Introduction
Periodontitis and malocclusion along with caries are
significant public oral diseases with extremely high incidence
rates. Epidemiologic studies show that more than 50% of adults
suffer from periodontitis and severe periodontitis is estimated to
occur in approximately 5–20% of adults worldwide (1–3).
The prevalence of malocclusion among children is also more than
50%. These two oral diseases are major public health issues that
require attention. Periodontal ligament (PDL) is a complex tissue
with abundant blood vessels and cells, including periodontal
ligament cells (PDLCs), periodontal ligament stem cells,
fibroblasts, osteoblasts and osteoclasts, which play a vital role
in periodontal tissue regeneration (4). Periodontitis and orthodontic tooth
movement can cause hypoxia around the PDL, which triggers a series
of molecular responses in PDLCs for hypoxic adaptation (4). The most sensitive and important
molecule is functional hypoxia-inducible factor-1 (HIF-1), composed
of HIF-α (HIF-1α, HIF-2α and HIF-3α) and HIF-β subunits. HIF-1α is
the dominant functional subunit that is unstable and tends to be
degraded under normoxia, but it can be highly increased in
short-term hypoxia (5,6). After translocating into the nucleus
and dimerizing with HIF-β, HIF-1α binds to hypoxia response
elements (HREs) of >100 genes and regulates their production,
such as vascular endothelial growth factor (VEGF), transforming
growth factor-β, erythropoietin (EPO) and microRNAs (7,8).
Meanwhile, HIF-1α has been confirmed to regulate osteogenic
differentiation, yet there is still controversy over whether HIF-1α
promotes or inhibits osteogenic differentiation (9–12).
Since HIF-1α was discovered, its regulatory
mechanism has aroused much interest. It is known that HIF-1α
protein can be rapidly degraded under normoxia by the Von
Hippel-Lindau protein-mediated ubiquitin-proteasome pathway. The
degradation of HIF-1α protein is regulated by many factors, such as
mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3
kinase (13–16). However, the regulatory mechanism
of HIF-1α mRNA has not been well studied. Thrash-Bingham and Tartof
(17) firstly named an antisense
non-coding RNA as aHIF that originated from the 3′ region of the
hif-1α gene and could bind to the HIF-1α mRNA 3′
untranslated region (UTR), and aHIF was also named as HIF1A
antisense RNA 2 (HIF1A-AS2) or 3′αHIF-1α. Then in 2010, Baranello
et al (18) identified
another antisense non-coding RNA that originated from the 5′ region
of the hif-1α gene, named HIF1A-AS1 or 5′aHIF-1α. Both
HIF1A-AS1 and HIF1A-AS2 are longer than 200 nt and belong to the
family of long non-coding RNAs (lncRNAs). Yet, they are different
in structure and loci. HIF1A-AS1 has both 5′cap and poly (A+) tail,
while HIF1A-AS2 has neither of them. HIF1A-AS1 accumulates at the
nuclear membrane, while HIF1A-AS2 locates only in the nucleus
(19).
HIF1A-AS2 can downregulate HIF-1α mRNA so that
during long-term hypoxia HIF-1α protein is suppressed (20). Moreover, the putative HIF-1α
protein binding sites-HREs are found in the HIF1A-AS2 promoter
region by analyzing its RNA sequence, which indicates that HIF-1α
might also regulate HIF1A-AS2, but this is not yet confirmed
(21). Overall, HIF1A-AS2 and
HIF-1α have a complicated regulatory mechanism. For the first time,
we demonstrated that HIF1A-AS1 and HIF1A-AS2 exist in PDLCs. We
then explored the differences between HIF1A-AS1 and HIF1A-AS2 in
regulating HIF-1α and the osteogenic differentiation of PDLCs under
hypoxia. Runt-related transcription factor 2 (Runx2) is a key
factor initiating and regulating the early osteogenesis and late
mineralization of bone (22).
Alkaline phosphatase (ALP) activity can also describe the early
cell differentiation of osteoblastic cells (23). Therefore, Runx2 and ALP activity
were selected as the osteogenic biomarkers in the present study.
Given that the osteogenic differentiation mechanism in PDLCs is
essential to periodontal tissue regeneration, the present research
can provide a theoretical basis for promoting periodontal tissue
remodeling, regeneration and repair during orthodontic tooth
movement and periodontitis.
Materials and methods
Bioinformatic analysis
UCSC Genome Bioinformatics (http://genome.ucsc.edu/) was used to locate the
HIF-1α, HIF1A-AS1 and HIF1A-AS2 genes, as well as to obtain HIF-1α
mRNA, HIF1A-AS1 RNA and HIF1A-AS2 RNA sequences. In addition, we
used the basic local alignment search tool to obtain the
complementary regions between HIF1A-AS1 and HIF-1α mRNA, as well as
HIF1A-AS2 and HIF-1α mRNA. Furthermore, we examined the promoter
regions of HIF1A-AS1 and HIF1A-AS2 to determine whether they have
putative HRE sequences.
Sample collection and cell culture
Healthy premolars or third-molars were collected
from patients (<25 years of age) at the Hospital of Stomatology,
Sun Yat-sen University. Informed consent from each patient was
obtained, and the present study was approved by the Ethics
Committee of the Hospital of Stomatology, Sun Yat-sen University.
PDL was separated from the middle-third of the root surface, washed
with phosphate-buffered saline (PBS; Life Technologies, Grand
Island, NY, USA), digested using type I collagenases (Life
Technologies), and then cultured in primary culture medium
containing 78% Modified Eagle's Medium α (MEMα), 20% fetal bovine
serum (FBS) (both from Gibco BRL, Gaithersburg, MD, USA), and 2%
antibiotics. Cells passaged to P4 were used in the
following experiments. For the hypoxia group, cells were incubated
in a three-air hypoxia chamber (Galaxy 170R; Eppendorf Co., Ltd.,
Hamburg, Germany) with 2% O2, 5% CO2 and 93%
N2 at 37°C, while the normoxia group was incubated in a
chamber with 20% O2, 5% CO2 and 75%
N2 at 37°C (Shellab 2323-2; Shellab, Cornelius, OR,
USA). Each group was divided into four subgroups (6, 12, 24 and 48
h).
Reverse transcription-quantitative
reverse transcriptase polymerase chain reaction (RT-qPCR)
Total RNA was extracted from PDLCs using TRIzol
reagent (Life Technologies) at each time point. The quality and
quantity of RNA were measured. Total RNA (1.0 μg) was
reverse transcribed for cDNA synthesis (ImProm-II™ Reverse
Transcription system; Promega Corp., Madison, WI, USA). Then cDNA
was mixed with 2X SYBR-Green master mix (Life Technologies) and
gene-specific primers in a final volume of 20 μl, and the
real- time PCR was performed on the LightCycler® 480 platform
(Roche, Basel, Switzerland) with the following reaction: 50°C for 2
min and 95°C for 2 min, followed by 40 cycles at 95°C for 15 sec
and 60°C for 32 sec. The primers used in the present study are
listed in Table I. U6 snRNA was
used to normalize the expression level of HIF1A-AS1 and HIF1A-AS2,
and β-actin was used to normalize the expression level of HIF-1α
and Runx2.
 | Table IPrimer sequences used in this
study. |
Table I
Primer sequences used in this
study.
| Gene name | Primer sequence
(forward and reverse) | Product size
(bp) |
|---|
| HIF1A-AS1 (F) |
5′-AGGCAGAGACGAGATGAACA-3′ | 100 |
| HIF1A-AS1 (R) |
5′-AGGCAGAGACGAGATGAACA-3′ | |
| HIF1A-AS1 (RT) |
5′-CTGGGTCTGGCCATTTCATT-3′ | |
| HIF1A-AS2 (F) |
5′-GACCTAAGGCTCTGGCACTT-3′ | 80 |
| HIF1A-AS2 (R) |
5′-CACTATGAATCCCTGCACCT 3′ | |
| HIF1A-AS2 (RT) |
5′-CACTATGAATCCCTGCACCT-3′ | |
| U6 F |
5′-CTCGCTTCGGCAGCACA-3′ | 94 |
| U6 R |
5′-AACGCTTCACGAATTTGCGT-3′ | |
| U6 RT |
5′-AACGCTTCACGAATTTGCGT-3′ | |
| HIF-1α |
5′-GTGGATTACCACAGCTGA-3′ | 115 |
|
5′-GCTCAGTTAACTTGATCCA-3′ | |
| Runx2 |
5′-TCTAAATCGCCAGGCTTCAT-3′ | 250 |
|
5′-GAGGACCTACTCCCAAAGGA-3′ | |
| β-actin |
5′-TGGATCAGCAAGCAGGAGTA-3′ | 275 |
|
5′-TCGGCCACATTGTGAACTTT-3′ | |
Western blot analysis
PDLCs under 2% and 20% O2 at each time
point were lysed using lysis buffer (99% RIPA, 1% PMSF) and the
protein concentration was determined by a bicinchoninic acid (BCA)
protein assay kit (CWBio, Co., Ltd., Beijing, China). Equal amount
of protein was separated by electrophoresis on duplicate 10% sodium
dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE)
gels, then transferred to polyvinylidene fluoride membranes and
incubated with primary antibodies, mouse anti-human HIF-1α (1:1,000
dilution; cat. no. ab113642; Abcam, Cambridge, MA, USA) and rabbit
anti-human Runx2 (1:1,000 dilution; cat. no. 12556; Cell Signaling
Technology, Inc., Danvers, MA, USA), for 24 h at 4°C. After
washing, the membranes were incubated with the corresponding
secondary antibodies (anti-mouse IgG, cat. no. M281; anti-rabbit
IgG, cat. no. M283; both from Takara Bio, Inc., Otsu, Japan) for 1
h at 37°C. Chemiluminescence was imaged and then the band intensity
was quantified by ImageJ software [National Institutes of Health
(NIH), Bethesda, MD, USA]. The relative protein levels were
calculated as the ratio to the level of β-actin.
ALP activity
PDLCs were cultured in 6-well plates in 2 or 20%
O2. Cells were lysed by Triton X-100 (1% Triton X-100,
99% PBS). The protein concentration was determined using the BCA
protein assay kit. The supernatant was then used for the ALP assay
(Nanjing Jiancheng Bioengineering Institute, Nanjing, China), and
the absorbance was measured at a wavelength of 520 nm. The ALP
activity was determined according to the manufacturer's
instructions.
Transfection of small interfering RNAs
(siRNAs)
siRNAs targeting HIF1A-AS1, HIF1A-AS2, HIF-1α and
non-specific control siRNAs (NC si) were designed by Sigma (St.
Louis, MO, USA). NC si was used as a negative control. The
sequences are listed in Table
II. siRNA was dissolved to a concentration of 20 μM, and
then HIF1A-AS1 siRNA, HIF1A-AS2 siRNA and HIF-1α siRNA (5
μl) were transfected using Lipofectamine™ RNAiMAX into
PDLCs, respectively. In addition, HIF1A-AS1 siRNA (2.5 μl)
and HIF1A-AS2 siRNA (2.5 μl) were transfected at the same
time to knock down both HIF1A-AS1 and HIF1A-AS2. After transfection
for 48 h, PDLCs were cultured under hypoxia for 12 h, and then
total RNA and protein were collected for RT-qPCR, western blot
analysis and ALP activity.
 | Table IISequences of the siRNAs used in this
study. |
Table II
Sequences of the siRNAs used in this
study.
| Gene
name/siRNA | Sequences
(5′→3′) | Size (bp) |
|---|
| HIF1A-AS1 | | |
| siRNA1-sense |
AUGGCCAGACCCAGAUGUUdTdT | 21 |
|
siRNA1-antisense |
AACAUCUGGGUCUGGCCAUdTdT | |
| siRNA2-sense |
GCUCCAGAACGCAGAGGAAdTdT | 21 |
|
siRNA2-antisense |
UUCCUCUGCGUUCUGGAGCdTdT | |
| HIF1A-AS2 | | |
| siRNA1-sense |
CUUAAAUUGUUGGUAAACAdTdT | 21 |
|
siRNA1-antisense |
UGUUUACCAACAAUUUAAGdTdT | |
| siRNA2-sense |
GUAACAUUGUGACUAUAAUdTdT | 21 |
|
siRNA2-antisense |
AUUAUAGUCACAAUGUUACdTdT | |
| HIF-1α | | |
| siRNA1-sense |
CAAAGUUCACCUGAGCCUAdTdT | 21 |
|
siRNA1-antisense |
UAGGCUCAGGUGAACUUUGdTdT | |
| siRNA2-sense |
GAUUAACUCAGUUUGAACUdTdT | 21 |
|
siRNA2-antisense |
AGUUCAAACUGAGUUAAUCdTdT | |
| NC | | |
| siRNA-sense |
UUCUCCGAACGUGUCACGUTT | 21 |
|
siRNA-antisense |
ACGUGACACGUUCGGAGAATT | |
Statistical analysis
Statistical analysis was carried out using SPSS 13.0
(SPSS, Inc., Chicago, IL, USA). All of the results are expressed as
the mean ± SEM. Data for the groups were compared by two-tailed
Student's t-test and the significant level was set at
P<0.05.
Results
Bioinformatic analysis of HIF-1α,
HIF1A-AS1 and HIF1A-AS2
Gene locations for HIF1A-AS1 and HIF1A-AS2 in the
human were from chr14:61681041-61695823 and
chr14:61747039-61749089. HIF-1α had three transcripts, in which
transcript 1 and 2 were from chr14:61695401-61748259 and transcript
3 was from chr14:61697622-61748259. The relative locations of
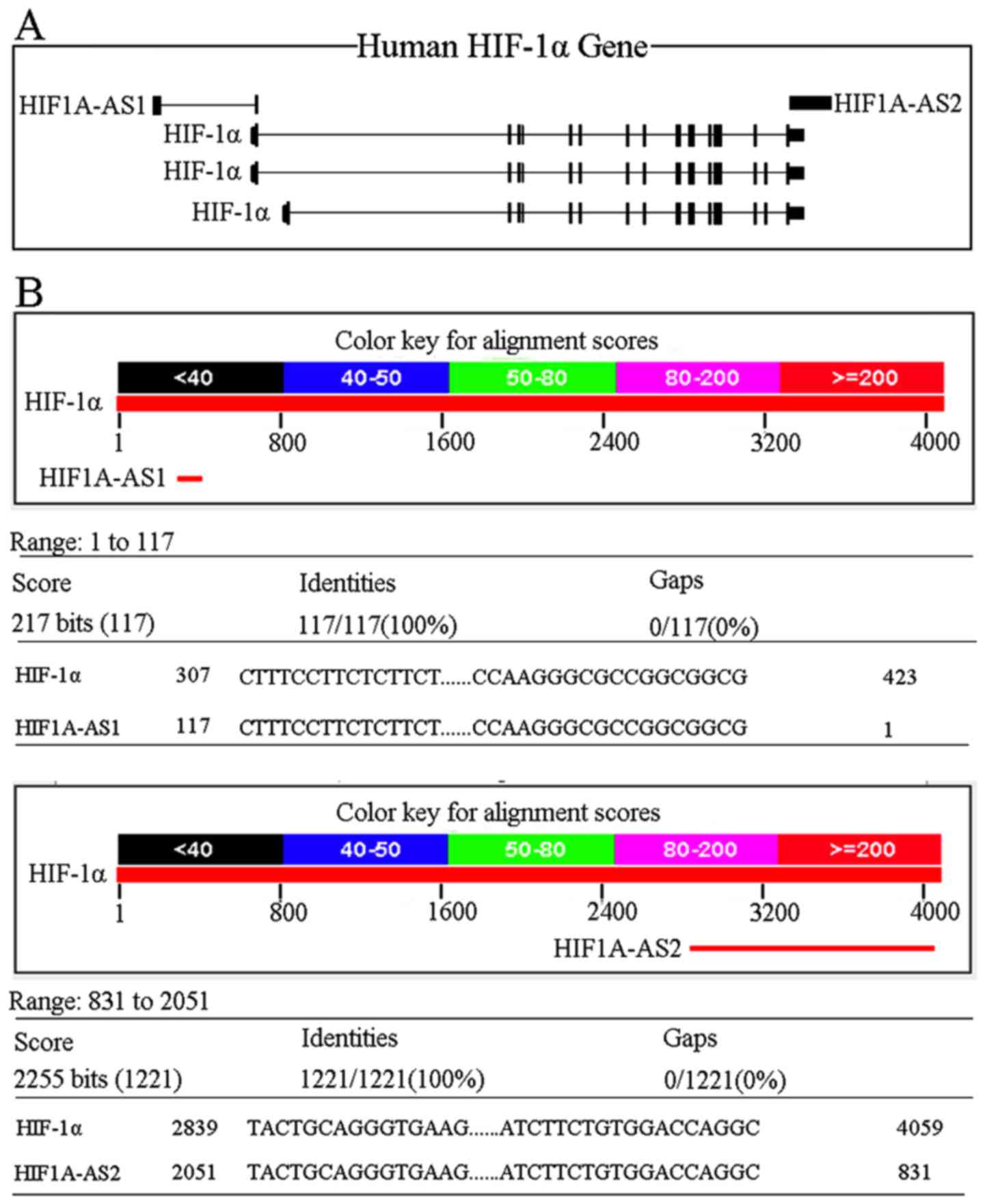
HIF-1α, HIF1A-AS1 and HIF1A-AS2 are shown in Fig. 1A.
The sizes of HIF1A-AS1 mRNA and HIF1A-AS2 mRNA were
652 and 2,051 nt. As shown in Fig.
1B, HIF1A-AS1 was complementary upon 117 nt to the HIF-1α mRNA
5′UTR (HIF1A-AS1: 117 to 1, HIF-1α mRNA: 307 to 423), while
HIF1A-AS2 was complementary upon 1,221 nt to HIF-1α mRNA 3′UTR
(HIF1A-AS2: 2,051 to 831, HIF-1α mRNA: 2,839 to 4,059). Consensus
putative HRE sequence was 5′-(A/G)GTG-3′, which has been identified
in many hypoxia-inducible genes, such as VEGF and EPO (24,25). Commonly, the promoter region was
from −2,000 to +200 bp from the RNA transcription initiation site.
After searching for HIF1A-AS1 and HIF1A-AS2 promoter, we identified
several putative HREs within the regions (HIF1A-AS1: -194, -492 and
-661, HIF1A-AS2: -63 and -600), indicating that HIF-1α may also
regulate the two long non-coding RNAs.
HIF1A-AS1, HIF1A-AS2, HIF-1α expression
and the osteogenic differentiation of PDLCs under hypoxia at
different times
Both HIF1A-AS1 and HIF1A-AS2 were expressed in the
PDLCs. Hypoxia increased the level of HIF1A-AS1 to 2.00-fold,
2.26-fold and 1.53-fold (Fig. 2A)
and HIF1A-AS2 to 6.33-fold, 8.77-fold and 5.79-fold (Fig. 2B) at 6, 12 and 24 h (P<0.05) in
the PDLCs when compared with levels in the normoxia groups. Both
HIF1A-AS1 and HIF1A-AS2 began to decrease under hypoxia after 12
h.
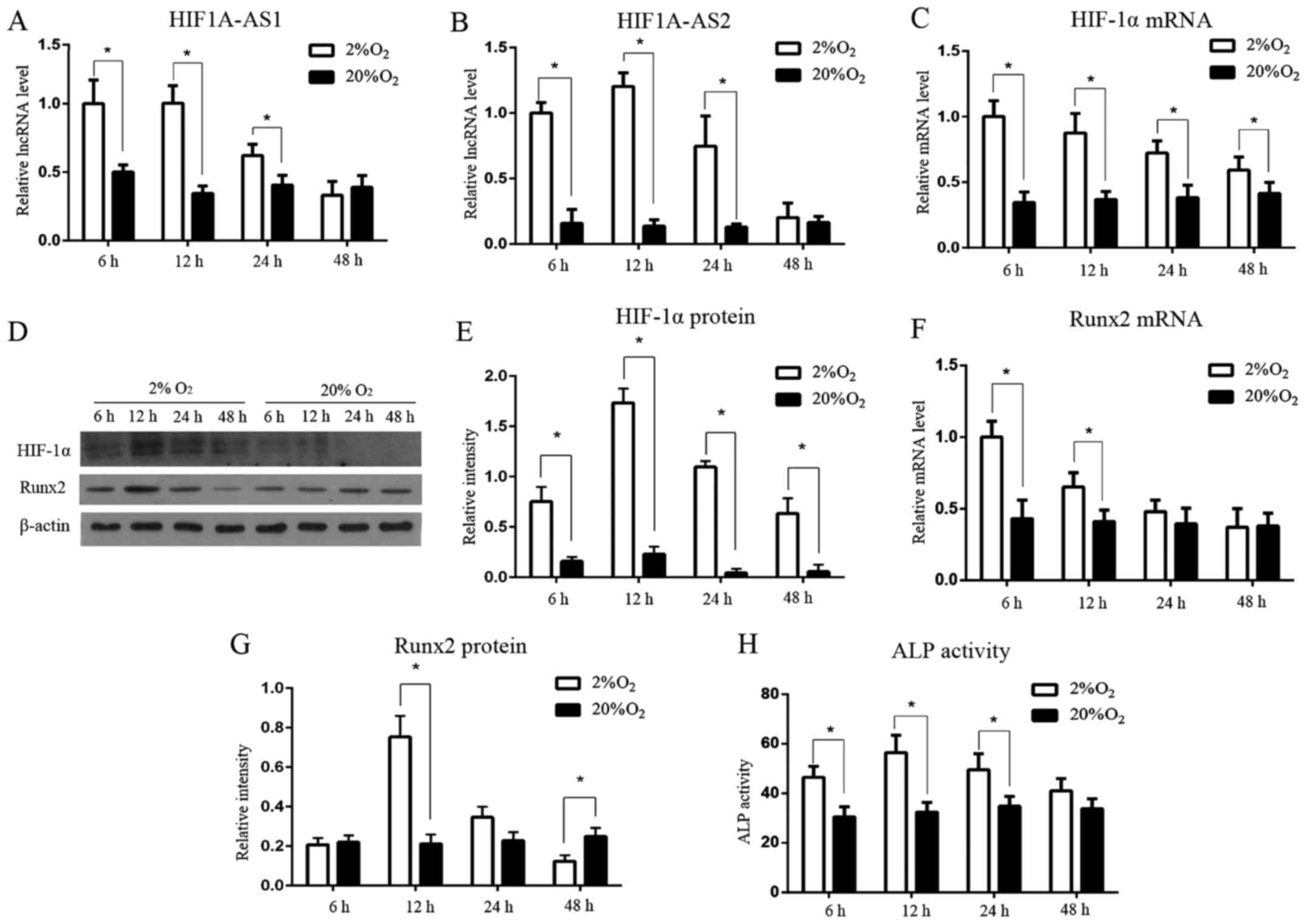 | Figure 2Expression patterns of HIF1A-AS1,
HIF1A-AS2, HIF-1α, ALP and Runx2 in PDLCs under hypoxia and
normoxia. (A and B) The lncRNA levels of HIF1A-AS1 and HIF1A-AS2
under hypoxia were significantly increased at 6, 12 and 24 h, and
peaked at 12 h. (C) The mRNA level of HIF-1α was significantly
higher under hypoxia than that under normoxia and gradually
decreased from 6 to 48 h. (D and E) The protein expression of
HIF-1α under hypoxia was notably higher than that under normoxia,
and peaked at 12 h. (F) The mRNA level of Runx2 was induced by
hypoxia before 12 h. (D and G) The protein expression of Runx2
under hypoxia was significantly induced at 12 h, and then was
suppressed at 48 h. (H) The ALP activity under hypoxia was induced
significantly at 6, 12 and 24 h, and peaked at 12 h.
*P<0.05 vs. the normoxia group, n=3. HIF1A-AS1, HIF1A
antisense RNA 1; HIF1A-AS2, HIF1A antisense RNA 2; HIF-1α,
hypoxia-inducible factor-1α; ALP, alkaline phosphatase; Runx2,
runt-related transcription factor 2; PDLCs, periodontal ligament
cells. |
HIF-1α mRNA (Fig.
2C) was significantly upregulated at 6 h, under hypoxia
(P<0.05), but it decreased gradually from 6 to 48 h. Meanwhile,
HIF-1α protein (Fig. 2D and E)
was barely detectable under normoxia, but it was sharply induced
under hypoxia from 6 h, peaked at 12 h, then gradually diminished
from 12 to 48 h, yet still higher than the normoxia groups
(P<0.05).
The expression level of Runx2 mRNA (Fig. 2F) under hypoxia was also higher
than the level in the normoxia groups at 6 and 12 h (P<0.05),
then it gradually decreased to the baseline at 24 and 48 h
(P>0.05). Runx2 protein (Fig. 2D
and G) was notably induced at 12 h under hypoxia, peaked at 12
h, and was suppressed at 48 h when compared with the normoxia group
(P<0.05). ALP activity (Fig.
2H) was facilitated significantly under hypoxia at 6, 12 and 24
h (P<0.05), reaching a peak at 12 h, but showing no significant
difference at 48 h (P>0.05).
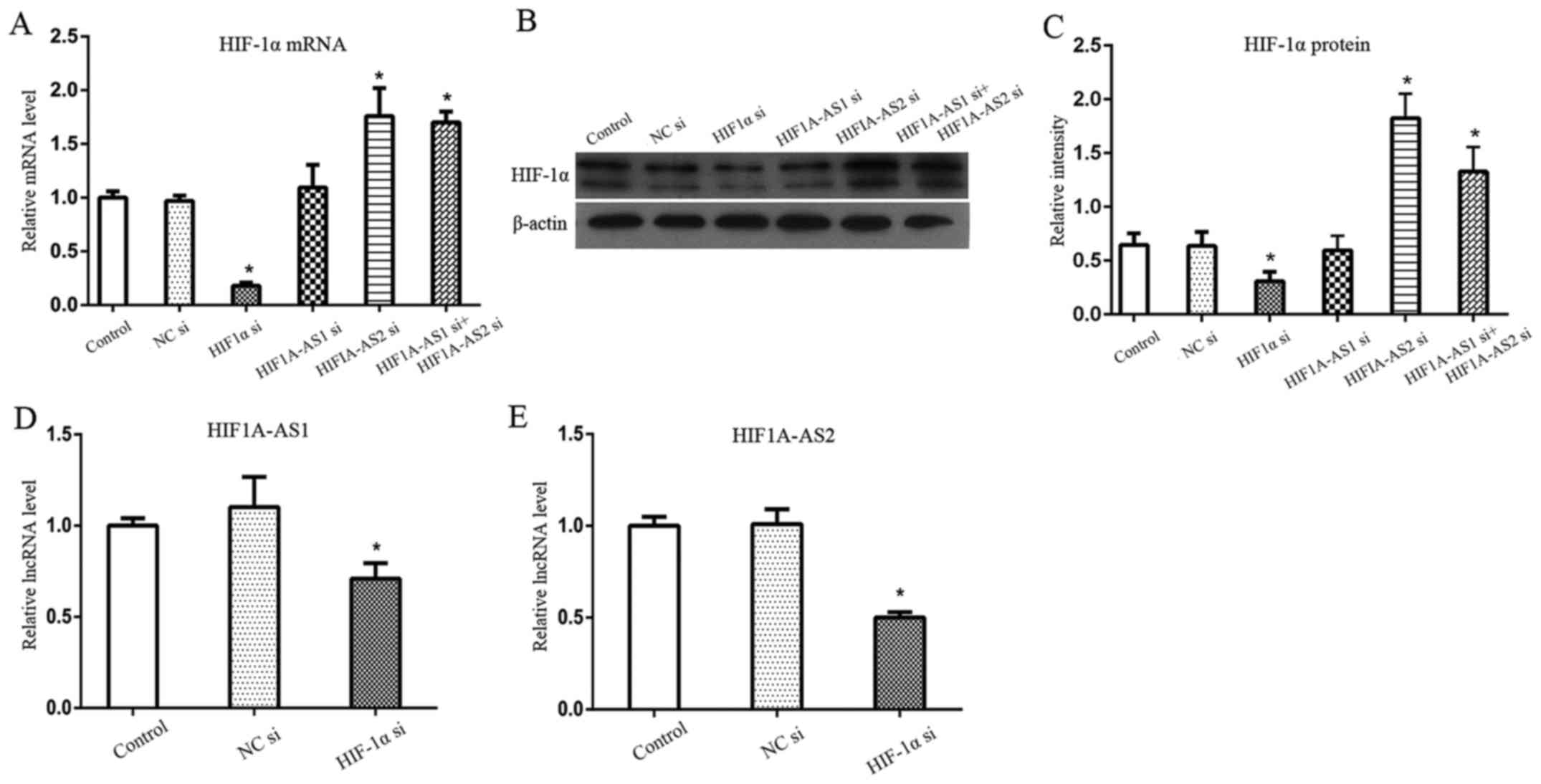
Silencing efficiency of HIF1A-AS1,
HIF1A-AS2 and HIF-1α
To further investigate the regulatory mechanism
among HIF1A-AS1, HIF1A-AS2 and HIF-1α, we silenced HIF1A-AS1,
HIF1A-AS2 and HIF-1α. The siRNAs of HIF1A-AS1, HIF1A-AS2, HIF-1α
and NC were successfully transfected into PDLCs. The transfection
of NC si is shown in Fig. 3A.
Knockdown efficiencies were detected by RT-qPCR for HIF1A-AS1,
HIF1A-AS2 and HIF-1α mRNA, and western blot analysis for HIF-1α
protein. Results showed that the silencing efficiencies of
HIF1A-AS1, HIF1A-AS2 and HIF-1α mRNA (Fig. 3B) were >90, 80 and 70%
respectively. Furthermore, HIF-1α protein (Fig. 3C) was also successfully inhibited
by HIF-1α siRNA under hypoxia.
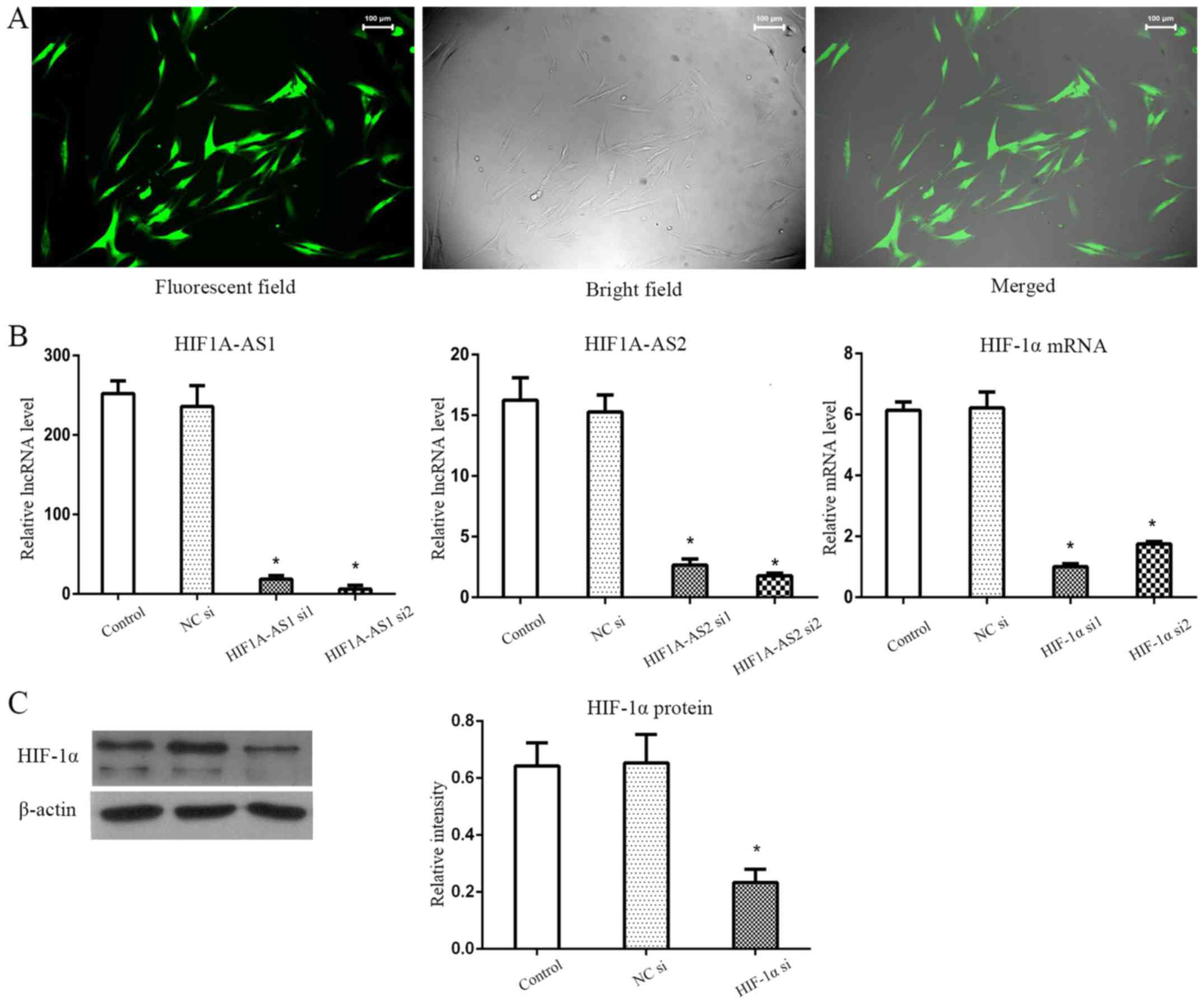 | Figure 3Silencing efficiencies of HIF1A-AS1,
HIF1A-AS2 and HIF-1α siRNAs. (A) NC si shown as an example was
successfully transfected into PDLCs. (B) HIF1A-AS1, HIF1A-AS2 and
HIF-1α mRNA were effectively reduced by >90, 80 and 70% in PDLCs
under hypoxia at 12 h, respectively. (C) HIF-1α protein was
significantly inhibited by HIF-1α siRNA in hypoxia.
*P<0.05 vs. the control group, n=3. HIF1A-AS1, HIF1A
antisense RNA 1; HIF1A-AS2, HIF1A antisense RNA 2; HIF-1α,
hypoxia-inducible factor-1α; NC si, non-specific control siRNA;
PDLCs, periodontal ligament cells. |
Effects of silencing HIF1A-AS1 and
HIF1A-AS2 on HIF-1α in PDLCs
Changes in HIF-1α mRNA (Fig. 4A) and HIF-1α protein (Fig. 4B and C) were consistent following
the silencing of HIF1A-AS1 and/or HIF1A-AS2. Silencing of HIF1A-AS1
did not significantly alter the expression of HIF-1α protein and
mRNA, whereas silencing of HIF1A-AS2 significantly induced the
expression of HIF-1α mRNA and protein. In order to determine
whether HIF1A-AS1 and HIF1A-AS2 had synergistic effects on HIF-1α
expression and osteogenic differentiation, we knocked down
HIF1A-AS1 and HIF1A-AS2 by co-transfecting PDLCs with HIF1A-AS1
siRNA and HIF1A-AS2 siRNA. Results showed that HIF-1α protein and
mRNA were significantly elevated, similar to silencing HIF1A-AS2
alone.
Effects of silencing HIF-1α on HIF1A-AS1
and HIF1A-AS2 in PDLCs
Since putative HRE sequences were identified within
the promoter regions of HIF1A-AS1 and HIF1A-AS2, we aimed to
ascertain whether HIF-1α regulates HIF1A-AS1 and HIF1A-AS2 by
silencing HIF-1α. Results demonstrated that both HIF1A-AS1
(Fig. 4D) and HIF1A-AS2 (Fig. 4E) were significantly inhibited by
HIF-1α siRNA (P<0.05), indicating that HIF-1α acts as an
upstream regulatory factor of HIF1A-AS1 and HIF1A-AS2.
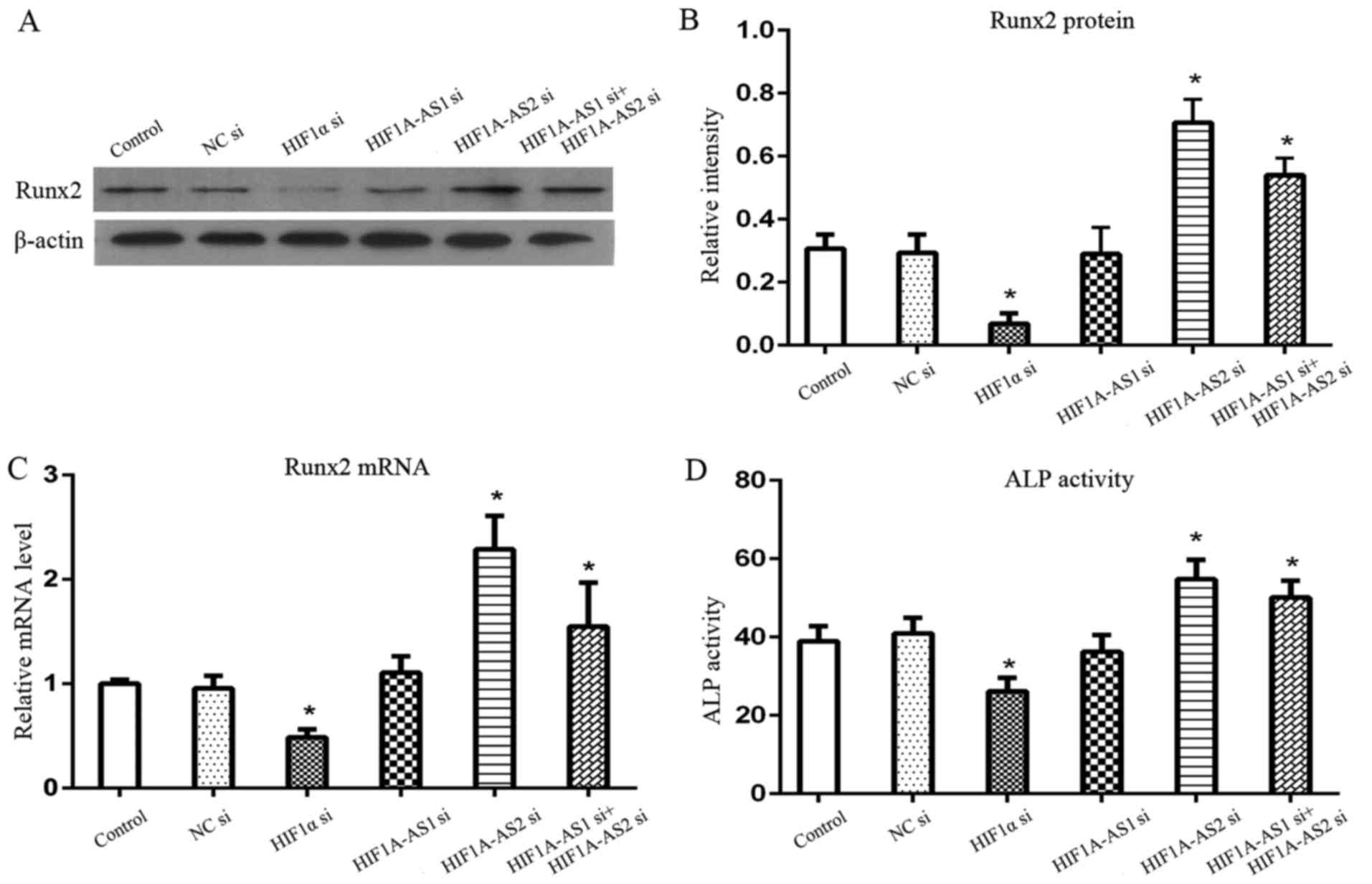
Effects of the silencing of HIF-1α,
HIF1A-AS1 and HIF1A-AS2 on the osteogenic biomarkers of PDLCs
To further investigate the effects of HIF-1α,
HIF1A-AS1 and HIF1A-AS2 on the osteogenic differentiation of PDLCs,
we investigated the changes in Runx2 mRNA, Runx2 protein and ALP
activity following the silencing of HIF-1α, HIF1A-AS1 and HIF1A-AS2
under hypoxia. The levels of osteogenic biomarkers (Fig. 5), Runx2 protein, Runx2 mRNA and
ALP activity, were significantly inhibited by the silencing of
HIF-1α and induced by the silencing of HIF1A-AS2 and co-silencing
of HIF1A-AS1 and HIF1A-AS2 (P<0.05), while there were no
significant changes following the silencing of HIF1A-AS1 alone
(P>0.05).
Discussion
The functions of HIF1A-AS1 and HIF1A-AS2 have not
been studied in PDLCs. HIF1A-AS1 and HIF1A-AS2 play key roles in
cell proliferation and apoptosis, and could be used as predictive
biomarkers for cancers (26–29). However, little is known concerning
HIF1A-AS1 and HIF1A-AS2 in periodontal tissues under hypoxia. Thus,
we explored the complex relationships among HIF1A-AS1, HIF1A-AS2
and HIF-1α and investigated whether they regulate the osteogenic
differentiation of PDLCs.
In the present study, we discovered that the two
antisense lncRNAs, HIF1A-AS1 and HIF1A-AS2, were expressed in
PDLCs. Our present findings showed that the expression levels of
HIF1A-AS1, HIF1A-AS2, HIF-1α and osteogenic biomarkers were altered
in a temporal manner under hypoxia. Both HIF1A-AS1 and HIF1A-AS2
were expressed at low levels under normoxia, but they were
obviously induced in short-term hypoxia, reaching a peak at 12 h.
Increased HIF1A-AS2 under hypoxia was found in the first few hours
by researchers in various cancer cells (17,30). The expression patterns of HIF-1α
protein and the osteogenic markers (Runx2 and ALP activity) were
consistent with that of HIF1A-AS1 and HIF1A-AS2. HIF-1α mRNA was
highly expressed under hypoxia and then declined gradually after 6
h. In addition, Huang et al (31) also observed that HIF-1α mRNA was
elevated at 1 h and peaked at 6 h under hypoxia in human
mesenchymal stem cells (hMSCs), and then the level was gradually
declined from 6 to 24 h. Uchida et al (20) found that HIF-1α protein was highly
induced during the first few hours in hypoxia, regulated by
translation or post-translation pathways, and inhibited in
prolonged hypoxia due to the decline in HIF-1α mRNA. With respect
to the osteogenic differentiation ability under hypoxia, Wu et
al (32) demonstrated that
Runx2 mRNA and protein were immediately enhanced from 1 h and ALP
activity was from 3 h in PDLSCs when exposed to hypoxia, and they
still remained higher than the normoxia group at 24 h. Furthermore,
Ding et al (33) found
that continuous hypoxia after 3 days impaired the osteogenenic
differentiation of hMSCs in hypoxia. Therefore, the above studies
indicated that HIF1A-AS1, HIF1A-AS2, HIF-1α and the osteogenic
biomarkers were altered but time-dependently under hypoxia. Our
research revealed that HIF1A-AS1, HIF1A-AS2 and HIF-1α might have
important roles in regulating the osteogenenic differentiation of
PDLCs under hypoxia.
Furthermore, we investigated whether both HIF1A-AS1
and HIF1A-AS2 participated in regulating HIF-1α mRNA by silencing
HIF1A-AS1 and HIF1A-AS2. Silencing of HIF1A-AS2 prominently
increased HIF-1α mRNA and protein, whereas the silencing of
HIF1A-AS1 did not affect HIF-1α mRNA and HIF-1α protein, which
indicated that only HIF1A-AS2 has a strong negative regulatory
function on HIF-1α in PDLCs. The increase in HIF-1α was slightly
lower following co-transfection of HIF1A-AS1 siRNA and HIF1A-AS2
siRNA than following transfection of HIF1A-AS2 siRNA alone, which
might be that the amount of HIF1A-AS2 siRNA was half when
co-transfecting HIF1A-AS1 siRNA (2.5 μl) and HIF1A-AS2 siRNA
(2.5 μl). In previous studies, HIF1A-AS2 has been reported
to be complementary on at least 1,027 nt of the HIF-1α mRNA 3′UTR
which was rich in AU elements and HIF1A-AS2 could expose these AU
rich elements to accelerate the degradation of HIF-1α mRNA
(21). Kumar and Carmichael
(34) speculated that HIF1A-AS2
suppressed HIF-1α mRNA translation by hybridizing to HIF-1α mRNA
3′UTR, therefore HIF-1α protein was suppressed. Our bioinformatic
analysis revealed that both HIF1A-AS1 and HIF1A-AS2 could be
strictly complementary to HIF-1α mRNA, in which HIF1A-AS1 was
complementary upon 117 nt and HIF1A-AS2 was 1,221 nt. In addition,
our experiment in vitro proved that only HIF1A-AS2
significantly downregulated HIF-1α. By analyzing the regulatory
mechanism of the HIF-system in human macrophages, elevated HIF-1α
mRNA and protein were also observed when silencing HIF1A-AS2, which
was in accordance with our results (35). Bertozzi et al (19) also reported that HIF1A-AS1 and
HIF1A-AS2 had different responses to different stimulations. Since
HIF1A-AS1 accumulates at the nuclear membrane, it might be
associated with the export of mRNA from the nucleus into the
cytoplasm. Therefore, we predicted that HIF1A-AS1 and HIF1A-AS2 may
be involved in different types of regulatory mechanisms.
HIF-1 was speculated as the upstream factor of the
antisense HIF-1α. Some evidence was provided by Uchida et al
(20) to support this hypothesis.
i) Cycloheximide, a protein synthesis inhibitor that restrains
hypoxia-induced HIF-1α increase, also inhibited the augmentation of
HIF1A-AS2 during long-term hypoxia; ii) HIF-1α was found to be
bound to the putative HRE sequence, which could be displaced by the
oligonucleotide sequence of HRE found in human HIF1A-AS2 gene
promoter, not by a mismatch of putative HRE sequence. Moreover, in
embryonic cells lacking the HIF-β subunit where HIF-1 could not be
formed, hypoxia-induced HIF1A-AS2 was attenuated, indicating that
HIF1A-AS2 was HIF-1 responsive (36,37). Consistently, the HIF1A-AS1
promoter also has putative HREs. To further investigate the effects
of HIF-1α on HIF1A-AS1 and HIF1A-AS2, we silenced HIF-1α, and the
results showed that HIF1A-AS1 and HIF1A-AS2 were downregulated,
which confirmed that HIF-1α has a positive regulatory effect on
both HIF1A-AS1 and HIF1A-AS2. This was also consistent with the
observation that HIF1A-AS1 and HIF1A-AS2 were decreased after 12 h
under hypoxia accompanied by the decline of HIF-1α protein.
HIF1A-AS1 and HIF1A-AS2 also had different roles in
regulating the osteogenic differentiation of PDLCs under hypoxia,
which might be related to HIF-1α. Inhibition of HIF-1α
significantly suppressed osteogenic biomarkers, which demonstrated
that HIF-1α positively regulated the osteogenic differentiation
ability of PDLCs. Typically, HIF-1α-induced downstream proteins
respond rapidly, usually occurring during the first several hours
(17,38). Zhou et al (32,39) found that hypoxia-simulated
osteogenesis was via ERK1/2 and p38-mediated HIF-1α pathway, in
which the ERK1/2 pathway was the dominant one. ERK1/2 and p38,
members of the MAPKs, were found to be involved not only in
maintaining the stability and promoting the transactivation of
HIF-1α, but also in regulating osteogenic differentiation.
Furthermore, we found that silencing of HIF1A-AS2 significantly
promoted the expression of osteogenic biomarkers, whereas silencing
of HIF1A-AS1 had no significant effects on the expressions of
osteogenic biomarkers. It should be noted that the changes in the
osteogenic biomarkers were in accordance with the changes in HIF-1α
following the silencing of HIF1A-AS2, since HIF-1α regulated the
osteogenic differentiation of PDLCs. Therefore, we speculated that
there may be an HIF1A-AS2/HIF-1α signaling pathway through which to
regulate the osteogenic differentiation of PDLCs.
In conclusion, we found that both HIF1A-AS1 and
HIF1A-AS2 were complementary to HIF-1α mRNA by bioinformatic
analysis, but the in vitro study revealed that only
HIF1A-AS2 had an inhibitory effect on HIF-1α in PDLCs. In addition,
HIF-1α also induced HIF1A-AS1 and HIF1A-AS2, which have putative
HREs in the promoters. Furthermore, HIF-1α promoted the osteogenic
differentiation of PDLCs and HIF1A-AS2 inhibited the osteogenic
differentiation of PDLCs.
Acknowledgments
The present study was supported by the Natural
Science Foundation of Guangdong Province (no. 2015A030313083) and
the Natural Science Foundation of China (no. 81400499).
References
|
1
|
Hong JW, Noh JH and Kim DJ: The prevalence
and associated factors of periodontitis according to fasting plasma
glucose in the Korean adults: The 2012–2013 Korea National Health
and Nutrition Examination Survey. Medicine (Baltimore).
95:e32262016. View Article : Google Scholar
|
|
2
|
Eke PI, Zhang X, Lu H, Wei L,
Thornton-Evans G, Greenlund KJ, Holt JB and Croft JB: Predicting
periodontitis at state and local levels in the United States. J
Dent Res. 95:515–522. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Eke PI, Dye BA, Wei L, Slade GD,
Thornton-Evans GO, Borgnakke WS, Taylor GW, Page RC, Beck JD and
Genco RJ: Update on prevalence of periodontitis in adults in the
United States: NHANES 2009 to 2012. J Periodontol. 86:611–622.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Niklas A, Proff P, Gosau M and Römer P:
The role of hypoxia in orthodontic tooth movement. Int J Dent.
2013:8418402013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hellwig-Bürgel T, Stiehl DP, Wagner AE,
Metzen E and Jelkmann W: Review: Hypoxia-inducible factor-1
(HIF-1): A novel transcription factor in immune reactions. J
Interferon Cytokine Res. 25:297–310. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Semenza GL and Wang GL: A nuclear factor
induced by hypoxia via de novo protein synthesis binds to the human
erythropoietin gene enhancer at a site required for transcriptional
activation. Mol Cell Biol. 12:5447–5454. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nallamshetty S, Chan SY and Loscalzo J:
Hypoxia: A master regulator of microRNA biogenesis and activity.
Free Radic Biol Med. 64:20–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang XJ and Si LB: Advances on hypoxia
inducible factor-1. Chin Med J (Engl). 126:3567–3571. 2013.
|
|
9
|
Holzwarth C, Vaegler M, Gieseke F, Pfister
SM, Handgretinger R, Kerst G and Müller I: Low physiologic oxygen
tensions reduce proliferation and differentiation of human
multipotent mesenchymal stromal cells. BMC Cell Biol. 11:112010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang DC, Yang MH, Tsai CC, Huang TF, Chen
YH and Hung SC: Hypoxia inhibits osteogenesis in human mesenchymal
stem cells through direct regulation of RUNX2 by TWIST. PLoS One.
6:e239652011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou Y, Fan W and Xiao Y: The effect of
hypoxia on the stemness and differentiation capacity of PDLC and
DPC. BioMed Res Int. 2014:8906752014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zou D, Han W, You S, Ye D, Wang L, Wang S,
Zhao J, Zhang W, Jiang X, Zhang X, et al: In vitro study of
enhanced osteogenesis induced by HIF-1α-transduced bone marrow stem
cells. Cell Prolif. 44:234–243. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Masoud GN and Li W: HIF-1α pathway: Role,
regulation and intervention for cancer therapy. Acta Pharm Sin B.
5:378–389. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Parsanejad M, Zhang Y, Qu D, Irrcher I,
Rousseaux MW, Aleyasin H, Kamkar F, Callaghan S, Slack RS, Mak TW,
et al: Regulation of the VHL/HIF-1 pathway by DJ-1. J Neurosci.
34:8043–8050. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Richard DE, Berra E, Gothié E, Roux D and
Pouysségur J: p42/p44 mitogen-activated protein kinases
phosphorylate hypoxia-inducible factor 1alpha (HIF-1alpha) and
enhance the transcriptional activity of HIF-1. J Biol Chem.
274:32631–32637. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhong H, Chiles K, Feldser D, Laughner E,
Hanrahan C, Georgescu MM, Simons JW and Semenza GL: Modulation of
hypoxia-inducible factor 1alpha expression by the epidermal growth
factor/phosphatidylinositol 3-kinase/PTEN/AKT/FRAP pathway in human
prostate cancer cells: implications for tumor angiogenesis and
therapeutics. Cancer Res. 60:1541–1545. 2000.PubMed/NCBI
|
|
17
|
Thrash-Bingham CA and Tartof KD: aHIF: A
natural antisense transcript overexpressed in human renal cancer
and during hypoxia. J Natl Cancer Inst. 91:143–151. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Baranello L, Bertozzi D, Fogli MV, Pommier
Y and Capranico G: DNA topoisomerase I inhibition by camptothecin
induces escape of RNA polymerase II from promoter-proximal pause
site, antisense transcription and histone acetylation at the human
HIF-1alpha gene locus. Nucleic Acids Res. 38:159–171. 2010.
View Article : Google Scholar
|
|
19
|
Bertozzi D, Iurlaro R, Sordet O, Marinello
J, Zaffaroni N and Capranico G: Characterization of novel antisense
HIF-1α transcripts in human cancers. Cell Cycle. 10:3189–3197.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Uchida T, Rossignol F, Matthay MA, Mounier
R, Couette S, Clottes E and Clerici C: Prolonged hypoxia
differentially regulates hypoxia-inducible factor (HIF)-1alpha and
HIF-2alpha expression in lung epithelial cells: implication of
natural antisense HIF-1alpha. J Biol Chem. 279:14871–14878. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rossignol F, Vaché C and Clottes E:
Natural antisense transcripts of hypoxia-inducible factor 1alpha
are detected in different normal and tumour human tissues. Gene.
299:135–140. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Komori T: Regulation of osteoblast
differentiation by Runx2. Adv Exp Med Biol. 658:43–49. 2010.
View Article : Google Scholar
|
|
23
|
Bhargavan B, Gautam AK, Singh D, Kumar A,
Chaurasia S, Tyagi AM, Yadav DK, Mishra JS, Singh AB, Sanyal S, et
al: Methoxylated isoflavones, cajanin and isoformononetin, have
non-estrogenic bone forming effect via differential mitogen
activated protein kinase (MAPK) signaling. J Cell Biochem.
108:388–399. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu Q, Geng H, Xue C, Beer TM and Qian DZ:
Functional regulation of hypoxia inducible factor-1α by SET9 lysine
methyltransferase. Biochim Biophys Acta. 1853:881–891. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rhim T, Lee DY and Lee M: Hypoxia as a
target for tissue specific gene therapy. J Control Release.
172:484–494. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen WM, Huang MD, Kong R, Xu TP, Zhang
EB, Xia R, Sun M, De W and Shu YQ: Antisense long non-coding RNA
HIF1A-AS2 is upregulated in gastric cancer and associated with poor
prognosis. Dig Dis Sci. 60:1655–1662. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
He Q, Tan J, Yu B, Shi W and Liang K: Long
non-coding RNA HIF1A-AS1A reduces apoptosis of vascular smooth
muscle cells: Implications for the pathogenesis of thoracoabdominal
aorta aneurysm. Pharmazie. 70:310–315. 2015.PubMed/NCBI
|
|
28
|
Tantai J, Hu D, Yang Y and Geng J:
Combined identification of long non-coding RNA XIST and HIF1A-AS1
in serum as an effective screening for non-small cell lung cancer.
Int J Clin Exp Pathol. 8:7887–7895. 2015.PubMed/NCBI
|
|
29
|
Wang S, Zhang X, Yuan Y, Tan M, Zhang L,
Xue X, Yan Y, Han L and Xu Z: BRG1 expression is increased in
thoracic aortic aneurysms and regulates proliferation and apoptosis
of vascular smooth muscle cells through the long non-coding RNA
HIF1A-AS1 in vitro. Eur J Cardiothorac Surg. 47:439–446. 2015.
View Article : Google Scholar
|
|
30
|
Rossignol F, de Laplanche E, Mounier R,
Bonnefont J, Cayre A, Godinot C, Simonnet H and Clottes E: Natural
antisense transcripts of HIF-1alpha are conserved in rodents. Gene.
339:121–130. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang J, Deng F, Wang L, Xiang XR, Zhou
WW, Hu N and Xu L: Hypoxia induces osteogenesis-related activities
and expression of core binding factor α1 in mesenchymal stem cells.
Tohoku J Exp Med. 224:7–12. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu Y, Yang Y, Yang P, Gu Y, Zhao Z, Tan L,
Zhao L, Tang T and Li Y: The osteogenic differentiation of PDLSCs
is mediated through MEK/ERK and p38 MAPK signalling under hypoxia.
Arch Oral Biol. 58:1357–1368. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ding H, Chen S, Yin JH, Xie XT, Zhu ZH,
Gao YS and Zhang CQ: Continuous hypoxia regulates the osteogenic
potential of mesenchymal stem cells in a time-dependent manner. Mol
Med Rep. 10:2184–2190. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kumar M and Carmichael GG: Nuclear
antisense RNA induces extensive adenosine modifications and nuclear
retention of target transcripts. Proc Natl Acad Sci USA.
94:3542–3547. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Poitz DM, Augstein A, Hesse K, Christoph
M, Ibrahim K, Braun-Dullaeus RC, Strasser RH and Schmeißer A:
Regulation of the HIF-system in human macrophages - differential
regulation of HIF-α subunits under sustained hypoxia. Mol Immunol.
57:226–235. 2014. View Article : Google Scholar
|
|
36
|
Maltepe E, Schmidt JV, Baunoch D,
Bradfield CA and Simon MC: Abnormal angiogenesis and responses to
glucose and oxygen deprivation in mice lacking the protein ARNT.
Nature. 386:403–407. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
37
|
Neckers LM: aHIF: The missing link between
HIF-1 and VHL? J Natl Cancer Inst. 91:106–107. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang GL, Jiang BH, Rue EA and Semenza GL:
Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS
heterodimer regulated by cellular O2 tension. Proc Natl
Acad Sci USA. 92:5510–5514. 1995. View Article : Google Scholar
|
|
39
|
Zhou Y, Guan X, Wang H, Zhu Z, Li C, Wu S
and Yu H: Hypoxia induces osteogenic/angiogenic responses of bone
marrow-derived mesenchymal stromal cells seeded on bone-derived
scaffolds via ERK1/2 and p38 pathways. Biotechnol Bioeng.
110:1794–1804. 2013. View Article : Google Scholar : PubMed/NCBI
|