.05 were
considered to indicate statistically significant differences. A
paired test was used for pair-wise sample comparisons.
Results
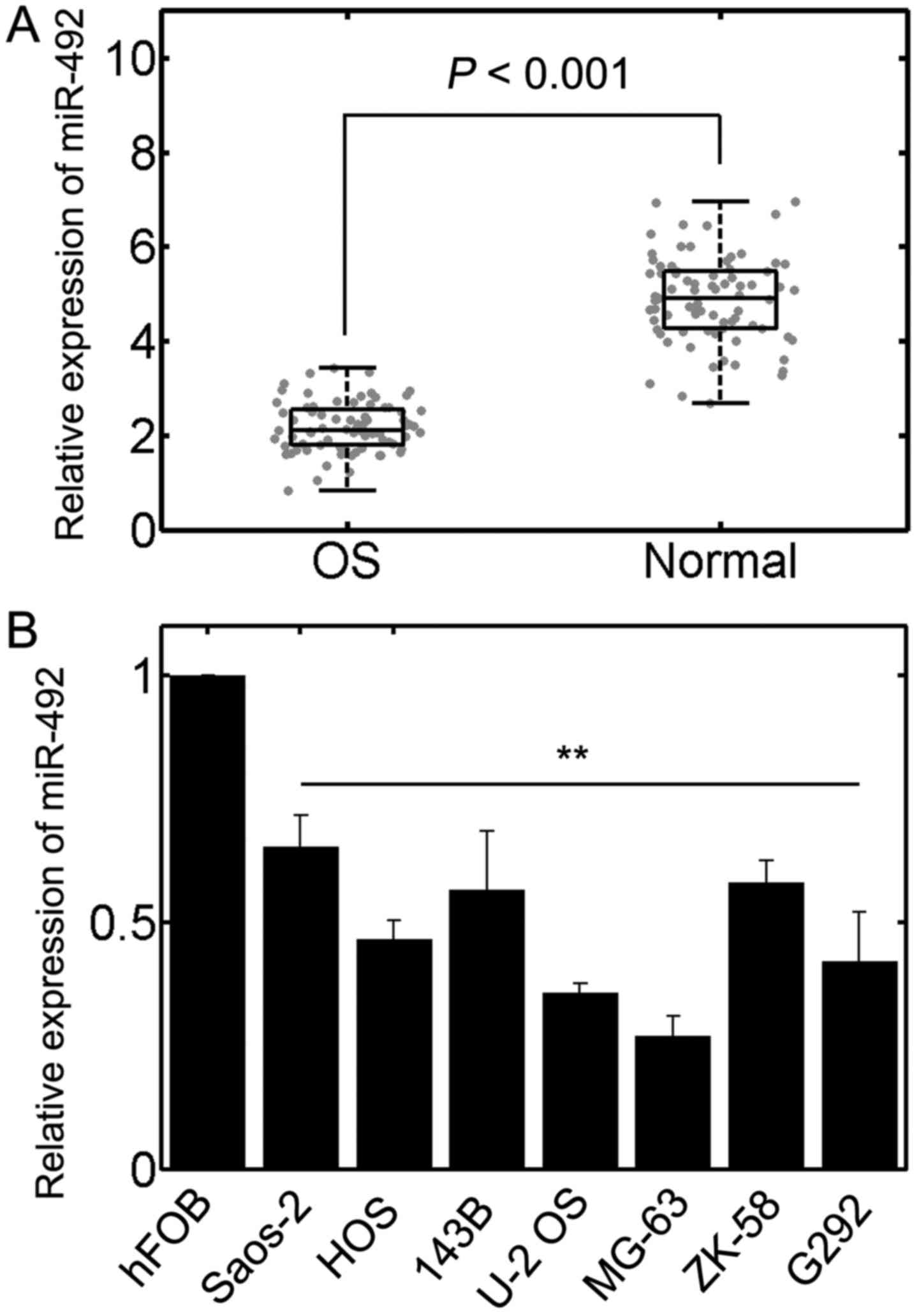
Expression of miR-492 is reduced in OS
samples and cell lines
To determine whether the expression of miR-492 is
altered in OS, RT-qPCR was performed. The results revealed that the
level of miR-492 was significantly downregulated in the OS samples
compared with the corresponding paired normal adjacent tissues
(n=77, P<0.01; Fig. 1A). In
addition, we also investigated the expression of miR-492 in
well-established OS cell lines. The results also confirmed that
miR-492 was markedly decreased in OS cell lines in contrast to the
normal osteoblast cell line, hFOB (Fig. 1B). These results implied that
miR-492 is downregulated in OS and may thus play a role in the
development of OS. Since the U-2 OS and MG-63 cells exhibited the
lowest miR-492 expression, we selected these 2 cell lines for
further analysis.
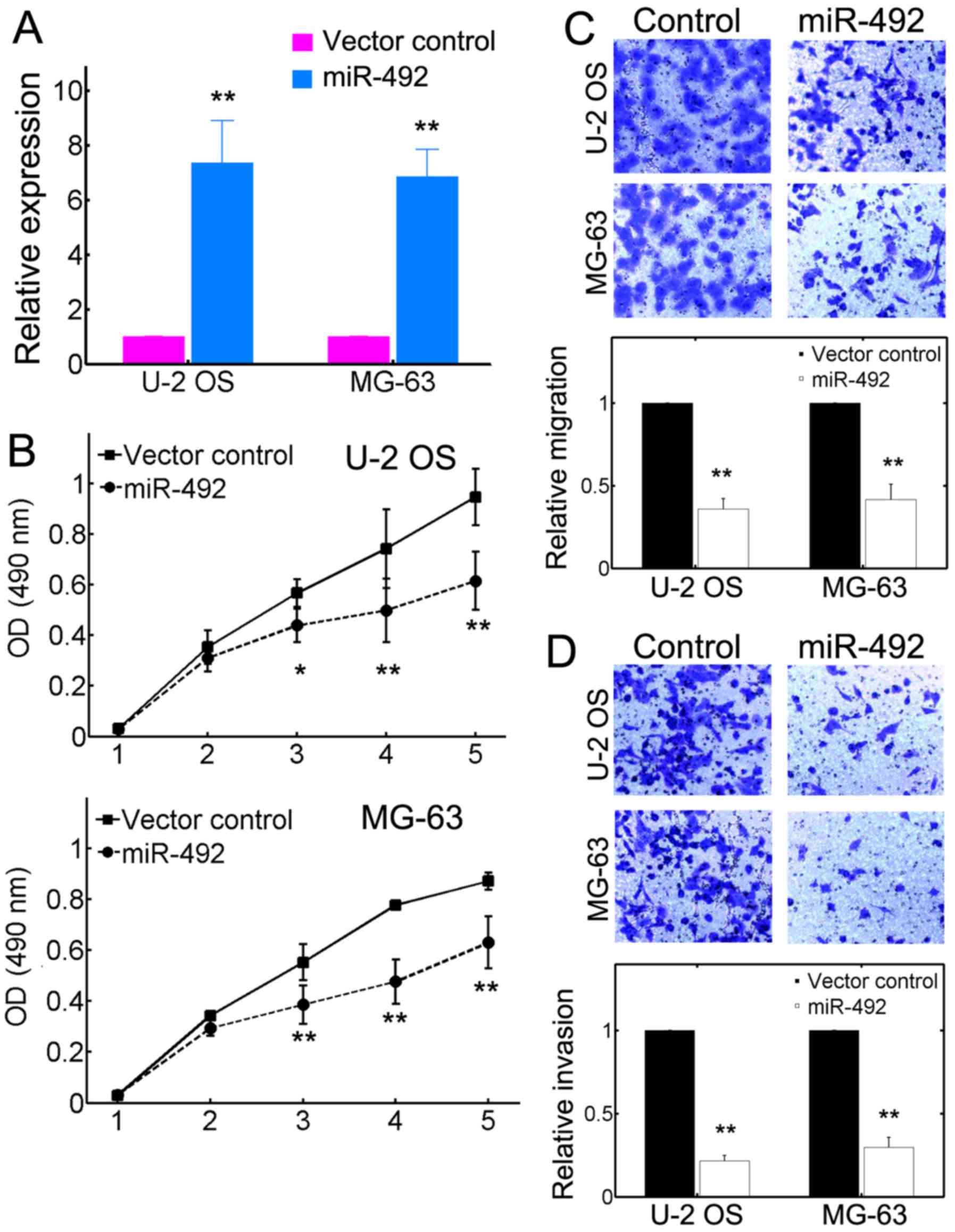
miR-492 inhibits the malignancy of OS in
vitro
We then performed experiments to access the role of
miR-492 in the proliferation, migration and invasion of OS cells.
The U-2 OS and MG-63 cells were transfected with either a control
vector or recombinant plasmid encoding miR-492 precursor. The
results of RT-qPCR revealed that miR-492 expression was
significantly elevated owing to miR-492 precursor transfection
(Fig. 2A). In addition, we found
that transfection with the miR-492 overexpression vector
substantially inhibited the proliferation of both U-2 OS and MG-63
cells (Fig. 2B). The difference
in proliferation was evident as early as 3 days in
miR-492-overexpressing cells (Fig.
2B). Furthermore, miR-492 also attenuated the migration of both
OS cell lines, as accessed by migration assays (Fig. 2C). The efficacy of inhibition was
even >50% (Fig. 2C, bottom
panels). Invasion assays also yielded similar results, which
suggested that miR-492 can significantly decrease the invasive
capacities of U-2 OS and MG-63 cell lines (Fig. 2D). These results collectively
demonstrated that miR-492 can potently inhibit the tumorigenic
potential of OS cell lines.
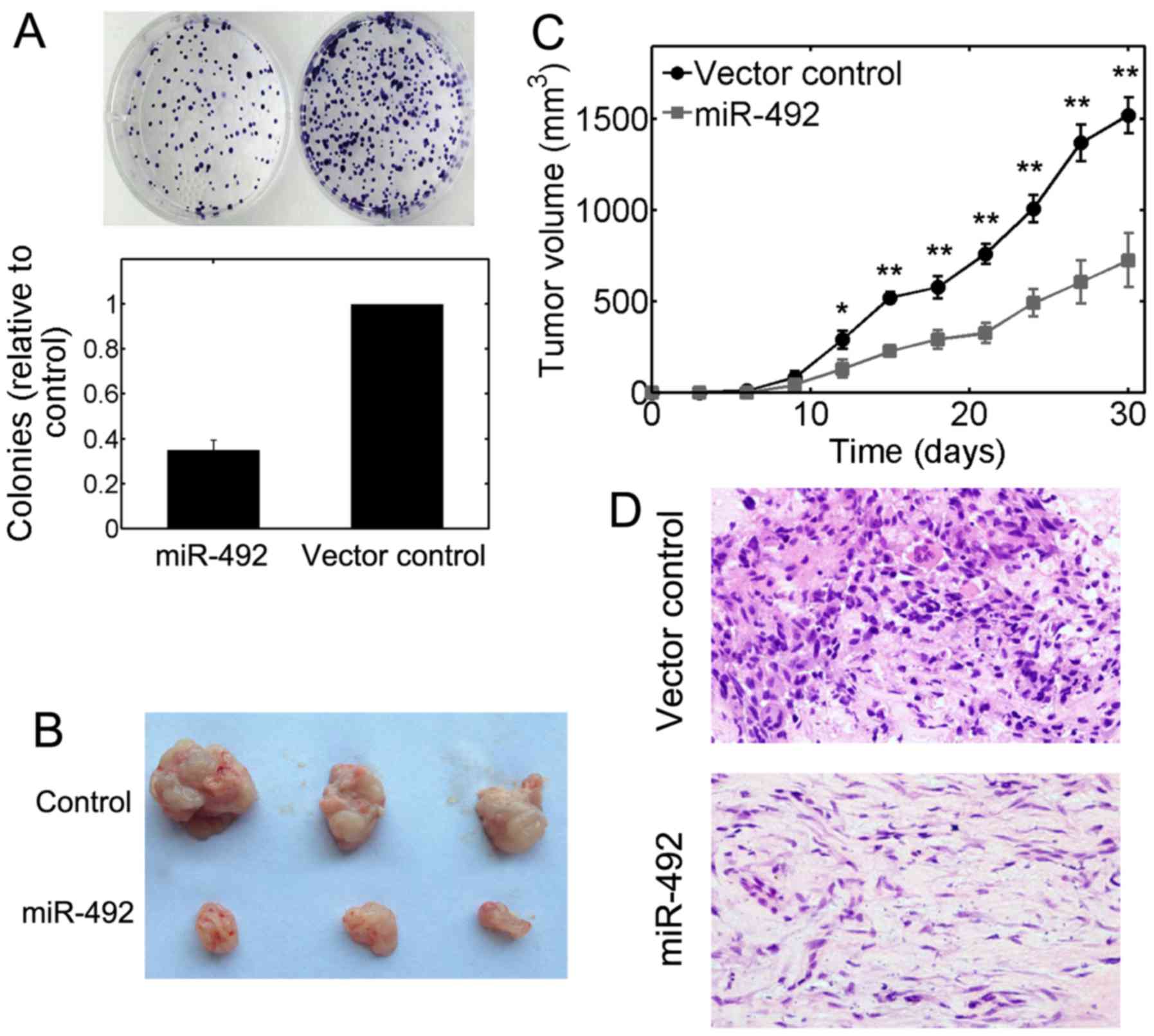
miR-492 modulates the growth of OS
tumors
We further used colony formation assays to access
the tumorigenic ability of OS cells. We found that the average
number of colonies in U-2 OS cells transfected with miR-492
precursor plasmids was significantly decreased (P<0.01; Fig. 3A). To investigate the role of
miR-492 in modulating tumor formation in vivo,
105 U-2 OS cells stably transfected with lentiviral
miR-492 or lentiviral controls were subcutaneously injected into
nude mice and the growth of solid tumors was evaluated every 3
days. The volume of solid tumors was significantly decreased in the
groups injected with miR-492-transfected cells (Fig. 3B). A smaller tumor size was also
evident in the groups injected with cells overexpressing miR-492
during a 30-day evaluation period (Fig. 3C). The Ki-67 immnostaining results
further indicated that the growth of OS tumors was strongly
inhibited by inducing miR-492 overexpression (Fig. 3D). Taken together, these results
suggest a tumor suppressive role for miR-492 in OS in
vivo.
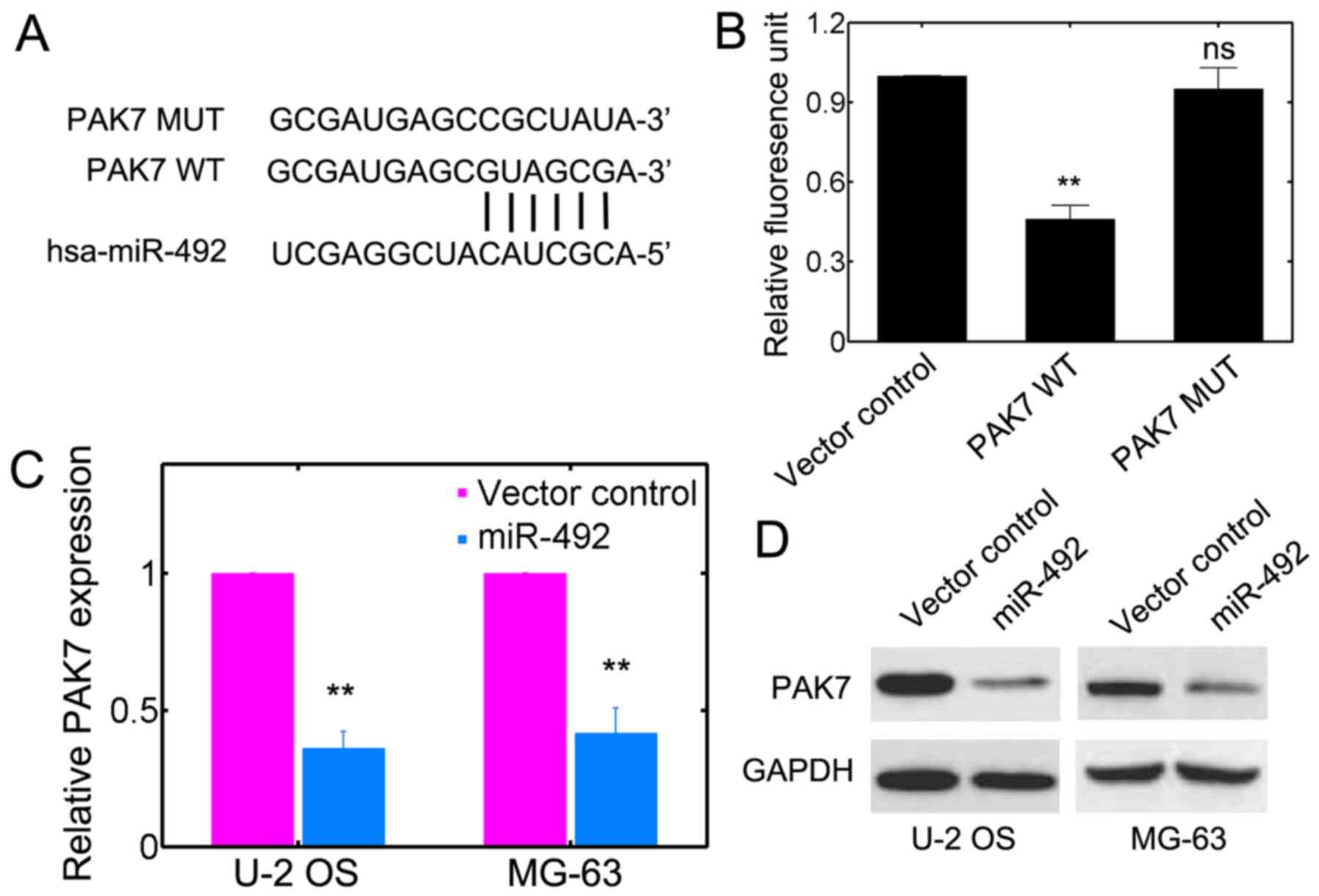
PAK7 is the target of miR-492
To unravel the potential target for miR-492, we used
online databases, such as microRNA.org
(http://www.microrna.org/microrna/home.do),
DIANA-microT (http://diana.imis.athenainnovation.gr/DianaTools/index.php?r=microT_CDS/index)
and MIRDB (www.mirdb.org). It was suggested that
PAK7 may be the target of miR-492 (Fig. 4A). We further investigated whether
miR-492 has a direct effect on PAK7 using luciferase
reporter assay. The complimentary sites on PAK7 for miR-492
base pairing were mutated (Fig.
4A). We noted that miR-492 transfection substantially lowered
the luciferase reporter activities in the PAK7 WT group
(Fig. 4B). However, miR-492
failed to inhibit the luciferase activity in cells harboring
PAK7 MUT (Fig. 4B).
Furthermore, the transcript level of PAK7 was significantly
downregulated by transfection iwth miR-492 precursor plasmids in
U-2 OS and MG-63 cells (Fig. 4C).
Western blot analysis further consolidated that the protein levels
of PAK7 were consistently reduced with miR-492 transfection
(Fig. 4D). These results support
the prediction that miR-492 targets PAK7 in OS cells.
Level of miR-492 inversely correlates
with PAK7 in OS
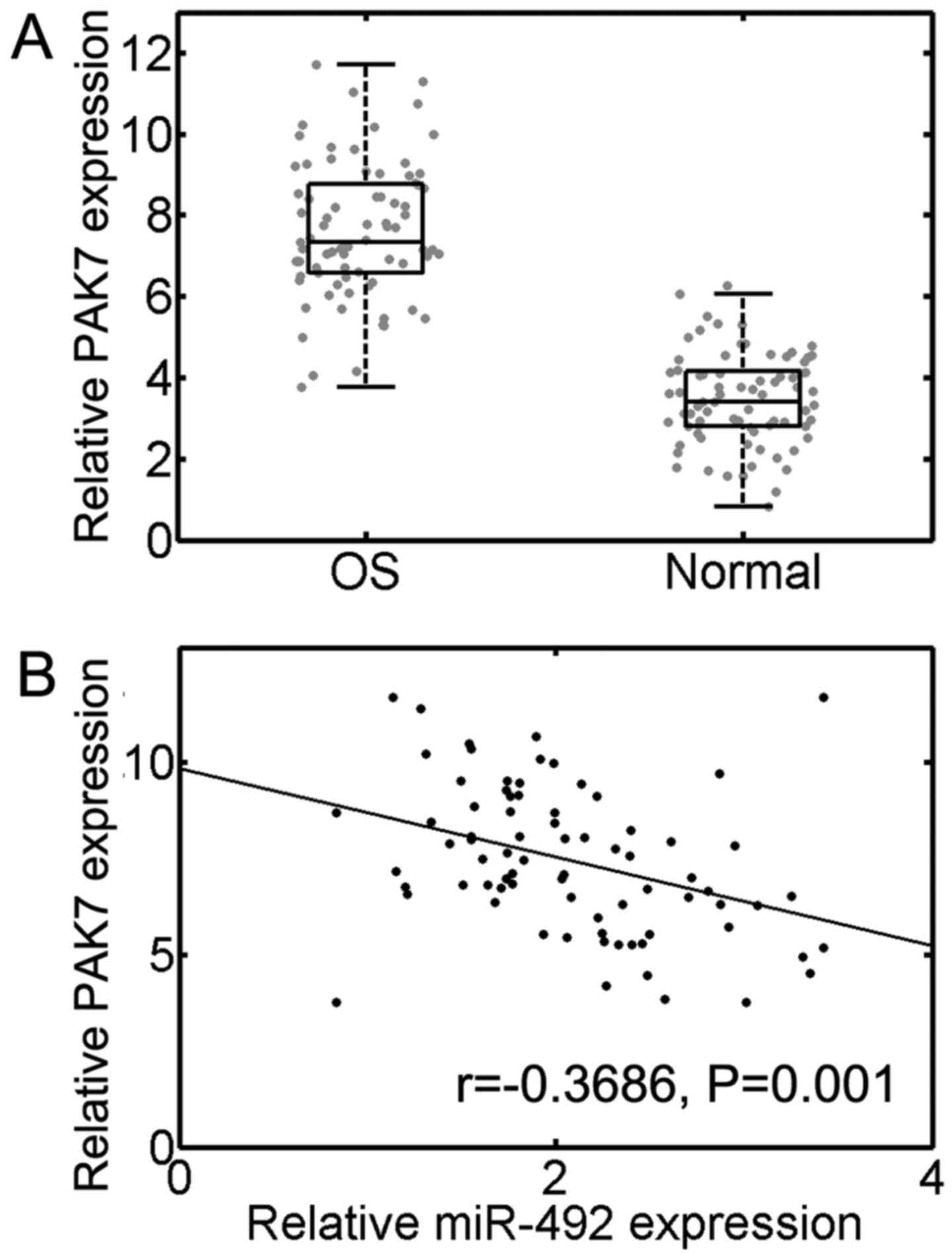
To further identify whether PAK7 is indeed
downregulated in OS specimens, we carried out RT-qPCR analysis. The
results suggested that PAK7 mRNA expression was
significantly upregulated in OS tissues compared to normal adjacent
tissues (n=77, P<0.001; Fig.
5A). Moreover, we further identified a significant inverse
correlation between miR-492 and PAK7 transcript levels in OS
samples (r=−0.3686, P=0.001; Fig.
5B). Taken together, our results suggest a tumorigenic role for
PAK7 and establish an inverse correlation between miR-492 and
PAK7.
Discussion
The occurrence of tumors is a complex process and is
associated with multiple factors. The dysregulation of miRs may
also render tumor cells with proliferative advantage, resulting in
uncontrolled growth, as well as other malignant characteristics
(31). The exact role of miRs in
tumorigenesis is undetermined and is possibly ascribed to the tumor
microenvironment (32). Another
reason may be due to the fact that miRs are usually located in the
fragile locus of genes (33).
Studies have focused on the role of miR in order to establish
significant linkage between miRs and carcinogenesis (34,35). Therefore, the identification of an
association between miRs and tumorigenic factors may not only
provide fruitful insight into diagnosis, but may also shed light on
more effective therapeutic interventions.
In present study, we found that miR-492 can function
as a tumor suppressor miR in OS. OS specimens usually showed
reduced miR-492 expression compared with normal osteoblast cell
lines. The ectopic expression of miR-492 using a lentiviral system
effectively inhibited tumor growth, and the migration and invasion
of OS cell lines. Further identification of miR-492 targets
confirmed that PAK7 may be the candidate which was then
experimentally verified. Moreover, PAK7 expression in OS also
showed a significantly inverse correlation with miR-492, therefore
further clarifying a tumor suppressor role for miR-492. The
function of miR-492 has been reported in some cancer types. For
instance, Jiang et al found that miR-492 promoted
tumorigenesis in HCC by targeting PTEN (36). The miR-492-mediated effect was
reversed by the addition of AKT inhibitor in HCC cell lines
(36). Another study showed that
miR-492 suppressed SOX7 expression and regulated the proliferation
of breast cancer cell lines (17). However, a lower miR-492 expression
contributes to oxaliplatin (a chemotherapeutic drug) resistance by
elevating CD147 expression in LS174T/L-OHP colon cancer cell lines,
implying that miR-492 could function similar to tumor suppressor
(37). Therefore, whether miR-492
plays a tumor suppressor role or acts as an oncogenic factor is
still elusive. Additionally, to date, there is no study focusing on
the role of miR-492 in OS. In the present study, we argued that
miR-492 can serve as a tumor suppressive factor as least in OS
cells. As a result, our experiments clarified a novel role of
miR-492 in OS and provide further knowledge about the functions of
miR-492 in tumorigenesis.
The serine/threonine protein kinase, PAK7, was later
identified as a putative miR-492 target suggesting that miR-492 can
exert its function by suppressing PAK7. PAK7 has recently been
predicted to be a potential biomarker in non-small cell lung cancer
and OS (11,38). Moreover, the aberrant expression
of PAK7 is also evident in gastric cancer, suggesting that PAK7 may
be critically important in promoting tumor progression (9). It has been previously demonstrated
that PAK7 can support cell mobility and proliferation by activating
survival pathways (11). The
apoptotic signaling is also inhibited due to the mitochondria
localization of PAK7 (40). In OS
cells, the knockdown of PAK7 can also favor the induction of
apoptosis. It can de-sensitize the apoptotic potential induced by
camptothecin and C2-ceramide by phosphorylating BAD, a
pro-apoptotic member of the BCL-2 family at Ser-112 (11). PAK7 has also been shown to mediate
cisplatin resistance and contributes to the progression of
esophageal cancer (40). The
mechanism was largely ascribed to the E2F1-induced transcriptional
upregulation of PAK7 via Aurora-A activation (40). PAK7 has also been shown to serve
as a biomarker for the proliferation of pancreatic cancer cell
xenografts (12). Taken together,
these findings strongly suggest an oncogenic role for PAK7 in
various types of cancers. Therefore, targeting PAK7 for
post-transcriptional regulation by miR-492 may be an effective
strategy to efficiently attenuate the development and progression
of OS.
In conclusion, in this study, we identified a tumor
suppressive function of miR-492, at least in OS, by targeting PAK7.
Since PAK7 has been shown to be extensively upregulated in various
types of tumor and substantially promotes tumor progression,
silencing PAK7 expression by miR-492 may prove to be an effective
strategy with which to suppress the malignancy of OS. The intricate
correlation between miR-492 and PAK7 needs to be investigated in
more detail in order to further unravel the hidden layer of
complexity in carcinogenesis and provide insight into diagnosis and
pharmaceutical intervention.
References
|
1
|
Friedrich P, Ortiz R, Strait K, Fuentes S,
Gamboa Y, Arambú I, Ah-Chu-Sanchez M, London W, Rodríguez-Galindo
C, Antillón-Klussmann F, et al Central American Association of
Pediatric Hematologists Oncologists AHOPCA: Pediatric sarcoma in
Central America: outcomes, challenges, and plans for improvement.
Cancer. 119:871–879. 2013. View Article : Google Scholar
|
|
2
|
Mirabello L, Troisi RJ and Savage SA:
International osteosarcoma incidence patterns in children and
adolescents, middle ages and elderly persons. Int J Cancer.
125:229–234. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Morrow JJ and Khanna C: Osteosarcoma
genetics and epigenetics: emerging biology and candidate therapies.
Crit Rev Oncog. 20:173–197. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Admassi D: Osteosarcoma of medial cuniform
bone. Ethiop Med J. 47:305–308. 2009.
|
|
5
|
Husmann K, Ducommun P, Sabile AA, Pedersen
EM, Born W and Fuchs B: Signal transduction and downregulation of
C-MET in HGF stimulated low and highly metastatic human
osteosarcoma cells. Biochem Biophys Res Commun. 464:1222–1227.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee JA, Kim MS, Kim DH, Lim JS, Park KD,
Cho WH, Song WS, Lee SY and Jeon DG: Postoperative infection and
survival in osteosarcoma patients. Ann Surg Oncol. 16:147–151.
2009. View Article : Google Scholar
|
|
7
|
Martin H, Mali RS, Ma P, Chatterjee A,
Ramdas B, Sims E, Munugalavadla V, Ghosh J, Mattingly RR, Visconte
V, et al: Pak and Rac GTPases promote oncogenic KIT-induced
neoplasms. J Clin Invest. 123:4449–4463. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Melzer J, Kraft KF, Urbach R and Raabe T:
The p21-activated kinase Mbt is a component of the apical protein
complex in central brain neuroblasts and controls cell
proliferation. Development. 140:1871–1881. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gu J, Li K, Li M, Wu X, Zhang L, Ding Q,
Wu W, Yang J, Mu J, Wen H, et al: A role for p21-activated kinase 7
in the development of gastric cancer. FEBS J. 280:46–55. 2013.
View Article : Google Scholar
|
|
10
|
Zhang HH, Zhang ZY, Che CL, Mei YF and Shi
YZ: Array analysis for potential biomarker of gemcitabine
identification in non-small cell lung cancer cell lines. Int J Clin
Exp Pathol. 6:1734–1746. 2013.PubMed/NCBI
|
|
11
|
Han K, Zhou Y, Gan ZH, Qi WX, Zhang JJ,
Fen T, Meng W, Jiang L, Shen Z and Min DL: p21-activated kinase 7
is an oncogene in human osteosarcoma. Cell Biol Int. 38:1394–1402.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Giroux V, Iovanna JL, Garcia S and Dagorn
JC: Combined inhibition of PAK7, MAP3K7 and CK2alpha kinases
inhibits the growth of MiaPaCa2 pancreatic cancer cell xenografts.
Cancer Gene Ther. 16:731–740. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gong W, An Z, Wang Y, Pan X, Fang W, Jiang
B and Zhang H: P21-activated kinase 5 is overexpressed during
colorectal cancer progression and regulates colorectal carcinoma
cell adhesion and migration. Int J Cancer. 125:548–555. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fromm B, Billipp T, Peck LE, Johansen M,
Tarver JE, King BL, Newcomb JM, Sempere LF, Flatmark K, Hovig E, et
al: A uniform system for the annotation of vertebrate microRNA
genes and the evolution of the human microRNAome. Annu Rev Genet.
49:213–242. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bartel DP: MicroRNAs: target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bai Y, Li J, Li J, Liu Y and Zhang B:
miR-615 inhibited cell proliferation and cell cycle of human breast
cancer cells by suppressing of AKT2 expression. Int J Clin Exp Med.
8:3801–3808. 2015.PubMed/NCBI
|
|
17
|
Shen F, Cai WS, Feng Z, Li JL, Chen JW,
Cao J and Xu B: miR-492 contributes to cell proliferation and cell
cycle of human breast cancer cells by suppressing SOX7 expression.
Tumour Biol. 36:1913–1921. 2015. View Article : Google Scholar
|
|
18
|
Cai K, Shen F, Cui JH, Yu Y and Pan HQ:
Expression of miR-221 in colon cancer correlates with prognosis.
Int J Clin Exp Med. 8:2794–2798. 2015.PubMed/NCBI
|
|
19
|
Cheng CJ, Bahal R, Babar IA, Pincus Z,
Barrera F, Liu C, Svoronos A, Braddock DT, Glazer PM, Engelman DM,
et al: MicroRNA silencing for cancer therapy targeted to the tumour
microenvironment. Nature. 518:107–110. 2015. View Article : Google Scholar
|
|
20
|
Yanaihara N, Caplen N, Bowman E, Seike M,
Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et
al: Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jiang J, Gusev Y, Aderca I, Mettler TA,
Nagorney DM, Brackett DJ, Roberts LR and Schmittgen TD: Association
of MicroRNA expression in hepatocellular carcinomas with hepatitis
infection, cirrhosis, and patient survival. Clin Cancer Res.
14:419–427. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen YJ, Wu H, Zhu JM, Li XD, Luo SW, Dong
L, Liu TT and Shen XZ: MicroRNA-18a modulates P53 expression by
targeting IRF2 in gastric cancer patients. J Gastroenterol Hepatol.
31:155–163. 2016. View Article : Google Scholar
|
|
23
|
Peng J: miR-638 a novel tumor suppressor
for triple-negative breast cancer. ProQuest/UMI. 2014.
|
|
24
|
Su J, Wang Q, Liu Y and Zhong M: miR-217
inhibits invasion of hepatocellular carcinoma cells through direct
suppression of E2F3. Mol Cell Biochem. 392:289–296. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
He C, Xiong J, Xu X, Lu W, Liu L, Xiao D
and Wang D: Functional elucidation of miR-34 in osteosarcoma cells
and primary tumor samples. Biochem Biophys Res Commun. 388:35–40.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li T, Lu YY, Zhao XD, Guo HQ, Liu CH, Li
H, Zhou L, Han YN, Wu KC, Nie YZ, et al: MicroRNA-296-5p increases
proliferation in gastric cancer through repression of
Caudal-related homeobox 1. Oncogene. 33:783–793. 2014. View Article : Google Scholar
|
|
27
|
Liu X, Zhang J, Xie B, Li H, Shen J and
Chen J: MicroRNA-200 family profile: a promising ancillary tool for
accurate cancer diagnosis. Am J Ther. 23:e388–e397. 2016.
View Article : Google Scholar :
|
|
28
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wong N and Wang X: miRDB: an online
resource for microRNA target prediction and functional annotations.
Nucleic Acids Res. 43:D146–D152. 2015. View Article : Google Scholar :
|
|
30
|
Maragkakis M, Reczko M, Simossis VA,
Alexiou P, Papadopoulos GL, Dalamagas T, Giannopoulos G, Goumas G,
Koukis E, Kourtis K, et al: DIANA-microT web server: elucidating
microRNA functions through target prediction. Nucleic Acids Res.
37:W273–276. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Leonardo TR, Schultheisz HL, Loring JF and
Laurent LC: The functions of microRNAs in pluripotency and
reprogramming. Nat Cell Biol. 14:1114–1121. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hayes J, Peruzzi PP and Lawler S:
MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol
Med. 20:460–469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Di Leva G, Garofalo M and Croce CM:
MicroRNAs in cancer. Annu Rev Pathol. 9:287–314. 2014. View Article : Google Scholar :
|
|
34
|
Roderburg C and Luedde T: Circulating
microRNAs as markers of liver inflammation, fibrosis and cancer. J
Hepatol. 61:1434–1437. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang Y, Xing Y, Liang C, Hu L, Xu F and
Chen Y: Crucial microRNAs and genes of human primary breast cancer
explored by microRNA-mRNA integrated analysis. Tumour Biol.
36:5571–5579. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jiang J, Zhang Y, Yu C, Li Z, Pan Y and
Sun C: MicroRNA-492 expression promotes the progression of hepatic
cancer by targeting PTEN. Cancer Cell Int. 14:952014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Peng L, Zhu H, Wang J, Sui H, Zhang H, Jin
C, Li L, Xu T and Miao R: miR-492 is functionally involved in
oxaliplatin resistance in colon cancer cells LS174T via its
regulating the expression of CD147. Mol Cell Biochem. 405:73–79.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Markou A, Sourvinou I, Vorkas PA, Yousef
GM and Lianidou E: Clinical evaluation of microRNA expression
profiling in non small cell lung cancer. Lung Cancer. 81:388–396.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wells CM and Jones GE: The emerging
importance of group II PAKs. Biochem J. 425:465–473. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
He S, Feng M, Liu M, Yang S, Yan S, Zhang
W, Wang Z, Hu C, Xu Q, Chen L, et al: P21-activated kinase 7
mediates cisplatin-resistance of esophageal squamous carcinoma
cells with Aurora-A overexpression. PLoS One. 9:e1139892014.
View Article : Google Scholar : PubMed/NCBI
|