Introduction
Osteoarthritis (OA) is a degenerative joint disease
characterized by the destruction of articular cartilage,
intraarticular inflammation and pathological alterations in
peri-articular and subchondral bone (1,2).
Various factors are involved in the pathogenesis of OA, including
age (3), a history of diabetes,
cancer or cardiovascular diseases (4), mechanical influences (5) and genetic factors (6). There is no disease-modifying
treatment for the onset or progression of OA and associated
structural damage, and the current treatments aim at relieving the
symptoms (7). Therefore, the
identification of novel molecules involved in the pathogenesis of
OA is urgently required, and will provide basis for the development
of therapies for OA.
MicroRNAs (miRNAs or miRs) are a category of
non-coding RNAs 22–25 nt in length (8). As the key gene regulators, miRNAs
directly bind to their target messenger RNAs (mRNAs) in a
sequence-specific manner to facilitate degradation of the
transcripts and to inhibit the protein translation (8). Differential expression profiles of
certain miRNAs in cancers at different stages suggests that miRNAs
are novel biomarkers for disease diagnostics (9). The application of microarray
technology enables the detection of the expression levels of
thousands of miRNAs simultaneously within tens of samples processed
in a single experiment (10). The
dysregulation of miRNAs has been found in tissue samples derived
from patients with OA in a number of previous studies, including
let-7 family miRNAs (11),
miR-149 (12), miR-21 (13) and miR-24 (14). Most of the earlier studies
compared miRNA expression in the injured cartilage and synovium
between patients with OA and normal controls (15–17); however, changes in cruciate
ligment have been less studied. The cruciate ligament is a
collagenous tissue for structural support and provides
proprioception to the body by mediating knee kinesthesia (18). Of note, the degradation of the
cruciate ligaments frequently occurs in osteoarthritic knees
(18,19). The present study was therefore
conducted to analyze the miRNA expression profiles in anterior
cruciate ligament (ACL) tissues surgically removed from patients
with OA and control subjects by using miRNA microarray analysis. In
addition, the biological functions and pathways affected by the
differentially expressed miRNAs were analyzed.
Materials and methods
Sample recruitment and RNA
extraction
Osteoarthritic ACL samples were surgically removed
from 3 patients (64.67±3.06 years of age, Kellgren-Lawrence grade
III–IV) during knee replacement surgery at Shengjing Hospital of
China Medical University, Shenyang, China. Samples derived from 3
patients without OA who encountered ACL rupture were used as
controls. The present research protocol was approved by the
Institutional Review Board of China Medical University, and written
informed consent was obtained from each participant prior to
obtaining the samples. Total RNA was extracted from the ACL tissue
samples using the total RNA purification kit (Norgen Biotek Corp.,
Thorold, ON, Canada), quantified on a NanoDrop ND-2100
spectrophotometer (Thermo Fisher Scientific, Inc., Pittsburgh, PA,
USA), and assessed on an Agilent 2100 Bioanalyzer (Agilent
Technologies, Inc., Santa Clara, CA, USA).
miRNA microarray procedures
The RNA samples were tailed with Poly(A) and labeled
with biotin using FlashTag™ biotin HSR ligation mix (Affymetrix,
Inc., Santa Clara, CA, USA) according to the manufacturer's
instructions. The labeled RNA samples were hybridized onto the
Affymetrix miRNA 4.0 arrays on a hybridization oven 645, washed and
stained on fluidics station 450, and then scanned with a Scanner
3000 (all from Affymetrix, Inc.).
Data analysis
Array images were analyzed with GeneChip Command
Console software (version 4.0; Affymetrix, Inc.) to generate raw
data. The obtained raw data were first normalized with robust
multi-array average (RMA) using Expression Console software
(version 1.3.1; Affymetrix, Inc.) and then analyzed with GeneSpring
software (version 12.5; Agilent Technologies, Inc.). Principal
component analysis (PCA) is a mathematical algorithm that is
performed to reduce the data dimensionality while retaining most of
the variation in the data set (20). Differentially expressed miRNAs
were identified by evaluating the fold change (FC). miRNAs with an
FC ≥2 and a P-value <0.05 (t-test) were considered as
differentially expressed. Hierarchical clustering was performed to
analyze the distinguishable miRNA expression patterns among the
samples. Genes targeted by the identified differentially expressed
miRNAs were shown as the intersection of Targetscan, PITA and
microRNA.org databases (GeneSpring software, version
12.5). These putative target genes were subjected to Gene Ontology
(GO) biological process annotation and Kyoto Encyclopedia of Genes
and Genomes (KEGG) pathway analysis using FunNet algorithm. The
P-value was calculated with a unilateral Fisher's exact test and
corrected by false discovery rate (FDR). GOs and pathways with
P-value <0.05 and FDR <0.05 were considered as significant.
To display the combinatorial interactions between miRNA pairs and
their shared targets, genes with P-value <0.05 (calculated by
hypergeometric distribution) were presented using Cytoscape
software.
Quantitative PCR
Quantitative PCR was performed on cDNA synthesized
from the same RNA samples used in the prior microarray analysis.
Primers used in this study were listed in Table I. The expression levels of 6
upregulated miRNAs (hsa-let-7f-5p, hsa-let-7g-5p, hsa-miR-146a-5p,
hsa-miR-146b-3p, hsa-miR-26b-5p and hsa-miR-335-5p) and 6
downregulated miRNAs (hsa-miR-18a-3p, hsa-miR-485-3p, hsa-miR-665,
hsa-miR-675-5p, hsa-miR-1207-5p and hsa-miR-138-5p) were determined
on the Exicycler™ 96 (Bioneer, Daejeon, Korea) using SYBR-Green
(Solarbio, Beijing, China). Triplicate reactions were performed.
The data were analyzed by the comparative threshold cycle (Ct)
method. U6 was used as the endogenous control.
 | Table IPrimers used in this work. |
Table I
Primers used in this work.
| MiRBase accession
number | Name | Sequence
information (5′→3′) |
|---|
| MIMAT0000067 | hsa-let-7f-5p | |
| RT primer |
GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACAACTAT |
| Forward real-time
PCR primer |
CGCGGCTGAGGTAGTAGATTGT |
| MIMAT0000414 | hsa-let-7g-5p | |
| RT primer |
GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACAACTGT |
| Forward real-time
PCR primer |
CGGTCGTGAGGTAGTAGTTTGT |
| MIMAT0000449 |
hsa-miR-146a-5p | |
| RT primer |
GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACAACCCA |
| Forward real-time
PCR primer |
GCGAGGTGAGAACTGAATTCCA |
| MIMAT0004766 |
hsa-miR-146b-3p | |
| RT primer |
GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACCCAGAA |
| Forward real-time
PCR primer |
GACTGCCCTGTGGACTCAGTTC |
| MIMAT0000083 | hsa-miR-26b-5p | |
| RT primer |
GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACACCTAT |
| Forward real-time
PCR primer |
CGCGGCTTCAAGTAATTCAGG |
| MIMAT0000765 | hsa-miR-335-5p | |
| RT primer |
GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACACATTT |
| Forward real-time
PCR primer |
CGCAGCTCAAGAGCAATAACGA |
| MIMAT0002891 | hsa-miR-18a-3p | |
| RT primer |
GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACCCAGAA |
| Forward real-time
PCR primer |
CGACTACTGCCCTAAGTGCTC |
| MIMAT0002176 | hsa-mir-485-3p | |
| RT primer |
GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACAGAGAG |
| Forward real-time
PCR primer |
CTGCTGTCATACACGGCTCTC |
| MIMAT0004952 | hsa-miR-665 | |
| RT primer |
GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACAGGGGC |
| Forward real-time
PCR primer |
CAGTTAACCAGGAGGCTGAGG |
| MIMAT0004284 | hsa-miR-675-5p | |
| RT primer |
GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACCACTGT |
| Forward real-time
PCR primer |
CTATAATGGTGCGGAGAGGGCC |
| MIMAT0005871 |
hsa-miR-1207-5p | |
| RT primer |
GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACCCCCTC |
| Forward real-time
PCR primer |
CTTATTGGCAGGGAGGCTG |
| MIMAT0000430 | hsa-miR-138-5p | |
| RT primer |
GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACCGGCCT |
| Forward real-time
PCR primer |
CGGTGCAGCTGGTGTTGTGAAT |
| Universal reverse
primer |
GTGCAGGGTCCGAGGTATTC |
Results
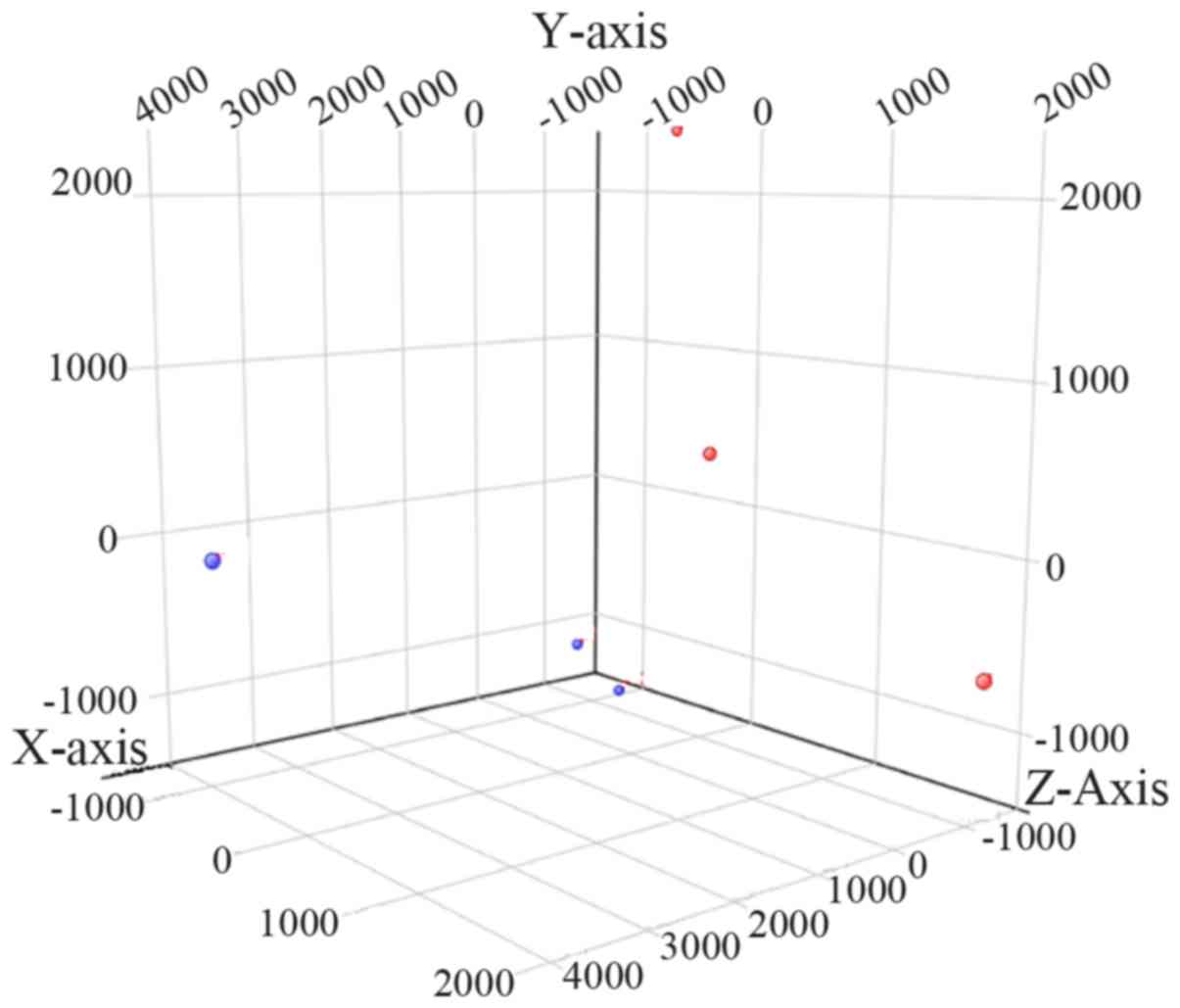
PCA distinguishes patients with OA from
control subjects
PCA was performed in the 6 knee ACL tissues based on
the microarray data, and the corresponding results revealed that
the ACL samples from the patients with OA could be distinguished
from those of the control subjects (Fig. 1). The above results suggested that
the specimens used in this study were properly prepared and could
be classified into two distinct groups.
Identification of differentially
expressed miRNAs in knee ACL tissues
Our data indicated that 22 miRNAs were upregulated
and 17 miRNAs were downregulated in the osteoarthritic ACL tissues
(FC ≥2 and P-value <0.05). Twelve miRNAs were selected for
further validation regarding their expression levels (such as
hsa-miR-138-5p, hsa-miR-26b-5p, hsa-miR-665), their previous
correlations with OA (such as hsa-miR-146a-5p, hsa-let-7g-5p,
hsa-let-7f-5p), or data from the following analysis (such as
hsa-miR-146b-3p, hsa-miR-1207-5p). Log2 results of the miRNA
expression levels from the microarray analysis and the quantitative
PCR analysis revealed an excellent correlation
(R2=0.889; Fig. 2A).
All analyzed human miRNAs were presented in a volcano plot
(Fig. 2B), and the dysregulated
ones were assessed via hierarchical clustering analysis (Fig. 2C). Results from hierarchical
clustering analysis revealed that the ACL tissue samples were
divided into two distinct clusters based on their pathological
statuses. Collectively, these results implied that the microarray
data reflected the reliable miRNA expression patterns in ACL
tissues and that the samples from same situation clustered
together.
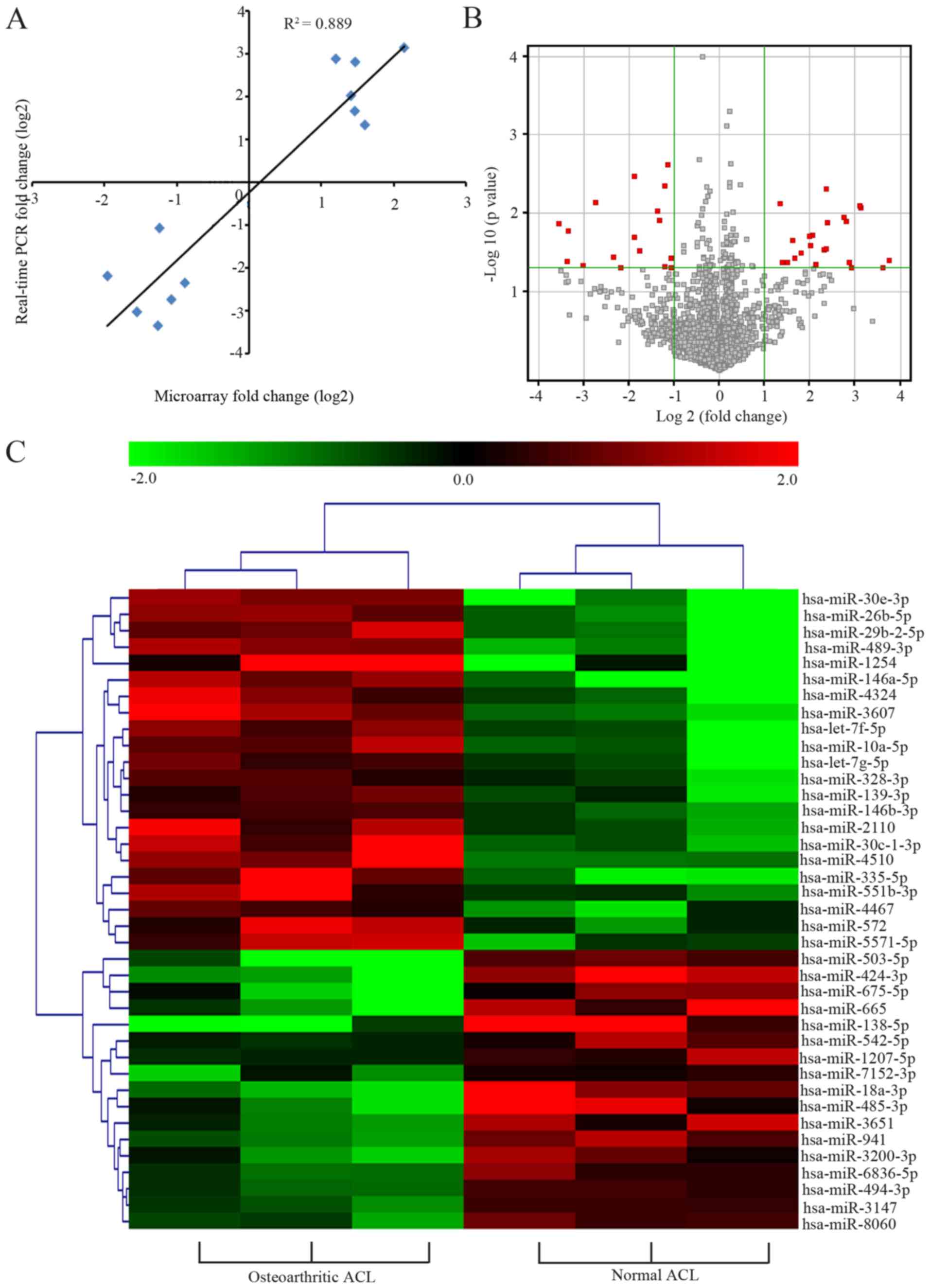 | Figure 2Differentially expressed miRNAs in
knee ACL tissues of patients with OA. (A) Log2 FC for microarray
data (y-axis) is plotted against log2 FC for quantitative PCR data
(x-axis) for each gene. (B) Volcano plots of miRNA microarray data
(x-axis, log2 FC; y-axis, negative log10 P-value). The red plots
depict differentially expressed miRNAs with an FC ≥2 and P<0.05
between the two groups. (C) Hierarchical cluster analysis of the
miRNA microarray data. Each column depicts a single tissue sample
and each row represented a miRNA. Red and green indicate high and
low expression levels, respectively, whereas black indicates the
mean expression levels. Distance metric, pearson centered; linkage
rule, average. miRNA, microRNA; ACL, anterior cruciate ligament;
FC, fold change; OA, osteoarthritis. |
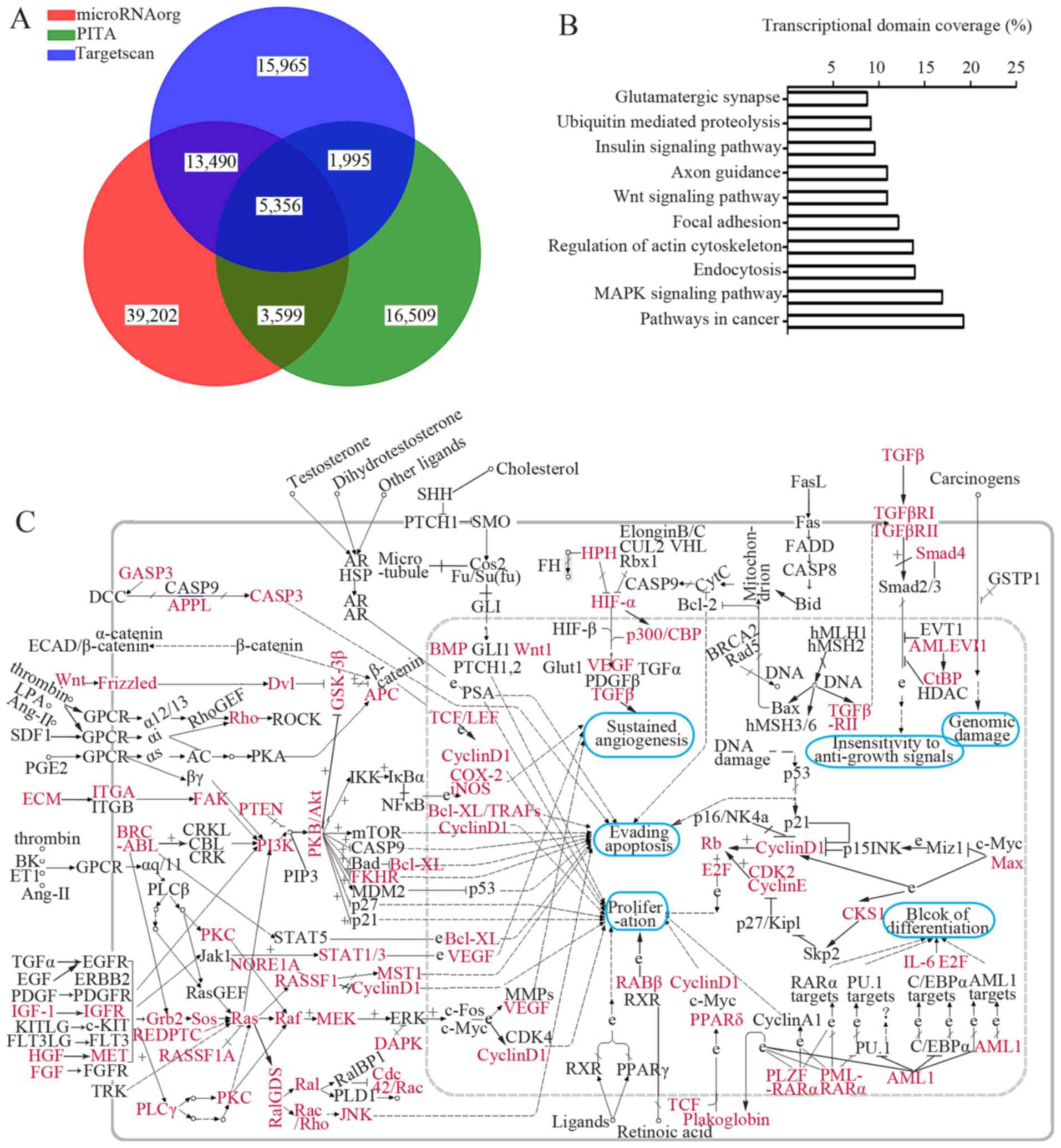
Microarray-based GO and KEGG pathway
annotations
Three data bases predicted a total of 5,356 genes as
putative targets for the differentially expressed miRNAs (Fig. 3A). Genes involved in cartilage
remodeling (21), collagen
biosynthesis (22), extracellular
matrix (ECM) homeostasis (23)
and inflammation (24) are
summarized in Table II.
Moreover, the GO annotation (at the biological process level) and
KEGG pathway analysis of all putative genes revealed that these
genes were enriched in 41 GO items and 23 KEGG pathways (data not
shown). As indicated in Table
III, several essential biological processes, including
DNA-dependent regulation of transcription, signal transduction,
multicellular organismal development, were affected by the
differentially expressed miRNAs. Additionally, a large set of genes
implicated in mitogen-activated protein kinase (MAPK), vascular
endothelial growth factor (VEGF), protein kinase C
(PKC)-mitogen-activated protein kinase (MEK), phosphatidylinositol
3-kinase (PI3K)-protein kinase B (AKT), as well as the WNT
signaling pathway were mediated by the dysregulated miRNAs
(Fig. 3B and C; pathway ID, 05200
for Fig. 3C). The above results
provided useful information for us to understand how cruciate
ligament injuries develop in OA patients.
 | Table IIPrediction of target genes
potentially related to osteoarthritis. |
Table II
Prediction of target genes
potentially related to osteoarthritis.
| miRNAs | Gene symbols | Function |
|---|
|
hsa-let-7f/7g-5p | BMP2, COL1A1,
COL1A2, COL3A1, COL4A1, COL4A6, COL5A2, COL14A1, COL15A1, COL24A1,
TGFBR1, ADAMTS8, IL6, IL10, IL13, HMGA1, HMGA2 | Cartilage
development and remodeling |
| hsa-miR-10a-5p | COL4A4, COL24A1,
ADAMTS4 | |
| hsa-miR-26b-5p | CILP, COL1A2,
COL9A1, COL10A1, COL11A1, ADAMTS19, HMGA1, HMGA2, IL6, IL1RAP | Collagen
biosynthesis and degradation |
| hsa-miR-138-5p | MMP16, ADAMTSL3,
IL6R, IL1RAP | |
|
hsa-miR-146a-5p | CCL5, CXCR7,
ADAMTS3, ADAMTS18 | |
|
hsa-miR-146b-3p | COL11A1, MMP24,
VEGFA | |
| hsa-miR-335-5p | COL5A1, COL6A3,
COL19A1, ADAMTS19, CCL5 | ECM
homeostasis |
| hsa-miR-542-5p | ADAMTS8 | |
| hsa-miR-485-3p | COL12A1, TGFB3,
MMP20, ADAMTS3 | Inflammatory
response |
| hsa-miR-572 | COL7A1 | |
| hsa-miR-665 | COL8A2, TGFBR1,
TGFBR2, ADAMTS8, CXCL11, CXCL12 | |
|
hsa-miR-1207-5p | COL9A2, TGFBR1,
ADAMTS10, ADAMTS19 | |
| hsa-miR-1254 | RUNX2, TGFBR3,
ADAMTSL5 | |
 | Table IIIIdentified biological process GO
terms for the differentially expressed miRNAs (top 10). |
Table III
Identified biological process GO
terms for the differentially expressed miRNAs (top 10).
| GO ID | GO term | List hits | P-value |
|---|
| GO:0006355 | Regulation of
transcription, DNA-dependent | 491 | 5.38E-09 |
| GO:0007165 | Signal
transduction | 289 | 8.11E-05 |
| GO:0007275 | Multicellular
organismal development | 237 | 5.38E-05 |
| GO:0006351 | Transcription,
DNA-dependent | 166 | 1.24E-08 |
| GO:0006468 | Protein
phosphorylation | 153 | 1.25E-08 |
| GO:0007155 | Cell adhesion | 151 | 3.54E-05 |
| GO:0045944 | Positive regulation
of transcription from RNA polymerase II promoter | 138 | 4.43E-06 |
| GO:0007399 | Nervous system
development | 131 | 8.66E-10 |
| GO:0007049 | Cell cycle | 121 | 5.31E-06 |
| GO:0007411 | Axon guidance | 114 | 4.42E-12 |
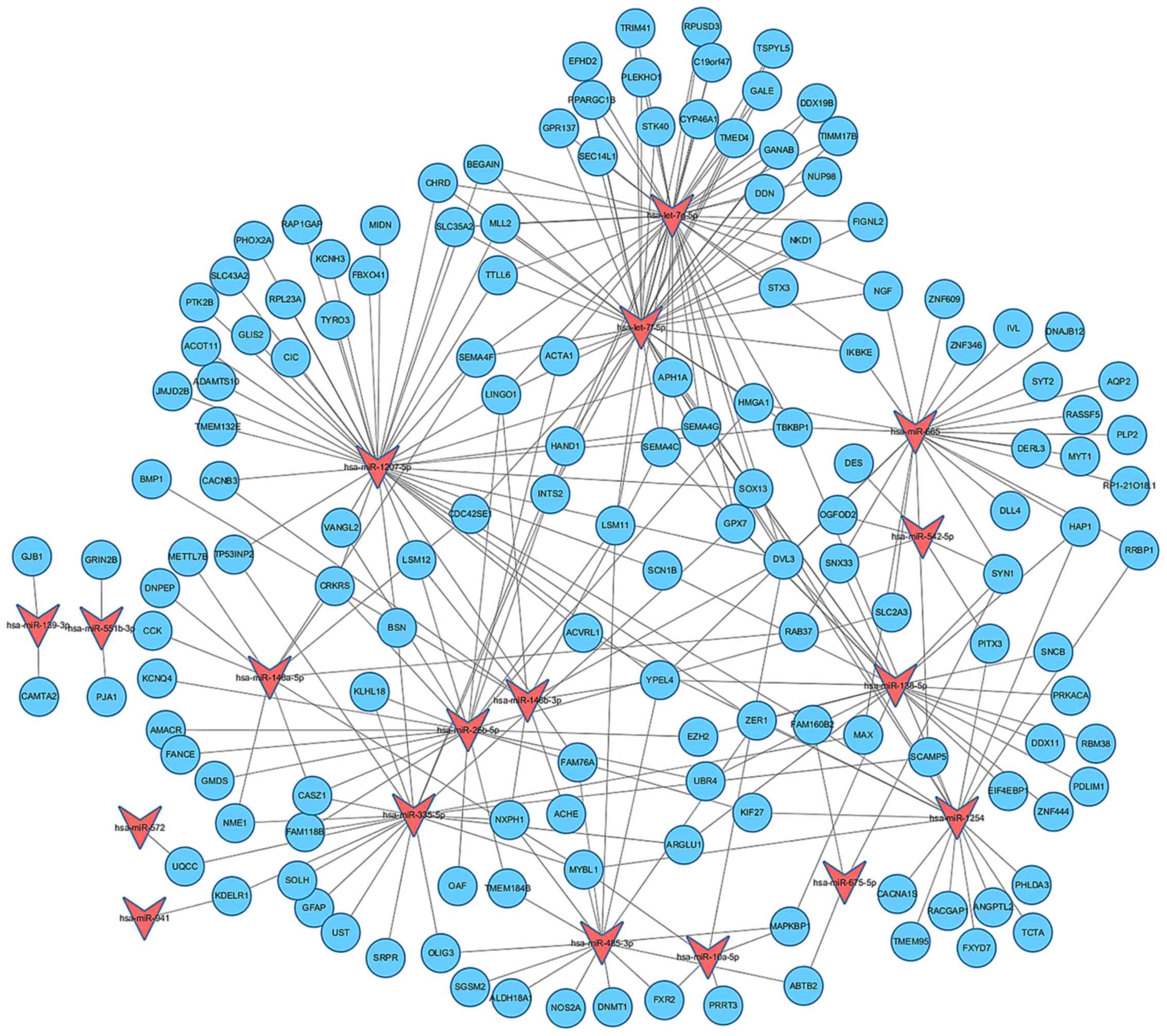
Establishment of miRNA-gene regulatory
network
A large set of genes were predicated as targets for
the differentially expressed miRNAs through the Targetscan, PITA
and microRNA.org data bases. In order to visualize
and integrate the interactions between the dysregulated miRNAs and
their targets, a miRNA-gene regulatory network was established by
using Cytoscape software (Fig.
4). Our results revealed that the differentially expressed
miRNAs may function in combination to exert effects on their target
genes.
Discussion
miRNAs play crucial roles in mediating
chondrogenesis, and are considered to link to the pathogenesis of
cartilage-related diseases, including OA (25). In this study, miRNA microarray was
performed to compare the miRNA expression levels in knee ACL
tissues from patients with OA to those of the controls. Appropriate
grouping of the 6 ACL samples was confirmed by PCA and heatmap
data. We found that 22 miRNAs were upregulated and 17 were
downregulated in the osteoarthritic ACL tissues. Additional
bioinformatics was performed to analyze the biological processes
and pathways that were affected by the identified differentially
expressed miRNAs. The obtained data enhanced our understanding of
the roles of the dysregulated miRNAs in OA pathogenesis.
Reportedly, let-7 miRNAs can regulate skeletal
development by orchestrating the proliferation and differentiation
of chondrocytes (11). The
enforced overexpression of Lin28a, a let-7 inhibitor, has been
shown to accelerate cartilage regrowth in a model of tissue injury
(26). It is likely that the
abnormal upregulation of let-7 miRNAs contributes to the
degeneration of articular cartilage. In this study, to the best of
our knowledge, we demonstrate for the first time that the
expression of let-7f-5p and let-7g-5p was increased by 2.04- and
1.68-fold (log2FC) in the osteoarthritic ACL tissues,
respectively. Though all let-7 family members share the identical
seed region (GAGGUAG) (27), only
these two let-7 members were identified to be dysregulated in
OA-affected ligaments. Bone morphogenetic protein (BMP)2 has been
reported to promote osteogenesis (28). Apart from its role in bone
formation, the pre-injection of recombinant human BMP2 in the
semitendinosus tendon enables successful ACL reconstruction
following injury (29),
suggesting a beneficial role of BMP2 in ligament injury. miR-140-5p
is a potent regulator of BMP2 (30), and its expression is markedly
reduced in osteoarthritic articular cartilage tissues (31), but not in ACL tissues, as
evidenced by our microarray data. Of note, we found that BMP2 is a
possible target for let-7f/7g-5p (Table II), although the interaction
between them has not been entirely clarified. Moreover, apart from
BMP2, other factors related to collagen biosynthesis and
degradation and inflammatory response, such as transforming growth
factor β receptor 1 (TGFβR1), various types of collagens (COL1A1
and COL1A2) and interleukins (IL)-6 were also putative targets for
let-7f/7g-5p. To address the roles of let-7f/7g in osteoarthritic
ligament lesion, their targets should also be taken into
consideration.
Formation and degradation of collagens and ECM
proteins are mediated by miR-26 family members (32). miR-26b is suggested to contribute
to rheumatoid arthritis regarding to its elevation in IL-17
producing T cells (33). Such
findings indicate that miR-26b may participate in inflammatory
diseases in the joints. A significant upregulation of miR-26b-5p
(previously miR-26b) was found in osteoarthritic ACL tissues in the
present study. Although several putative targets of miRNA-26b, such
as high mobility group AT-hook 1 (HMGA1 and HMGA2), cartilage
intermediate layer protein (CILP), as well as a variety of
collagens (COLs) are implicated in the development and progression
of OA (34–36), the direct correlation of miRNA-26b
dysregulation with OA has not been fully elucidated, and requires
for further exploration.
miR-146a controls knee joint homeostasis by
balancing inflammatory responses in cartilage (37). Its expression is increased in
articular cartilage and/or synovium derived from patients with OA
(38,39). Our results were consistent with
these earlier findings by showing a significant upregulation of
miR-146a-5p (previously miR-146a) in osteoarthritic knee ACL
tissues. Studies on the correlation between OA pathogenesis and
miR-146b-3p are limited. Our data indicated that miR-146b-3p was
also overexpressed in osteoarthritic ACL tissues. Several genes
associated to ECM homeostasis and inflammation such as matrix
metalloproteinase (MMP)24 and VEGFA in OA (40,41) were predicted as targets for
miR-146b-3p.
A disintegrin and metalloproteinase domain with
thrombospondin type-1 motifs (ADAMTS) are a new family of
metalloproteases that play important roles in physiological and
pathological conditions (42,43). Previous studies have demonstrated
that ADAMTS7 overexpression leading to the increased expression of
tumor necrosis factor (TNF)-α and MMPs contributes OA development
(44), while the knockdown or
knockout of ADAMTS4 and/or ADAMTS5 prevents OA progression
(45,46). These studies suggest that ADAMTS
may be the potential molecular targets for the prevention and
treatment of OA. In this study, we found that ADAMTS3, 4, 8, 10,
18, 19, ADAMTS-like-3, -5 were the putative targets for several
differentially expressed miRNAs, including let-7f/7g-5p,
miR-146a-5p, miR-1207-5p (Table
II). Investigations of the interaction between these
dysregulated miRNAs and their target ADAMTS will help to understand
the mechanisms through which OA develops and progresses.
Several essential biological processes, such as the
DNA-depen dent regulation of transcription, signal transduction and
multicellular organismal development, are affected by miRNAs with
differential expression levels in OA as indicated in GO annotation.
To provide an overall understanding of the association between the
dysregulated miRNAs and OA pathogenesis, KEGG pathway analysis was
further performed. We found that several pathways enriched by the
putative target genes were essential for OA pathogenesis. For
instance, a study from Prasadam et al demonstrated that p38
MAPK phosphorylation was decreased in OA-affected chondrocytes as
compared to normal chondrocytes, and that the inactivation of p38
signaling leads to OA-like changes in rats (47). In addition, activation of VEGF
signaling has been suggested to contribute to synovial inflammation
during the progression of OA (48).
In conclusion, our study revealed that 39 miRNAs
were differentially expressed in knee ACL tissues from patients
with OA. The functional bioinformatic analyses suggest that the
dysregulated miRNAs may regulate cartilage development and
remodeling, collagen biosynthesis and degradation, ECM homeostasis
and pathology by interacting with their targets. Collectively, our
study provides novel insight into the ligament injury-related miRNA
dysregulation in patients with OA.
Acknowledgments
The present study was supported by grants from the
Natural Science Foundation of Liaoning Province (no. 2014021011)
and the National Natural Science Foundation of China (no.
81171716).
References
|
1
|
Goldring MB and Goldring SR:
Osteoarthritis. J Cell Physiol. 213:626–634. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Johnson K, Zhu S, Tremblay MS, Payette JN,
Wang J, Bouchez LC, Meeusen S, Althage A, Cho CY, Wu X, et al: A
stem cell-based approach to cartilage repair. Science. 336:717–721.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Buckwalter JA and Martin JA:
Osteoarthritis. Adv Drug Deliv Rev. 58:150–167. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nüesch E, Dieppe P, Reichenbach S,
Williams S, Iff S and Jüni P: All cause and disease specific
mortality in patients with knee or hip osteoarthritis: population
based cohort study. BMJ. 342:d11652011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Riordan EA, Little C and Hunter D:
Pathogenesis of post-traumatic OA with a view to intervention. Best
Pract Res Clin Rheumatol. 28:17–30. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chapman K and Valdes AM: Genetic factors
in OA pathogenesis. Bone. 51:258–264. 2012. View Article : Google Scholar
|
|
7
|
Matthews GL and Hunter DJ: Emerging drugs
for osteoarthritis. Expert Opin Emerg Drugs. 16:479–491. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang B and Farwell MA: MicroRNAs: a new
emerging class of players for disease diagnostics and gene therapy.
J Cell Mol Med. 12:3–21. 2008. View Article : Google Scholar
|
|
9
|
Ryan BM, Robles AI and Harris CC: Genetic
variation in microRNA networks: the implications for cancer
research. Nat Rev Cancer. 10:389–402. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Love C and Dave S: MicroRNA expression
profiling using microarrays. Methods Mol Biol. 999:285–296. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Papaioannou G, Inloes JB, Nakamura Y,
Paltrinieri E and Kobayashi T: let-7 and miR-140 microRNAs
coordinately regulate skeletal development. Proc Natl Acad Sci USA.
110:E3291–E3300. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Santini P, Politi L, Vedova PD, Scandurra
R and Scotto d'Abusco A: The inflammatory circuitry of miR-149 as a
pathological mechanism in osteoarthritis. Rheumatol Int.
34:711–716. 2014. View Article : Google Scholar
|
|
13
|
Zhang Y, Jia J, Yang S, Liu X, Ye S and
Tian H: MicroRNA-21 controls the development of osteoarthritis by
targeting GDF-5 in chondrocytes. Exp Mol Med. 46:e792014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Philipot D, Guérit D, Platano D, Chuchana
P, Olivotto E, Espinoza F, Dorandeu A, Pers YM, Piette J, Borzi RM,
et al: p16INK4a and its regulator miR-24 link senescence
and chondrocyte terminal differentiation-associated matrix
remodeling in osteoarthritis. Arthritis Res Ther. 16:R582014.
View Article : Google Scholar
|
|
15
|
Iliopoulos D, Malizos KN, Oikonomou P and
Tsezou A: Integrative microRNA and proteomic approaches identify
novel osteoarthritis genes and their collaborative metabolic and
inflammatory networks. PLoS One. 3:e37402008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mirzamohammadi F, Papaioannou G and
Kobayashi T: MicroRNAs in cartilage development, homeostasis, and
disease. Curr Osteoporos Rep. 12:410–419. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Qi Y, Ma N, Yan F, Yu Z, Wu G, Qiao Y, Han
D, Xiang Y, Li F, Wang W, et al: The expression of intronic miRNAs,
miR-483 and miR-483*, and their host gene, Igf2, in
murine osteoarthritis cartilage. Int J Biol Macromol. 61:43–49.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rajgopal A, Vasdev N, Pathak A, Gautam D
and Vasdev A: Histological changes and neural elements in the
posterior cruciate ligament in osteoarthritic knees. J Orthop Surg
(Hong Kong). 22:142–145. 2014. View Article : Google Scholar
|
|
19
|
Svoboda SJ: ACL injury and posttraumatic
osteoarthritis. Clin Sports Med. 33:633–640. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ringnér M: What is principal component
analysis? Nat Biotechnol. 26:303–304. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li Y and Xu L: Advances in understanding
cartilage remodeling. F1000Res 4 (F1000 Faculty Rev). 642:2015.
|
|
22
|
Henrotin Y, Addison S, Kraus V and Deberg
M: Type II collagen markers in osteoarthritis: what do they
indicate? Curr Opin Rheumatol. 19:444–450. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fuhrmann IK, Steinhagen J, Rüther W and
Schumacher U: Comparative immunohistochemical evaluation of the
zonal distribution of extracellular matrix and inflammation markers
in human meniscus in osteoarthritis and rheumatoid arthritis. Acta
Histochem. 117:243–254. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Berenbaum F, Eymard F and Houard X:
Osteoarthritis, inflammation and obesity. Curr Opin Rheumatol.
25:114–118. 2013. View Article : Google Scholar
|
|
25
|
Shang J, Liu H and Zhou Y: Roles of
microRNAs in prenatal chondrogenesis, postnatal chondrogenesis and
cartilage-related diseases. J Cell Mol Med. 17:1515–1524. 2013.
View Article : Google Scholar
|
|
26
|
Shyh-Chang N, Zhu H, Yvanka de Soysa T,
Shinoda G, Seligson MT, Tsanov KM, Nguyen L, Asara JM, Cantley LC
and Daley GQ: Lin28 enhances tissue repair by reprogramming
cellular metabolism. Cell. 155:778–792. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang X, Rutnam ZJ, Jiao C, Wei D, Xie Y,
Du J, Zhong L and Yang BB: An anti-let-7 sponge decoys and decays
endogenous let-7 functions. Cell Cycle. 11:3097–3108. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gugala Z, Davis AR, Fouletier-Dilling CM,
Gannon FH, Lindsey RW and Olmsted-Davis EA: Adenovirus BMP2-induced
osteogenesis in combination with collagen carriers. Biomaterials.
28:4469–4479. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hashimoto Y, Yoshida G, Toyoda H and
Takaoka K: Generation of tendon-to-bone interface 'enthesis' with
use of recombinant BMP-2 in a rabbit model. J Orthop Res.
25:1415–1424. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hwang S, Park SK, Lee HY, Kim SW, Lee JS,
Choi EK, You D, Kim CS and Suh N: miR-140-5p suppresses
BMP2-mediated osteogenesis in undifferentiated human mesenchymal
stem cells. FEBS Lett. 588:2957–2963. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Miyaki S, Sato T, Inoue A, Otsuki S, Ito
Y, Yokoyama S, Kato Y, Takemoto F, Nakasa T, Yamashita S, et al:
MicroRNA-140 plays dual roles in both cartilage development and
homeostasis. Genes Dev. 24:1173–1185. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li Z, Hassan MQ, Jafferji M, Aqeilan RI,
Garzon R, Croce CM, van Wijnen AJ, Stein JL, Stein GS and Lian JB:
Biological functions of miR-29b contribute to positive regulation
of osteoblast differentiation. J Biol Chem. 284:15676–15684. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Niimoto T, Nakasa T, Ishikawa M, Okuhara
A, Izumi B, Deie M, Suzuki O, Adachi N and Ochi M: MicroRNA-146a
expresses in interleukin-17 producing T cells in rheumatoid
arthritis patients. BMC Musculoskelet Disord. 11:2092010.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Valdes AM, Van Oene M, Hart DJ, Surdulescu
GL, Loughlin J, Doherty M and Spector TD: Reproducible genetic
associations between candidate genes and clinical knee
osteoarthritis in men and women. Arthritis Rheum. 54:533–539. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Amin AR and Islam AB: Genomic analysis and
differential expression of HMG and S100A family in human arthritis:
upregulated expression of chemokines, IL-8 and nitric oxide by
HMGB1. DNA Cell Biol. 33:550–565. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gasparini G, De Gori M, Paonessa F,
Chiefari E, Brunetti A and Galasso O: Functional relationship
between high mobility group A1 (HMGA1) protein and insulin-like
growth factor-binding protein 3 (IGFBP-3) in human chondrocytes.
Arthritis Res Ther. 14:R2072012. View
Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li X, Gibson G, Kim JS, Kroin J, Xu S, van
Wijnen AJ and Im HJ: MicroRNA-146a is linked to pain-related
pathophysiology of osteoarthritis. Gene. 480:34–41. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jin L, Zhao J, Jing W, Yan S, Wang X, Xiao
C and Ma B: Role of miR-146a in human chondrocyte apoptosis in
response to mechanical pressure injury in vitro. Int J Mol Med.
34:451–463. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yamasaki K, Nakasa T, Miyaki S, Ishikawa
M, Deie M, Adachi N, Yasunaga Y, Asahara H and Ochi M: Expression
of microRNA-146a in osteoarthritis cartilage. Arthritis Rheum.
60:1035–1041. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Leijten JC, Bos SD, Landman EB, Georgi N,
Jahr H, Meulenbelt I, Post JN, van Blitterswijk CA and Karperien M:
GREM1, FRZB and DKK1 mRNA levels correlate with osteoarthritis and
are regulated by osteoarthritis-associated factors. Arthritis Res
Ther. 15:R1262013. View
Article : Google Scholar : PubMed/NCBI
|
|
41
|
Borgonio Cuadra VM, González-Huerta NC,
Romero-Córdoba S, Hidalgo-Miranda A and Miranda-Duarte A: Altered
expression of circulating microRNA in plasma of patients with
primary osteoarthritis and in silico analysis of their pathways.
PLoS One. 9:e976902014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cal S, Obaya AJ, Llamazares M, Garabaya C,
Quesada V and López-Otín C: Cloning, expression analysis, and
structural characterization of seven novel human ADAMTSs, a family
of metalloproteinases with disintegrin and thrombospondin-1
domains. Gene. 283:49–62. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Durham TB, Klimkowski VJ, Rito CJ,
Marimuthu J, Toth JL, Liu C, Durbin JD, Stout SL, Adams L,
Swearingen C, et al: Identification of potent and selective
hydantoin inhibitors of aggrecanase-1 and aggrecanase-2 that are
efficacious in both chemical and surgical models of osteoarthritis.
J Med Chem. 57:10476–10485. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lai Y, Bai X, Zhao Y, Tian Q, Liu B, Lin
EA, Chen Y, Lee B, Appleton CT, Beier F, et al: ADAMTS-7 forms a
positive feedback loop with TNF-α in the pathogenesis of
osteoarthritis. Ann Rheum Dis. 73:1575–1584. 2014. View Article : Google Scholar
|
|
45
|
Majumdar MK, Askew R, Schelling S, Stedman
N, Blanchet T, Hopkins B, Morris EA and Glasson SS: Double-knockout
of ADAMTS-4 and ADAMTS-5 in mice results in physiologically normal
animals and prevents the progression of osteoarthritis. Arthritis
Rheum. 56:3670–3674. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chu X, You H, Yuan X, Zhao W, Li W and Guo
X: Protective effect of lentivirus-mediated siRNA targeting
ADAMTS-5 on cartilage degradation in a rat model of osteoarthritis.
Int J Mol Med. 31:1222–1228. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Prasadam I, Mao X, Wang Y, Shi W, Crawford
R and Xiao Y: Inhibition of p38 pathway leads to OA-like changes in
a rat animal model. Rheumatology (Oxford). 51:813–823. 2012.
View Article : Google Scholar
|
|
48
|
Almasry SM, Soliman HM, El-Tarhouny SA,
Algaidi SA and Ragab EM: Platelet rich plasma enhances the
immuno-histochemical expression of platelet derived growth factor
and vascular endothelial growth factor in the synovium of the
meniscectomized rat models of osteoarthritis. Ann Anat. 197:38–49.
2015. View Article : Google Scholar
|